Clinical Application of Comprehensive Genomic Profiling Tests for Diffuse Gliomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
- (1)
- Termination of standard treatment because of local progression, including those expecting termination.
- (2)
- Eligibility for alternate drug therapy after CGPT was confirmed by an attending physician after an examination.
- (3)
- A Karnofsky Performance Status ≥70. This criterion implicates that they can attend outpatient clinics.
2.2. Tissue Diagnosis
2.3. Preparation of Tumor Tissue for CGPT
2.4. Analysis of the CGPT Results
2.5. Molecular Tumor Board, Clinical Actionability, and Germline Mutations
3. Results
3.1. Demographics of the Enrolled Patients
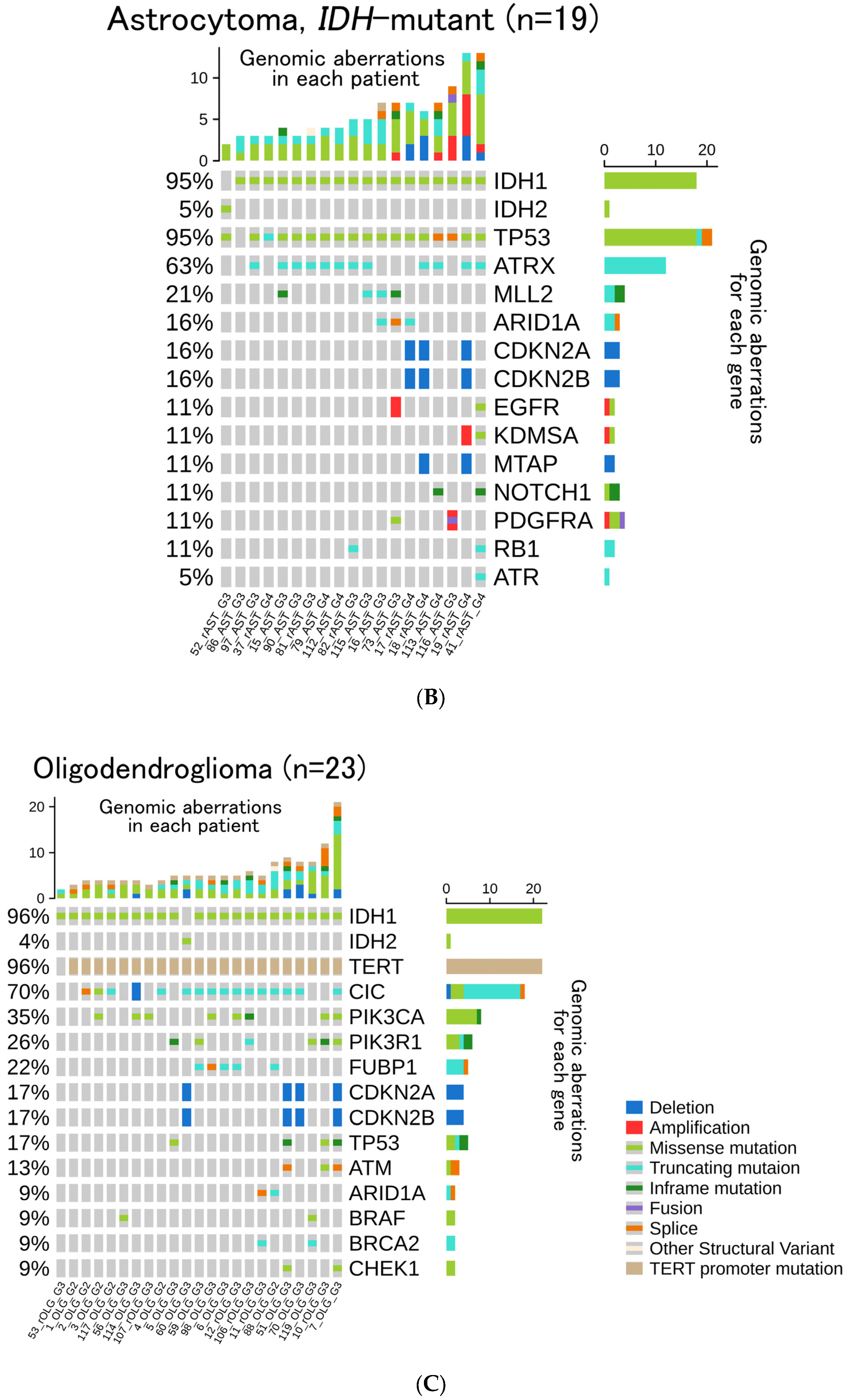
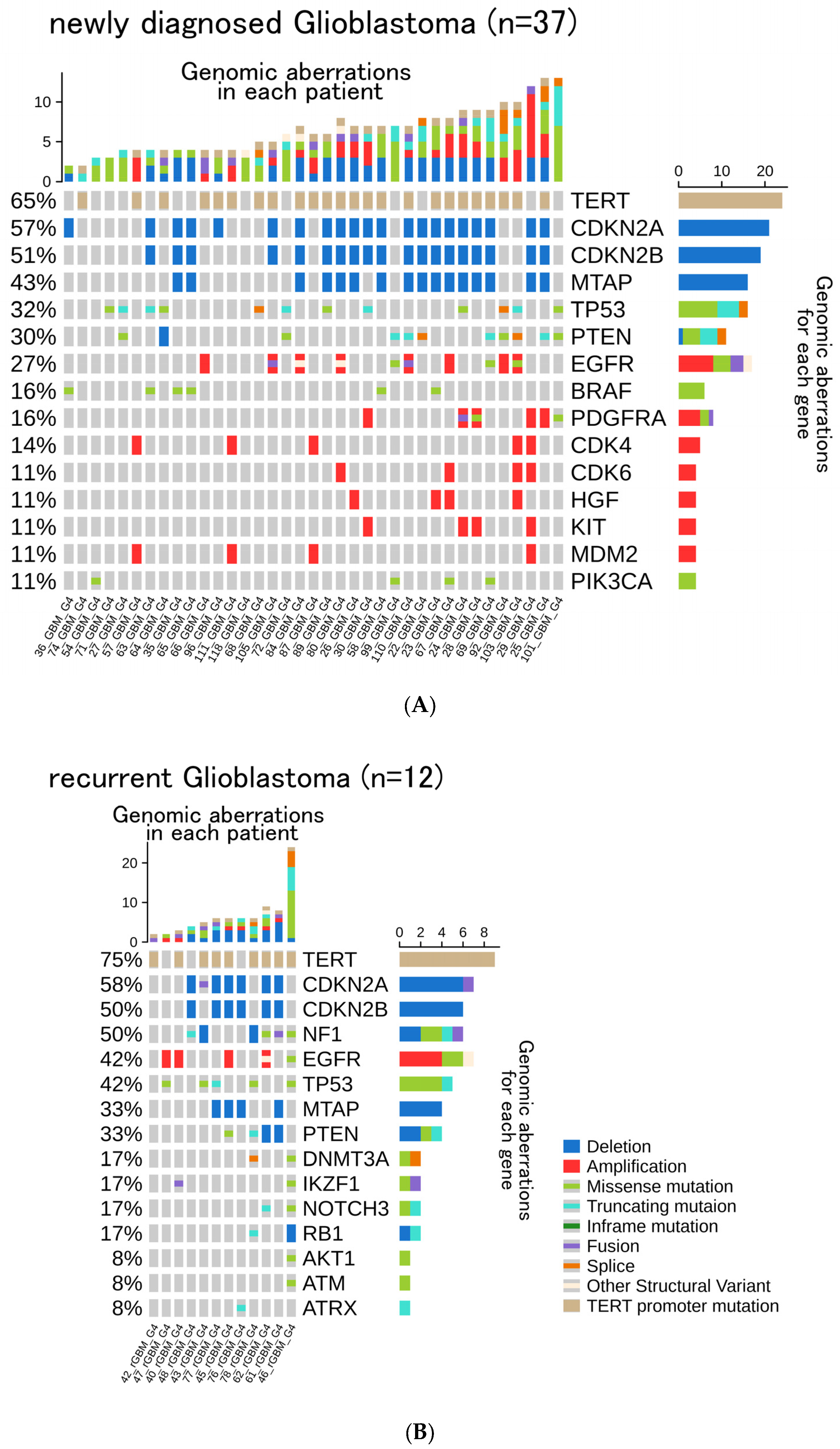
3.2. Genomic Aberrations, TMB, and MSI
3.3. Frequency of Gene Aberrations and Histological Classification
3.4. Histological Re-Classification Based on CGPTs
3.5. Clinical Actionability
3.6. Germline Mutations and Subsequent Genetic Counseling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brain Tumor Registry of Japan (2005–2008). Neurol Med. Chir. 2017, 57, 9–102. [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro. Oncol. 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Herrlinger, U.; Tzaridis, T.; Mack, F.; Steinbach, J.P.; Schlegel, U.; Sabel, M.; Hau, P.; Kortmann, R.D.; Krex, D.; Grauer, O.; et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): A randomised, open-label, phase 3 trial. Lancet 2019, 393, 678–688. [Google Scholar] [CrossRef]
- Blumenthal, D.T.; Dvir, A.; Lossos, A.; Tzuk-Shina, T.; Lior, T.; Limon, D.; Yust-Katz, S.; Lokiec, A.; Ram, Z.; Ross, J.S.; et al. Clinical utility and treatment outcome of comprehensive genomic profiling in high grade glioma patients. J. Neurooncol. 2016, 130, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Ebi, H.; Bando, H. Precision Oncology and the Universal Health Coverage System in Japan. JCO Precis. Oncol. 2019, 3, 19.00291. [Google Scholar] [CrossRef]
- Sunami, K.; Ichikawa, H.; Kubo, T.; Kato, M.; Fujiwara, Y.; Shimomura, A.; Koyama, T.; Kakishima, H.; Kitami, M.; Matsushita, H.; et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer. Sci. 2019, 110, 1480–1490. [Google Scholar] [CrossRef] [Green Version]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro. Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Satomi, K.; Ohno, M.; Matsushita, Y.; Takahashi, M.; Miyakita, Y.; Narita, Y.; Ichimura, K.; Yoshida, A. Utility of methylthioadenosine phosphorylase immunohistochemical deficiency as a surrogate forCDKN2Ahomozygous deletion in the assessment of adult-type infiltrating astrocytoma. Mod. Pathol. 2021, 34, 688–700. [Google Scholar] [CrossRef]
- Gu, Z.G.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Marcus, L.; Lemery, S.J.; Keegan, P.; Pazdur, R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin. Cancer Res. 2019, 25, 3753–3758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishimaru, S.; Tatsunori, S.; Kuniko, S.; Taro, S.; Nobuko, O.; Mikio, M.; Sachie, K.; Natsuko, O.; Kenichi, N.; Noborum, Y. A Novel Approach for Improving Drug Access Using Patient-Proposed HealthcareService. Pediatric Blood Cancer 2019, 66 (Suppl. 5), S14. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; Penas-Prado, M.; Peters, K.B.; Burris, H.A., 3rd; Maher, E.A.; Janku, F.; Cote, G.M.; de la Fuente, M.I.; Clarke, J.L.; Ellingson, B.M.; et al. Vorasidenib, a Dual Inhibitor of Mutant IDH1/2, in Recurrent or Progressive Glioma; Results of a First-in-Human Phase I Trial. Clin. Cancer Res. 2021, 27, 4491–4499. [Google Scholar] [CrossRef] [PubMed]
- Wick, A.; Bahr, O.; Schuler, M.; Rohrberg, K.; Chawla, S.P.; Janku, F.; Schiff, D.; Heinemann, V.; Narita, Y.; Lenz, H.J.; et al. Phase I Assessment of Safety and Therapeutic Activity of BAY1436032 in Patients with IDH1-Mutant Solid Tumors. Clin. Cancer Res. 2021, 27, 2723–2733. [Google Scholar] [CrossRef]
- Natsume, A.; Wakabayashi, T.; Miyakita, Y.; Narita, Y.; Mineharu, Y.; Arakawa, Y.; Yamasaki, F.; Sugiyama, K.; Hata, N.; Muragaki, Y.; et al. Phase I study of a brain penetrant mutant IDH1 inhibitor DS-1001b in patients with recurrent or progressive IDH1 mutant gliomas. J. Clin. Oncol. 2019, 37, 2004. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.J.; Chen, A.P.; Li, S.L.; McShane, L.M.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Iafrate, A.J.; Sklar, J.; et al. Molecular Landscape and Actionable Alterations in a Genomically Guided Cancer Clinical Trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J. Clin. Oncol. 2020, 38, 3883–3894. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Fujimoto, K.; Arita, H.; Satomi, K.; Yamasaki, K.; Matsushita, Y.; Nakamura, T.; Miyakita, Y.; Umehara, T.; Kobayashi, K.; Tamura, K.; et al. TERT promoter mutation status is necessary and sufficient to diagnose IDH-wildtype diffuse astrocytic glioma with molecular features of glioblastoma. Acta Neuropathol. 2021, 142, 323–338. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.; Musket, A.; Musich, P.R.; Schweitzer, J.B.; Xie, Q. Receptor tyrosine kinases as druggable targets in glioblastoma: Do signaling pathways matter? Neurooncol. Adv. 2021, 3, vdab133. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Pascual, A.; Siebzehnrubl, F.A. Fibroblast Growth Factor Receptor Functions in Glioblastoma. Cells 2019, 8, 715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, R.; Carneiro, B.A.; Taxter, T.; Tavora, F.A.; Kalyan, A.; Pai, S.A.; Chae, Y.K.; Giles, F.J. FGFR3-TACC3 fusion in solid tumors: Mini review. Oncotarget 2016, 7, 55924–55938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuma, H.S.; Yonemori, K.; Narita, S.N.; Sukigara, T.; Hirakawa, A.; Shimizu, T.; Shibata, T.; Kawai, A.; Yamamoto, N.; Nakamura, K.; et al. MASTER KEY Project: Powering Clinical Development for Rare Cancers Through a Platform Trial. Clin. Pharmacol. Ther. 2020, 108, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, K.; Shibata, T. Regulatory changes after the enforcement of the new Clinical Trials Act in Japan. Jpn. J. Clin. Oncol. 2020, 50, 399–404. [Google Scholar] [CrossRef]
- Leung, S.Y.; Chan, T.L.; Chung, L.P.; Chan, A.S.; Fan, Y.W.; Hung, K.N.; Kwong, W.K.; Ho, J.W.; Yuen, S.T. Microsatellite instability and mutation of DNA mismatch repair genes in gliomas. Am. J. Pathol. 1998, 153, 1181–1188. [Google Scholar] [CrossRef] [Green Version]
- Hause, R.J.; Pritchard, C.C.; Shendure, J.; Salipante, S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016, 22, 1342–1350. [Google Scholar] [CrossRef]
- Martinez, R.; Schackert, H.K.; Plaschke, J.; Baretton, G.; Appelt, H.; Schackert, G. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology 2004, 66, 395–403. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.M.; Su, F.Y.; Kalyana-Sundaram, S.; Khazanov, N.; Ateeq, B.; Cao, X.H.; Lonigro, R.J.; Vats, P.; Wang, R.; Lin, S.F.; et al. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discov. 2013, 3, 636–647. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Yu, Y.; Grimmer, M.R.; Wahl, M.; Chang, S.M.; Costello, J.F. Temozolomide-associated hypermutation in gliomas. Neuro Oncol. 2018, 20, 1300–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Characteristic | Subset | n Total (Recurrent) | Median Age (Range) [y] |
|---|---|---|---|
| Gender | male | 63 (15) | 49 (24–91) |
| female | 41 (11) | 59 (22–72) | |
| Genomic Profiling Test | FoundationOne CDx Cancer Genomic Profile | 94 (24) | 49.5 (24–91) |
| OncoGuide NCC OncoPanel System | 10 (2) | 49 (22–72) | |
| Diagnosis (WHO 2021) | Glioblastoma, IDH-wildtype | 49 (12) | 56 (27–91) |
| Diffuse Astrocytoma, IDH-wildtype, NEC | 6 (0) | 49 (37–64) | |
| Astrocytoma, IDH-mutant | |||
| Grade 4 | 8 (5) | 40.5 (31–52) | |
| Grade 3 | 11 (3) | 46 (29–74) | |
| Oligodendroglioma, IDH-mutant, and 1p/19q-codeleted | |||
| Grade 3 | 17 (6) | 46 (29–74) | |
| Grade 2 | 6 (0) | 49 (35–72) | |
| Diffuse midline glioma, H3 K27-altered | 6 (0) | 36.5 (30–45) | |
| Pilocytic astrocytoma | 1 (0) | 22 (NA) |
| No. | ID (in this Cohort) | CGPT | Diagnosis | Age | Gender | Acctionable Gene Aberration | Therapeutic Agents | Drug Type |
|---|---|---|---|---|---|---|---|---|
| 1 | 58_GBM | F-One | GBM, IDH-wt | 40 | M | BRAF V600E | Dabrafenib/Trametinib | clinical trial |
| 2 | 63_GBM | F-One | GBM, IDH-wt | 62 | F | BRAF V600E | Dabrafenib/Trametinib | clinical trial |
| 3 | 65_GBM | F-One | GBM, IDH-wt | 52 | F | BRAF V600E | Dabrafenib/Trametinib | clinical trial |
| 4 | 71_GBM | F-One | GBM, IDH-wt | 55 | F | FGFR1 K656E | FGFR inhibitor | Investigational drug * |
| 5 | 87_GBM | F-One | GBM, IDH-wt | 59 | M | FGFR3 FGFR3-TACC3 fusion | FGFR inhibitor | Investigational drug * |
| 6 | 101_GBM | F-One | GBM, IDH-wt | 59 | F | MSH6 C694fs*4, MSH6 I795fs*15 | Pembrolizumab | clinical trial * |
| 7 | 111_GBM | F-One | GBM, IDH-wt | 53 | M | FGFR3 FGFR3-TACC3 fusion | FGFR inhibitor | Investigational drug * |
| No. | ID (in this Cohort) | CGPT | Diagnosis | Age | Gender | Gene Aberration | Blood Sample Diagnosis | Therapeutic Drug Accesibility |
|---|---|---|---|---|---|---|---|---|
| 1 | 92_GBM | F-One | GBM, IDH-wt | 91 | M | BRCA2 R2318* | no | no |
| 2 | 101_GBM | F-One | GBM, IDH-wt | 59 | F | MSH6 C694fs*4, MSH6 I795fs*15 | no | Yes (Pembrolizumab) * |
| 3 | 104_DA_NEC | NOP | Diffuse Astrocytoma, IDH-wt, NEC | 51 | F | NF1 Q2434* | Yes | no |
| 4 | 115_AST | F-One | Astrocytoma, IDH-mt | 51 | M | MSH6 Y524fs*46 | no | no |
| 5 | 119_OLG | F-One | Oligodendroglioma, IDH-mt | 52 | F | BRCA2 I682fs*48 | no | Yes (Pembrolizumab/Olaparib) * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omura, T.; Takahashi, M.; Ohno, M.; Miyakita, Y.; Yanagisawa, S.; Tamura, Y.; Kikuchi, M.; Kawauchi, D.; Nakano, T.; Hosoya, T.; et al. Clinical Application of Comprehensive Genomic Profiling Tests for Diffuse Gliomas. Cancers 2022, 14, 2454. https://doi.org/10.3390/cancers14102454
Omura T, Takahashi M, Ohno M, Miyakita Y, Yanagisawa S, Tamura Y, Kikuchi M, Kawauchi D, Nakano T, Hosoya T, et al. Clinical Application of Comprehensive Genomic Profiling Tests for Diffuse Gliomas. Cancers. 2022; 14(10):2454. https://doi.org/10.3390/cancers14102454
Chicago/Turabian StyleOmura, Takaki, Masamichi Takahashi, Makoto Ohno, Yasuji Miyakita, Shunsuke Yanagisawa, Yukie Tamura, Miyu Kikuchi, Daisuke Kawauchi, Tomoyuki Nakano, Tomohiro Hosoya, and et al. 2022. "Clinical Application of Comprehensive Genomic Profiling Tests for Diffuse Gliomas" Cancers 14, no. 10: 2454. https://doi.org/10.3390/cancers14102454
APA StyleOmura, T., Takahashi, M., Ohno, M., Miyakita, Y., Yanagisawa, S., Tamura, Y., Kikuchi, M., Kawauchi, D., Nakano, T., Hosoya, T., Igaki, H., Satomi, K., Yoshida, A., Sunami, K., Hirata, M., Shimoi, T., Sudo, K., Okuma, H. S., Yonemori, K., ... Narita, Y. (2022). Clinical Application of Comprehensive Genomic Profiling Tests for Diffuse Gliomas. Cancers, 14(10), 2454. https://doi.org/10.3390/cancers14102454







