Prognostic MicroRNA Panel for HCV-Associated HCC: Integrating Computational Biology and Clinical Validation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis
2.2. Patients and Samples
2.2.1. Inclusion and Exclusion Criteria
2.2.2. Sampling and Serum Preparation
2.2.3. Liver Function Testing and HCV Antibodies Testing
2.3. RNA Isolation and cDNA Synthesis
2.4. Real Time PCR Amplification of miRNAs
- hsa-miR-142-3p; 5′UGUAGUGUUUCCUACUUUAUGGA3′,
- hsa-miR-150-5p; 5′UCUCCCAACCCUUGUACCAGUG3′,
- hsa-miR-183-5p; 5′UAUGGCACUGGUAGAAUUCACU3′,
- hsa-miR-199a-3p; 3′ACAGUAGUCUGCACAUUGGUUA5′,
- hsa-miR-215-5p; 5′AUGACCUAUGAAUUGACAGAC3′,
- hsa-miR-217-5p, 5′UACUGCAUCAGGAACUGAUUGGA3′,
- hsa-miR-224-5p; 5′UCAAGUCACUAGUGGUUCCGUUUAG3′,
- hsa-miR-424-5p; 5′CAGCAGCAAUUCAUGUUUUGAA3′ and
- hsa-miR-3607-5p; 5′GCAUGUGAUGAAGCAAAUCAGU3′.
2.5. Data Analysis
2.6. Logistic Regression Analysis
2.7. Pathway Analysis
2.8. Statistics
3. Results
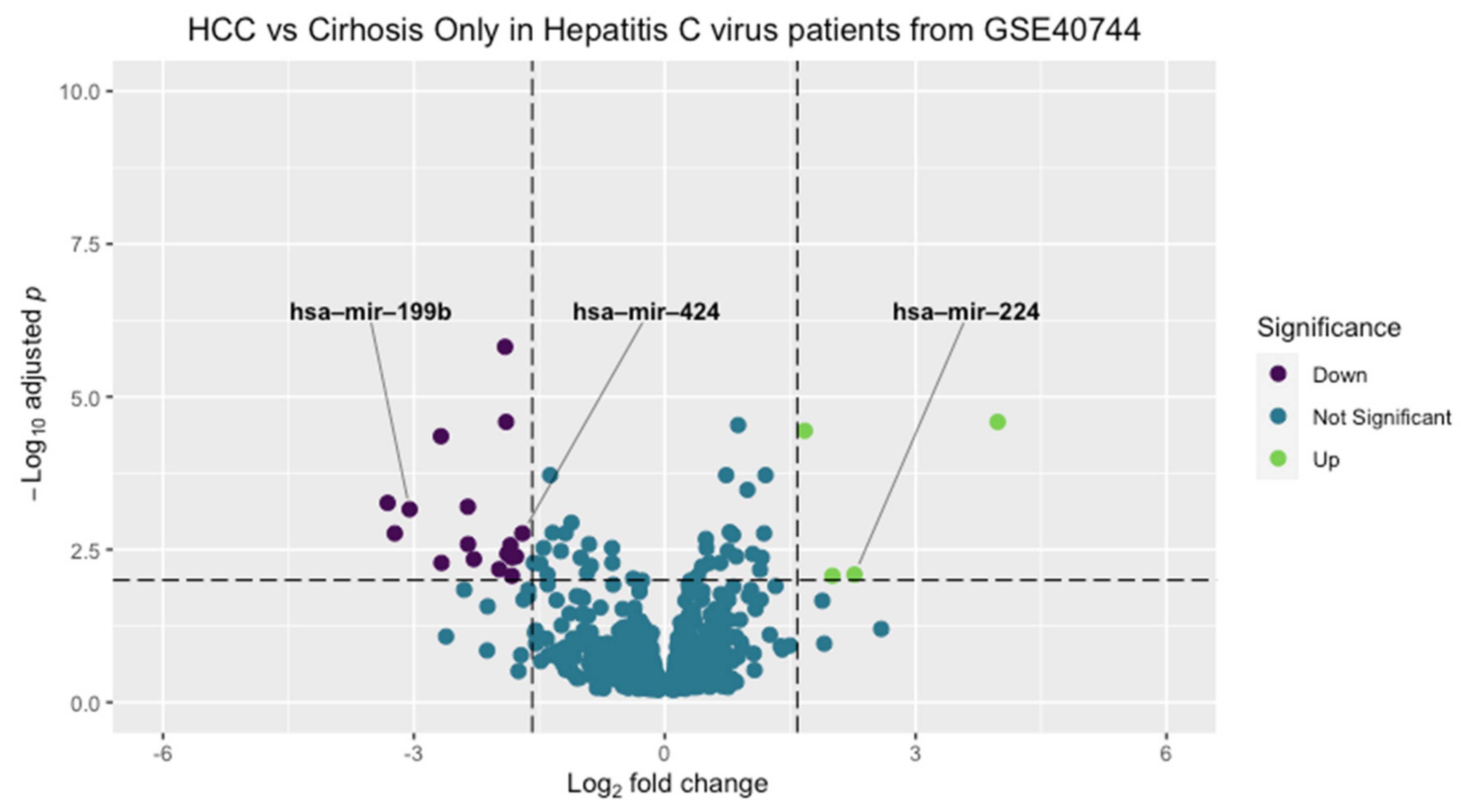
3.1. Microarray Bioinformatic Analysis
3.2. RNA Sequencing Bioinformatic Analysis
3.3. Description of the Study Population
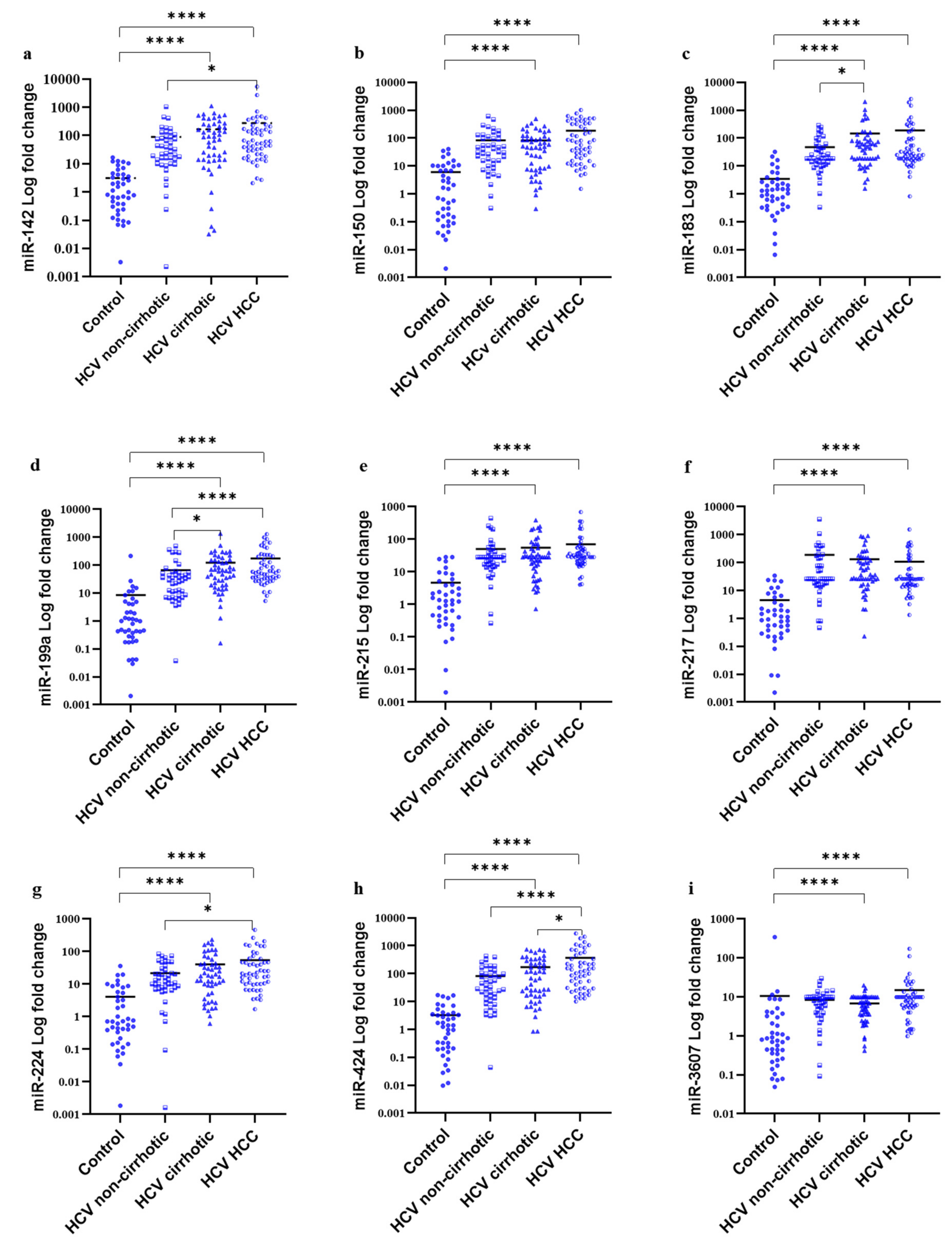
3.4. MiRNAs Serum Signature in the Study Groups
3.5. Correlation between the Studied miRNAs
3.6. Diagnostic Potential of the DE-miRNAs
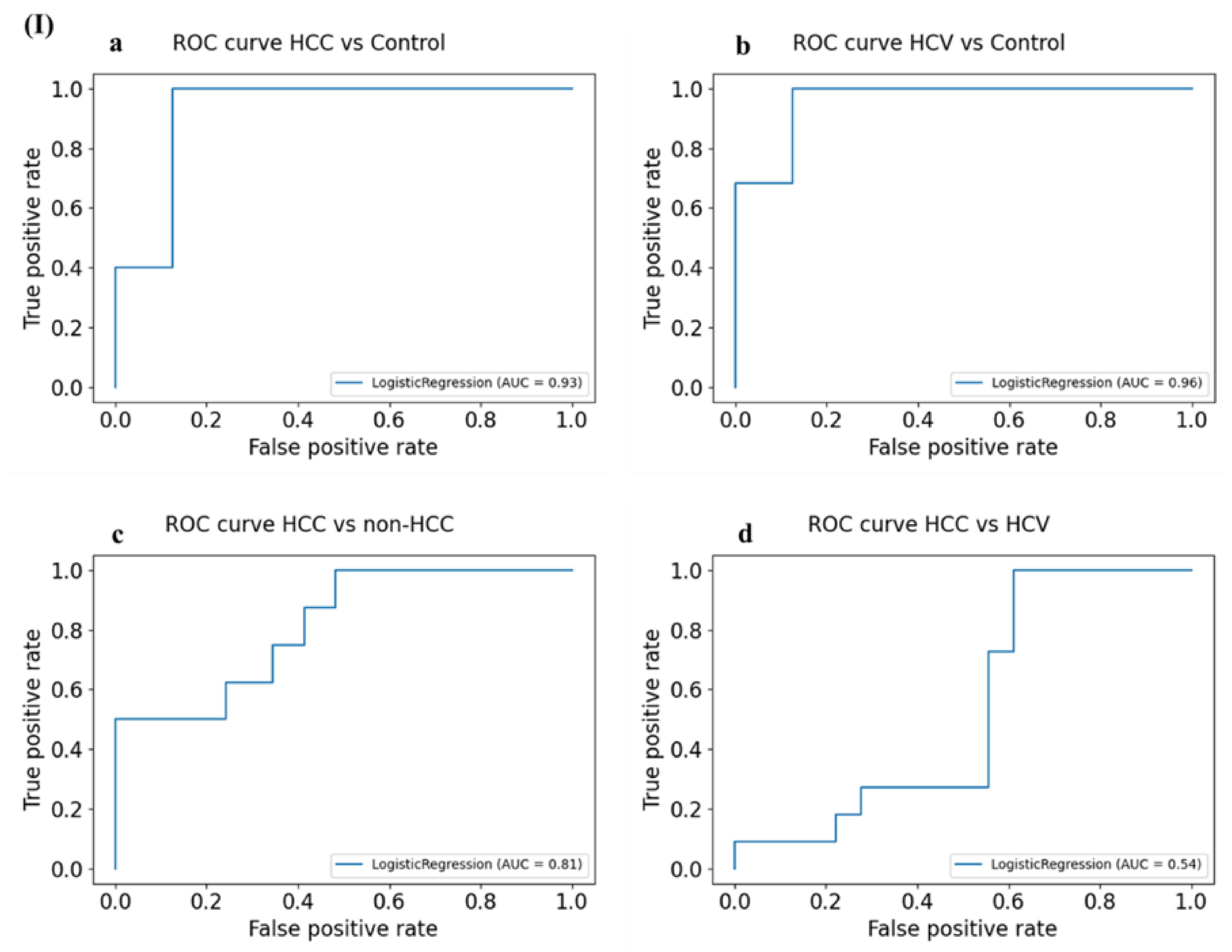
3.6.1. Diagnostic Potential of the DE-miRNAs in HCC Patients Compared to Healthy Individuals
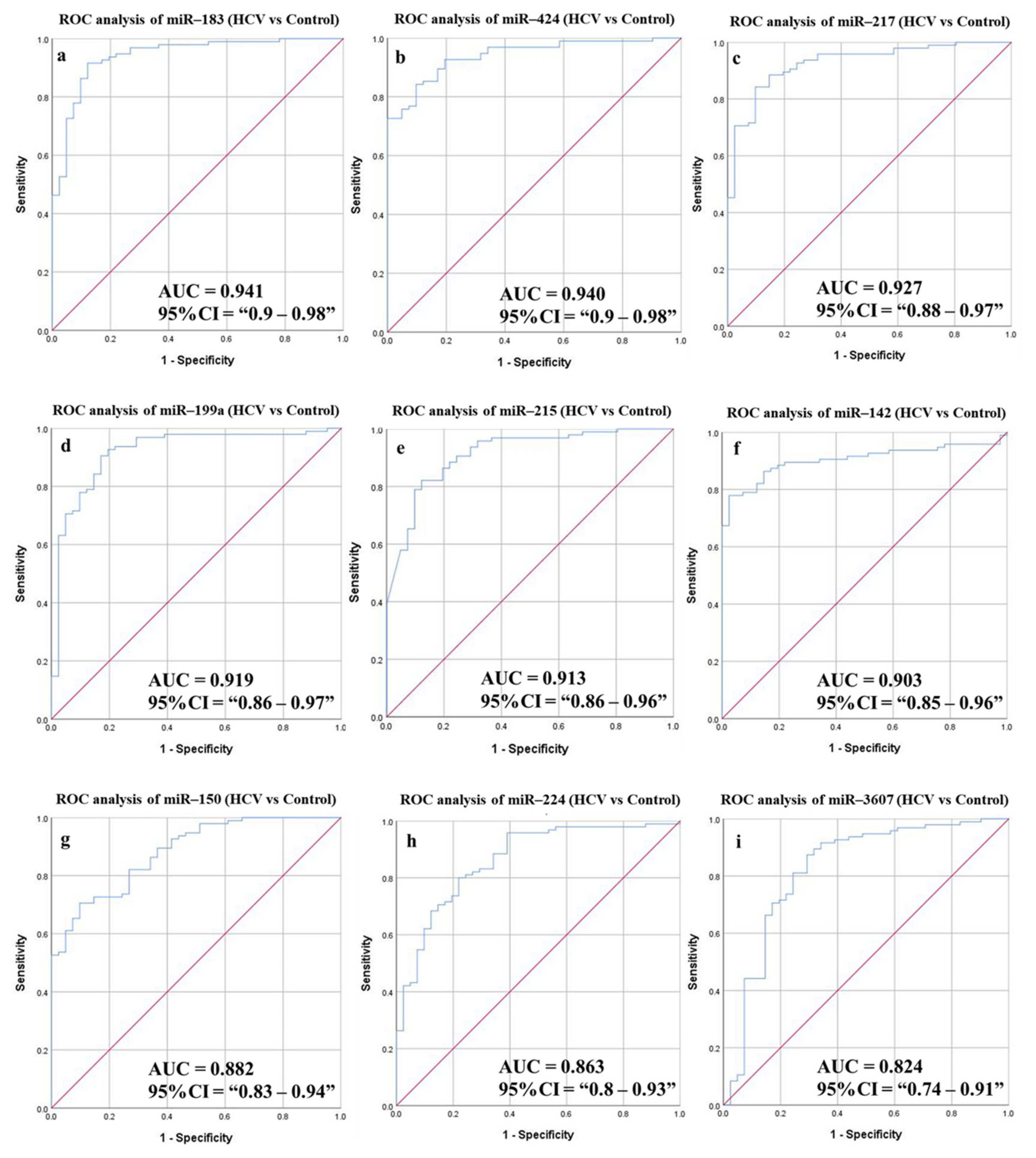
3.6.2. Diagnostic Potential of the DE-miRNAs in HCV Patients Compared to Healthy Individuals
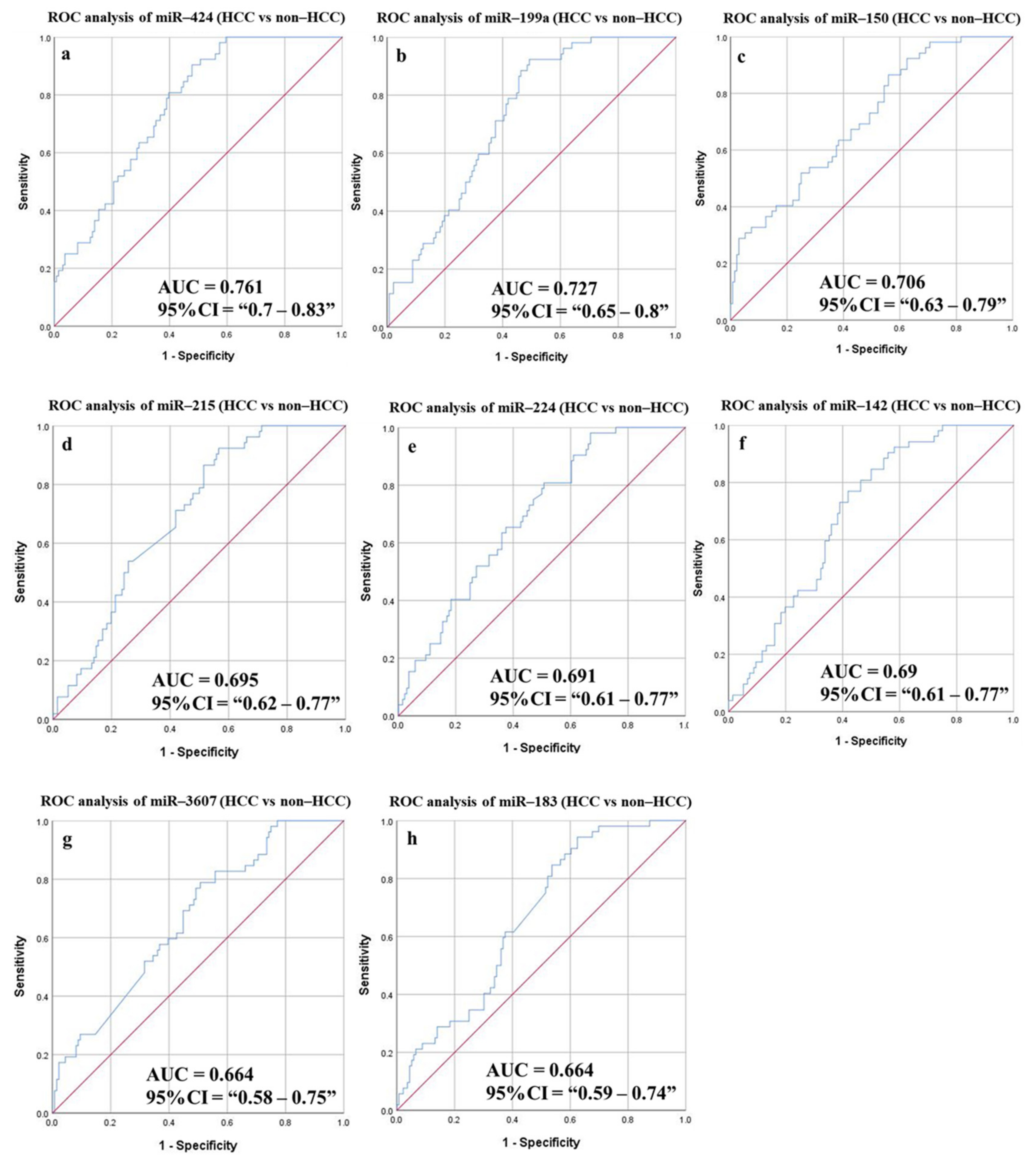
3.6.3. Diagnostic Potential of the DE-miRNAs in HCC Patients Compared to Non-HCC Individuals
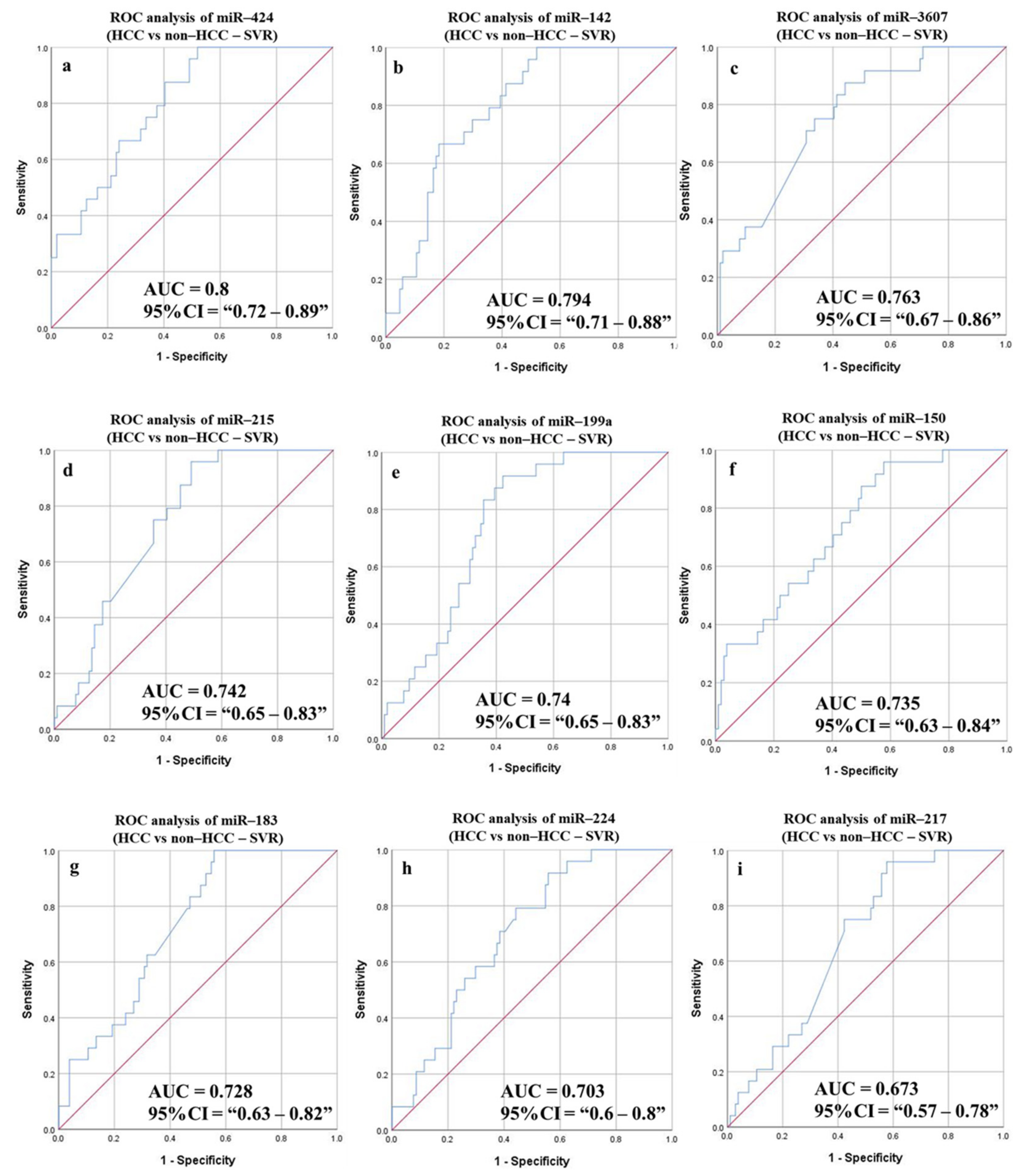
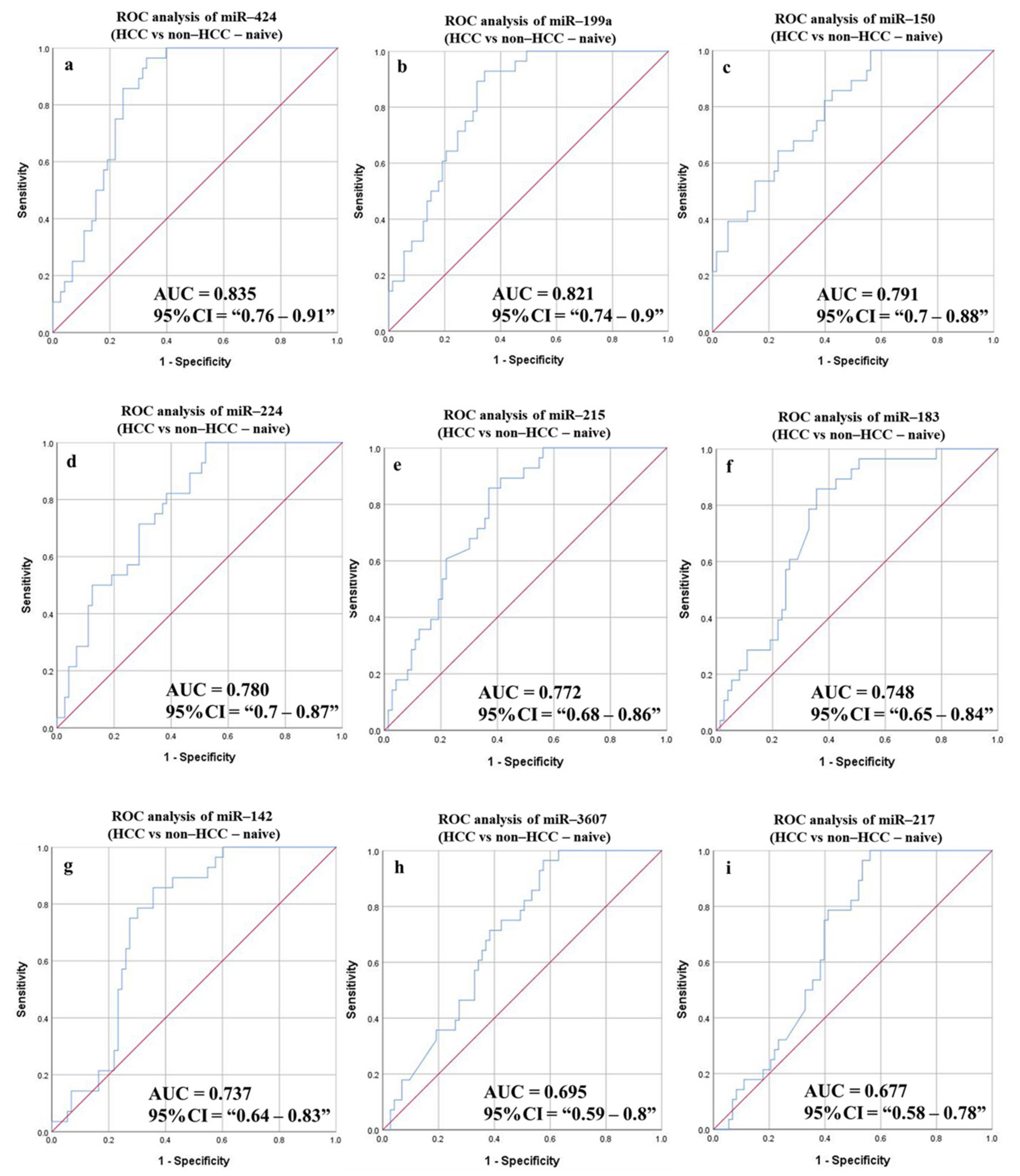
3.6.4. Diagnostic Potential of the DE-miRNAs in HCC (Sustained Virological Response (SVR)/Treatment Naïve) Patients Compared to Non-HCC (SVR/Treatment Naïve) Patients
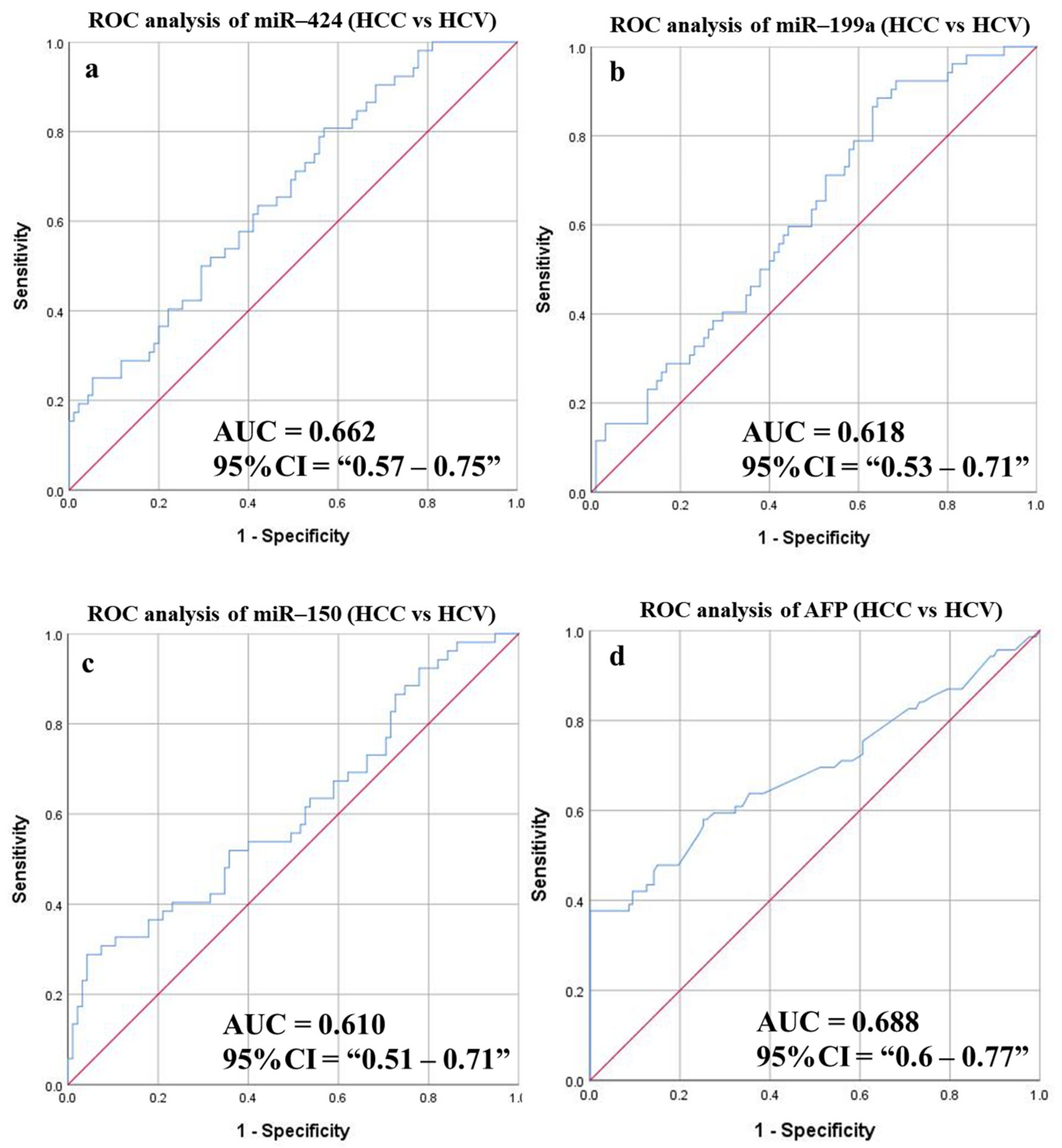
3.6.5. Diagnostic Potential of the DE-miRNAs in HCC Patients Compared to Patients with HCV
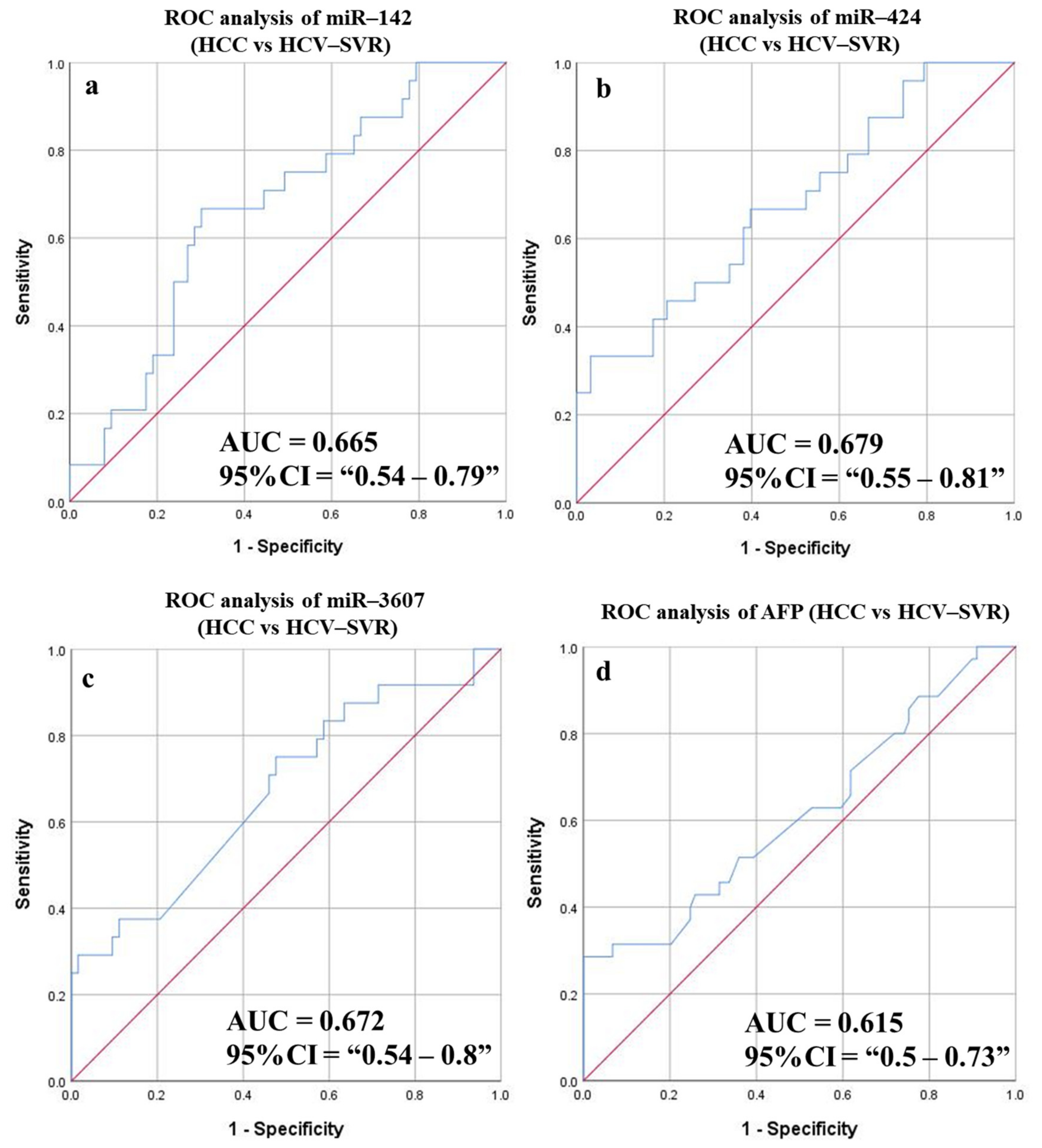
3.6.6. Diagnostic Potential of the DE-miRNAs in HCC (SVR) Patients Compared to Patients with HCV (SVR)
3.7. Establishment of the miRNAs’ Predictive Ability Using Logistic Regression Analysis
3.7.1. Normalized Logistic Regression (LR) Weights
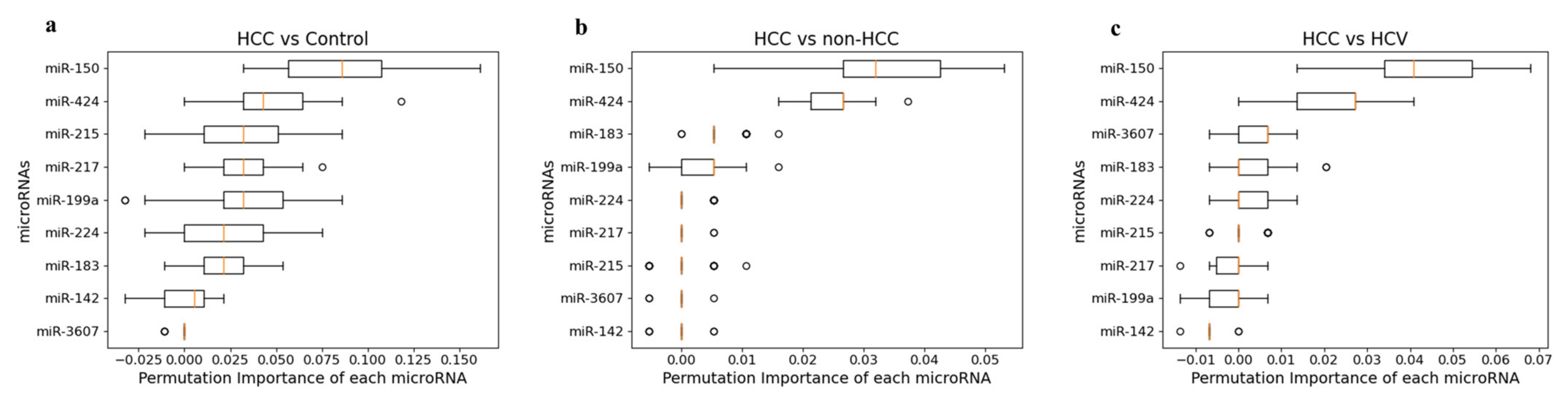
3.7.2. Permutation Feature Importance Measurement
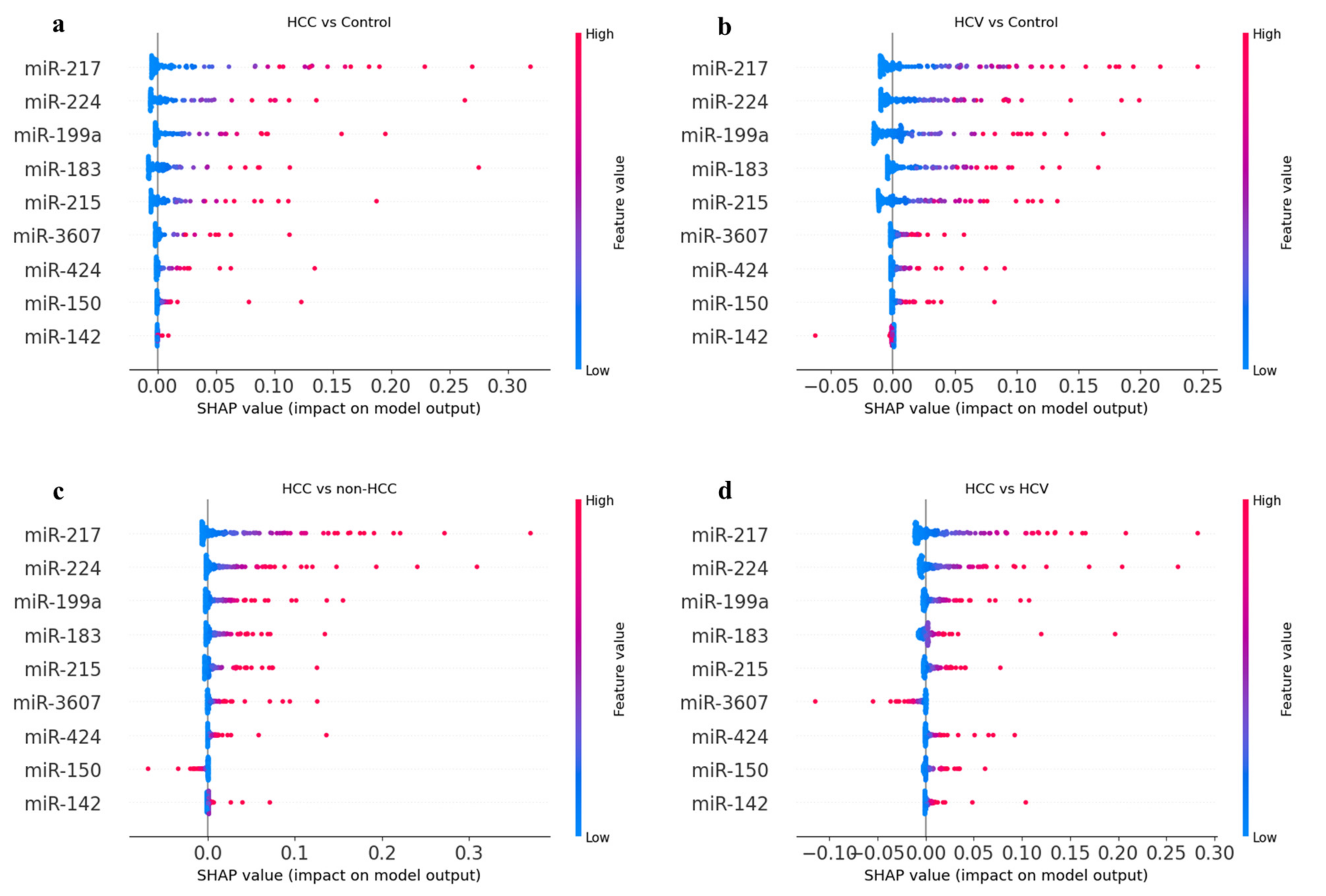
3.7.3. SHapley Additive exPlanation (SHAP) Analysis
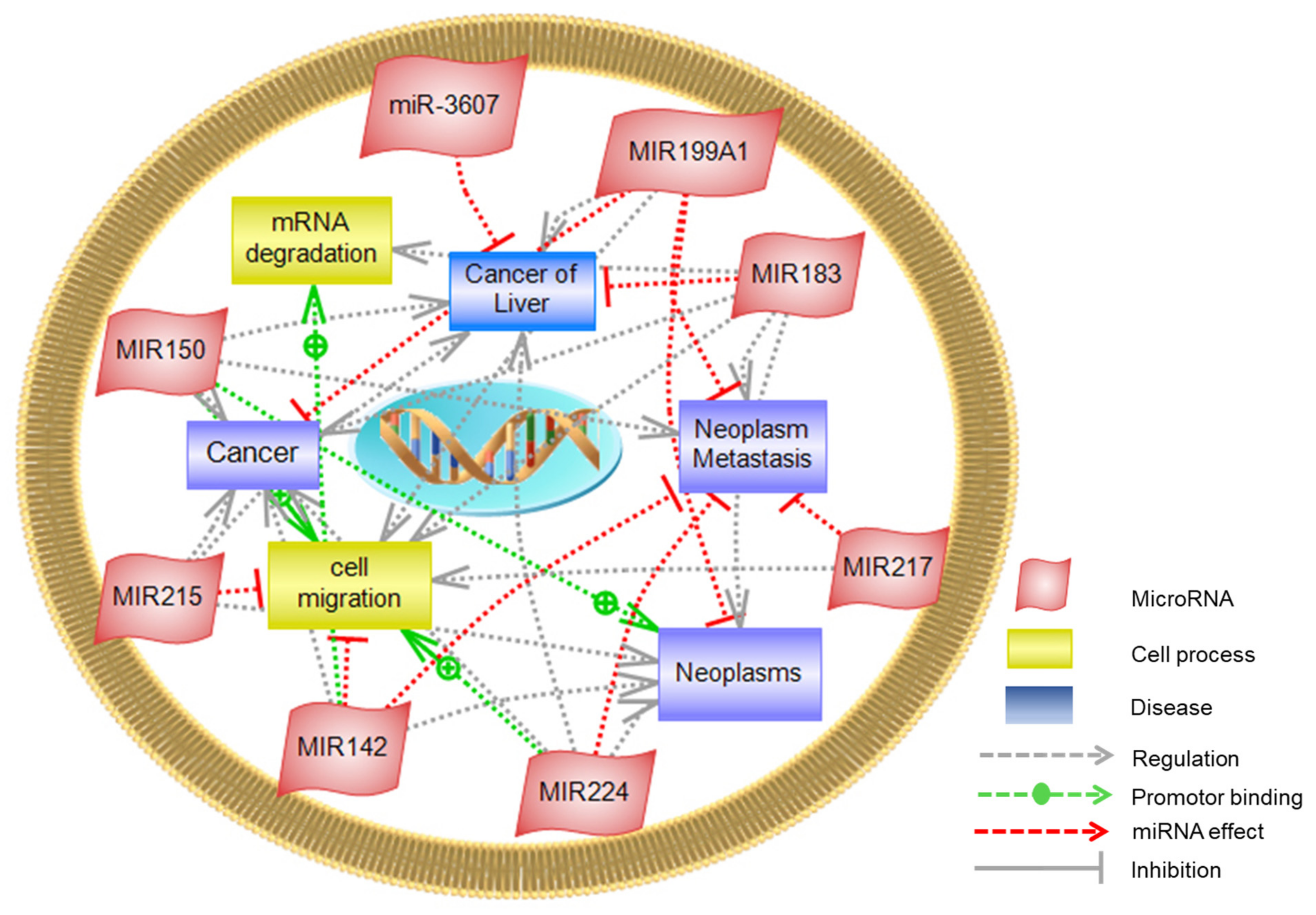
3.8. Pathway Studio
4. Discussion
Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petrick, J.L.; McGlynn, K.A. The Changing Epidemiology of Primary Liver Cancer. Curr. Epidemiol. Rep. 2019, 6, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- World Health Organization International Agency for Research on Cancer Latest Global Cancer Data 2018. Available online: https://www.who.int/cancer/PRGlobocanFinal.pdf (accessed on 3 May 2021).
- Dasgupta, P.; Henshaw, C.; Youlden, D.R.; Clark, P.J.; Aitken, J.F.; Baade, P.D. Global Trends in Incidence Rates of Primary Adult Liver Cancers: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 171. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization GLOBOCAN 2020: Bladder Cancer 10th Most Commonly Diagnosed Worldwide—World Bladder Cancer Patient Coalition. Available online: https://worldbladdercancer.org/news_events/globocan-2020-bladder-cancer-10th-most-commonly-diagnosed-worldwide/ (accessed on 3 May 2021).
- El-Zayadi, A.R.; Badran, H.M.; Barakat, E.M.F.; Attia, M.E.D.; Shawky, S.; Mohamed, M.K.; Selim, O.; Saed, A. Hepatocellular carcinoma in Egypt: A single center study over a decade. World J. Gastroenterol. 2005, 11, 5193–5198. [Google Scholar] [CrossRef]
- Demerdash, H.M.; Hussien, H.M.; Hassouna, E.; Arida, E.A. Detection of MicroRNA in Hepatic Cirrhosis and Hepatocellular Carcinoma in Hepatitis C Genotype-4 in Egyptian Patients. Biomed. Res. Int. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Fiorino, S.; Bacchi-Reggiani, M.L.; Visani, M.; Acquaviva, G.; Fornelli, A.; Masetti, M.; Tura, A.; Grizzi, F.; Zanello, M.; Mastrangelo, L.; et al. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis b-and c-related-hepatocellularcarcinoma. World J. Gastroenterol. 2016, 22, 3907–3936. [Google Scholar] [CrossRef]
- Trevisani, F.; D’Intino, P.E.; Morselli-Labate, A.M.; Mazzella, G.; Accogli, E.; Caraceni, P.; Domenicali, M.; De Notariis, S.; Roda, E.; Bernardi, M. Serum α-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: Influence of HBsAg and anti-HCV status. J. Hepatol. 2001, 34, 570–575. [Google Scholar] [CrossRef]
- Stefaniuk, P.; Cianciara, J.; Wiercinska-Drapalo, A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 2010, 16, 418–424. [Google Scholar] [CrossRef]
- Chen, S.; Chen, H.; Gao, S.; Qiu, S.; Zhou, H.; Yu, M.; Tu, J. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol. Res. 2017, 47, 312–320. [Google Scholar] [CrossRef]
- Han, L.L.; Lv, Y.; Guo, H.; Ruan, Z.P.; Nan, K.J. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J. Gastroenterol. 2014, 20, 10249–10261. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef] [PubMed]
- Dimitroulis, D.; Damaskos, C.; Valsami, S.; Davakis, S.; Garmpis, N.; Spartalis, E.; Athanasiou, A.; Moris, D.; Sakellariou, S.; Kykalos, S.; et al. From diagnosis to treatment of hepatocellular carcinoma: An epidemic problem for both developed and developing world. World J. Gastroenterol. 2017, 23, 5282–5294. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Liu, M.; Liu, J.; Wang, L.; Wu, H.; Zhou, C.; Zhu, H.; Xu, N.; Xie, Y. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS ONE 2014, 9, e0109347. [Google Scholar] [CrossRef]
- Giordano, S.; Columbano, A. MicroRNAs: New tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 2013, 57, 840–847. [Google Scholar] [CrossRef]
- Mizuguchi, Y.; Takizawa, T.; Yoshida, H.; Uchida, E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol. Res. 2016, 46, 391–406. [Google Scholar] [CrossRef]
- Morishita, A.; Fujita, K.; Iwama, H.; Chiyo, T.; Fujihara, S.; Oura, K.; Tadokoro, T.; Mimura, S.; Nomura, T.; Tani, J.; et al. Role of microRNA-210-3p in hepatitis B virus-related hepatocellular carcinoma. Am. J. Physiol.-Gastrointest. Liver Physiol. 2020, 318, G401–G409. [Google Scholar] [CrossRef]
- Pezzuto, F.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. The role of circulating free DNA and microRNA in non-invasive diagnosis of HBV- and HCV-related hepatocellular carcinoma. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Wu, J.; Wu, Y.; Luo, Y.; Li, X.; Lin, N.; Yang, X.; Lin, Y.; Chen, M. Circulating miRNA-199a and miRNA-122 Levels as Potential Diagnostic and Prognostic Biomarkers for Hepatocellular Carcinoma. Available online:https://pubmed.ncbi.nlm.nih.gov/32366560/ (accessed on 3 May 2021).
- Li, L.M.; Hu, Z.B.; Zhou, Z.X.; Chen, X.; Liu, F.Y.; Zhang, J.F.; Shen, H.B.; Zhang, C.Y.; Zen, K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010, 70, 9798–9807. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, L.; Gao, X.; Hu, J.; Wang, J.; Dai, Z.; Wang, J.F.; Zhang, Z.; Lu, S.; Huang, X.; et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 2011, 29, 4781–4788. [Google Scholar] [CrossRef] [PubMed]
- National Center of Biotechnology Information Gene Expression Omnibus GEO Accession Viewer. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40744 (accessed on 3 May 2021).
- The Cancer Genome Atlas The Cancer Genome Atlas—Hepatocellular Carcinoma Study—National Cancer Institute. Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/liver (accessed on 3 May 2021).
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Parikh, N.D.; Rich, N.E.; John, B.V.; Pillai, A. Hepatocellular Carcinoma Surveillance and Staging. In Hepatocellular Carcinoma; Spring: Berlin/Heidelberg, Germany, 2019; pp. 27–51. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McCall, M.N.; McMurray, H.R.; Land, H.; Almudevar, A. On non-detects in qPCR data. Bioinformatics 2014, 30, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Sherina, V.; McMurray, H.R.; Powers, W.; Land, H.; Love, T.M.T.; McCall, M.N. Multiple imputation and direct estimation for qPCR data with non-detects. BMC Bioinform. 2020, 21, 545. [Google Scholar] [CrossRef]
- Gevaert, A.B.; Witvrouwen, I.; Vrints, C.J.; Heidbuchel, H.; Van Craenenbroeck, E.M.; Van Laere, S.J.; Van Craenenbroeck, A.H. MicroRNA profiling in plasma samples using qPCR arrays: Recommendations for correct analysis and interpretation. PLoS ONE 2018, 13, e0193173. [Google Scholar] [CrossRef]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef]
- Shapley, L. 17. A Value for n-Person Games. In Contributions to the Theory of Games (AM-28); Kuhn, H., Tucker, A., Eds.; Princeton University Press: Princeton, NJ, USA, 2016; Volume II, pp. 307–318. [Google Scholar] [CrossRef]
- Biological Research—Pathway Studio. Elsevier. Available online: https://www.elsevier.com/solutions/pathway-studio-biological-research (accessed on 9 January 2022).
- El-Garem, H.; Ammer, A.; Shehab, H.; Shaker, O.; Anwer, M.; El-Akel, W.; Omar, H. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J. Hepatol. 2014, 6, 818–824. [Google Scholar] [CrossRef]
- El-Nady, G.M.; Ling, R.; Harrison, T.J. Gene expression in HCV-associated hepatocellular carcinoma—Upregulation of a gene encoding a protein related to the ubiquitin-conjugating enzyme. Liver Int. 2003, 23, 329–337. [Google Scholar] [CrossRef]
- Alter, M.J. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 2007, 13, 2436–2441. [Google Scholar] [CrossRef]
- Lopez, P.M.; Villanueva, A.; Llovet, J.M. Systematic review: Evidence-based management of hepatocellular carcinoma—An updated analysis of randomized controlled trials. Aliment. Pharmacol. Ther. 2006, 23, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Tsai, H.I.; Lee, W.C.; Huang, S.W.; Lin, C.Y.; Hsieh, Y.C.; Kuo, T.; Chen, C.W.; Yu, M.C. Normal Alpha-Fetoprotein Hepatocellular Carcinoma: Are They Really Normal? J. Clin. Med. 2019, 8, 1736. [Google Scholar] [CrossRef] [PubMed]
- Carr, B.I.; Akkiz, H.; Üsküdar, O.; Yalçın, K.; Guerra, V.; Kuran, S.; Karaoğullarından, Ü.; Altıntaş, E.; Özakyol, A.; Tokmak, S.; et al. HCC with low- and normal-serum alpha-fetoprotein levels. Clin. Pract. 2018, 15, 453. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Z.; Gao, Y.; Zhai, D.; Rao, Q.; Shi, W.; Yang, B.; Jing, L.; Guo, H.; Liu, T.; et al. Factors influencing circulating microRNA level in the studies of hepatocellular carcinoma biomarker. Neoplasma 2015, 62, 798–804. [Google Scholar] [CrossRef][Green Version]
- Bharali, D.; Jebur, H.B.; Baishya, D.; Kumar, S.; Sarma, M.P.; Masroor, M.; Akhter, J.; Husain, S.A.; Kar, P. Expression analysis of serum microRNA-34a and microRNA-183 in hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2018, 19, 2561–2568. [Google Scholar] [CrossRef]
- Liu, A.M.; Yao, T.J.; Wang, W.; Wong, K.F.; Lee, N.P.; Fan, S.T.; Poon, R.T.P.; Gao, C.; Luk, J.M. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open 2012, 2, e000825. [Google Scholar] [CrossRef]
- Gui, J.; Tian, Y.; Wen, X.; Zhang, W.; Zhang, P.; Gao, J.; Run, W.; Tian, L.; Jia, X.; Gao, Y. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. 2011, 120, 183–193. [Google Scholar] [CrossRef]
- Li, J.; Jin, B.; Wang, T.; Li, W.; Wang, Z.; Zhang, H.; Song, Y.; Li, N. Serum microRNA expression profiling identifies serum biomarkers for HCV-related hepatocellular carcinoma. Cancer Biomark. 2019, 26, 501–512. [Google Scholar] [CrossRef]
- Yoon, E.L.; Yeon, J.E.; Ko, E.; Lee, H.J.; Je, J.H.; Yoo, Y.J.; Kang, S.H.; Suh, S.J.; Kim, J.H.; Seo, Y.S.; et al. An explorative analysis for the role of serum mir-10b-3p levels in predicting response to sorafenib in patients with advanced hepatocellular carcinoma. J. Korean Med. Sci. 2017, 32, 212–220. [Google Scholar] [CrossRef]
- Zhuang, L.P.; Meng, Z.Q. Serum miR-224 reflects stage of hepatocellular carcinoma and predicts survival. Biomed. Res. Int. 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, C.; Li, Y.; He, X.; He, M. MiR-224 is an early-stage biomarker of hepatocellular carcinoma with miR-224 and miR-125b as prognostic biomarkers. Biomark. Med. 2020, 14, 1485–1500. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.H.; Higazi, A.M.; Moness, H.M.; Farag, N.M.; Saad, Z.M.; Moukareb, H.A.; Soliman, W.; El Sagheer, G.; Abd El Hamid, S.R.; Hamid, H.A. Clinical significances and diagnostic utilities of both miR-215 and squamous cell carcinoma antigen-IgM versus alpha-fetoprotein in egyptian patients with hepatitis C virus-induced hepatocellular carcinoma. Clin. Exp. Gastroenterol. 2019, 12, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Mamdouh, S.; Khorshed, F.; Aboushousha, T.; Hamdy, H.; Diab, A.; Seleem, M.; Saber, M. Evaluation of Mir-224, Mir-215 and Mir-143 as Serum biomarkers for HCV associated Hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2017, 18, 3167–3171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Meng, H.; Wang, N.; Liang, L.N.; Liu, L.N.; Lu, S.M.; Luan, Y. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn. Pathol. 2014, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Alahmady, Z.; Yamany, H.; Aboulfottoh, A. Serum Expression Levels of miR-141 and miR-215 for Differentiation between Liver Cirrhosis, Chronic Hepatitis C and Hepatocellular Carcinoma Patients. Microbiol. Res. J. Int. 2017, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cabral, B.C.A.; Hoffmann, L.; Bottaro, T.; Costa, P.F.; Ramos, A.L.A.; Coelho, H.S.M.; Villela-Nogueira, C.A.; Ürményi, T.P.; Faffe, D.S.; Silva, R. Circulating microRNAs associated with liver fibrosis in chronic hepatitis C patients. Biochem. Biophys. Rep. 2020, 24, 100814. [Google Scholar] [CrossRef]
- Devhare, P.B.; Steele, R.; Bisceglie, A.M.D.; Kaplan, D.E.; Ray, R.B. Differential expression of MicroRNAs in hepatitis C virus-mediated liver disease between African Americans and Caucasians: Implications for racial health disparities. Gene Expr. 2017, 17, 89–98. [Google Scholar] [CrossRef]
- Shaheen, N.M.H.; Zayed, N.; Riad, N.M.; Tamim, H.H.; Shahin, R.M.H.; Labib, D.A.; ELsheikh, S.M.; Moneim, R.A.; Yosry, A.; khalifa, R.H. Role of circulating miR-182 and miR-150 as biomarkers for cirrhosis and hepatocellular carcinoma post HCV infection in Egyptian patients. Virus Res. 2018, 255, 77–84. [Google Scholar] [CrossRef]
- Zhang, S.; Ouyang, X.; Jiang, X.; Gu, D.; Lin, Y.; Kong, S.K.; Xie, W. Dysregulated serum microRNA expression profile and potential biomarkers in hepatitis C virus-infected patients. Int. J. Med. Sci. 2015, 12, 590–598. [Google Scholar] [CrossRef]
- Yao, H.; Liu, X.; Chen, S.; Xia, W.; Chen, X. Decreased expression of serum miR-424 correlates with poor prognosis of patients with hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 14830–14835. [Google Scholar] [PubMed]
- Yalcinkaya, B.; TANOĞLU, E.G.; Taştekin, D.; Pence, S. Role of mir-33a, mir-203b, mir-361-3p, mir-424 in hepatocellular carcinoma. TURKISH J. Med. Sci. 2021, 51, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.Z.; Zhang, K.; Li, H.; Afdhal, N.H.; Albitar, M. Circulating MicroRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 2011, 45, 355–360. [Google Scholar] [CrossRef]
- Kamel, R.R.; Amr, K.S.; Afify, M.; Elhosary, Y.A.; Hegazy, A.E.; Fahim, H.H.; Ezzat, W.M. Relation between microRNAs and apoptosis in hepatocellular carcinoma. Open Access Maced. J. Med. Sci. 2016, 4, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Amr, K.S.; Ezzat, W.M.; Elhosary, Y.A.; Hegazy, A.E.; Fahim, H.H.; Kamel, R.R. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene 2016, 575, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hou, P.; Wu, Z.; Wang, T.; Nie, Y. Circulating miR-375 and miR-199a-3p as potential biomarkers for the diagnosis of hepatocellular carcinoma. Tumor Biol. 2015, 36, 4501–4507. [Google Scholar] [CrossRef] [PubMed]
- Zekri, A.R.N.; Youssef, A.S.E.D.; El-Desouky, E.D.; Ahmed, O.S.; Lotfy, M.M.; Nassar, A.A.M.; Bahnassey, A.A. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumor Biol. 2016, 37, 12273–12286. [Google Scholar] [CrossRef]
- Murakami, Y.; Aly, H.H.; Tajima, A.; Inoue, I.; Shimotohno, K. Regulation of the hepatitis C virus genome replication by miR-199a*. J. Hepatol. 2009, 50, 453–460. [Google Scholar] [CrossRef]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef]
- Jia, X.Q.; Cheng, H.Q.; Qian, X.; Bian, C.X.; Shi, Z.M.; Zhang, J.P.; Jiang, B.H.; Feng, Z.Q. Lentivirus-Mediated Overexpression of MicroRNA-199a Inhibits Cell Proliferation of Human Hepatocellular Carcinoma. Cell Biochem. Biophys. 2012, 62, 237–244. [Google Scholar] [CrossRef]
- El-Guendy, N.M.; Helwa, R.; El-Halawany, M.S.; Ali, S.A.R.; Aly, M.T.; Alieldin, N.H.; Fouad, S.A.H.; Saeid, H.; Abdel-Wahab, A.H.A. The liver microRNA expression profiles associated with chronic hepatitis C virus (HCV) genotype-4 infection: A preliminary study. Hepat. Mon. 2016, 16, 33881. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.; Yang, M.; Su, Y.; Xie, R. Dysregulation of miR-3607 predicts prognosis of hepatocellular carcinoma and regulates tumor cell proliferation, migration and invasion. Diagn. Pathol. 2020, 15, 1–9. [Google Scholar] [CrossRef]
- Hu, W.Y.; Wei, H.Y.; Liu, L.Y.; Li, K.M.; Wang, R.B.; Xu, X.Q.; Feng, R. miR-3607, a biomarker of hepatocellular carcinoma invasion and aggressiveness: Its relationship with epithelial-mesenchymal transition process. IUBMB Life 2020, 72, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Chen, J.; Ding, B.; Fan, W. XIAP, commonly targeted by tumor suppressive miR-3607-5p and miR-3607-3p, promotes proliferation and inhibits apoptosis in hepatocellular carcinoma. Genomics 2021, 113, 933–945. [Google Scholar] [CrossRef] [PubMed]
- El-Tawdi, A.H.F.; Matboli, M.; Shehata, H.H.; Tash, F.; El-Khazragy, N.; Azazy, A.E.S.M.; Abdel-Rahman, O. Evaluation of Circulatory RNA-Based Biomarker Panel in Hepatocellular Carcinoma. Mol. Diagn. Ther. 2016, 20, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Scisciani, C.; Vossio, S.; Guerrieri, F.; Schinzari, V.; De Iaco, R.; D’Onorio De Meo, P.; Cervello, M.; Montalto, G.; Pollicino, T.; Raimondo, G.; et al. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J. Hepatol. 2012, 56, 855–861. [Google Scholar] [CrossRef]
- Huang, S.; He, X. The role of microRNAs in liver cancer progression. Br. J. Cancer 2011, 104, 235–240. [Google Scholar] [CrossRef]
- Li, J.; Fu, H.; Xu, C.; Tie, Y.; Xing, R.; Zhu, J.; Qin, Y.; Sun, Z.; Zheng, X. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer 2010, 10, 354. [Google Scholar] [CrossRef]





| Clinico- Pathological Features | No. of Participants (n = 245) | Groups | Statistics | |||||
|---|---|---|---|---|---|---|---|---|
| Control (n = 44) (% within Group) | HCV Non-Cirrhotic (n = 62) (% within Group) | HCV Cirrhotic (n = 67) (% within Group) | HCV-HCC (n = 72) (% within Group) | Test Statistics | p-Value (among the Groups) | p-Value (HCV vs. HCC) | ||
| Age | - | 54.6 ± 1.93 | 52.6 ± 1.253 | 59.2 ± 1.45 | 61.4 ± 0.96 | 26.53 a | <0.0001 | 0.388 d (N.S) |
| Mean age (≤57) | 136 | 25 (56.8%) | 48 (77.4%) | 36 (53.7%) | 27 (37.5) | 21.624 b | <0.0001 | - |
| Mean age (>57) | 109 | 19 (43.2%) | 14 (22.6%) | 31 (46.3%) | 45 (62.5%) | - | - | - |
| Gender | - | - | - | - | - | 43.775 b | <0.0001 | <0.0001 |
| Male | 122 | 30 (68.2%) | 15 (24.2%) | 24 (35.8%) | 53 (73.6%) | - | - | - |
| Female | 123 | 14 (31.8%) | 47 (75.8%) | 43 (64.2%) | 19 (26.4%) | - | - | - |
| HCV infection | - | - | - | - | 245 b | NA | 0.005 | |
| Negative | 44 | 44 (100%) | 0 | 0 | 0 | - | - | - |
| Positive (SVR) | 126 | 0 | 48 (77.4%) | 42 (62.7%) | 36 (50%) | - | - | - |
| Positive (Treatment naïve) | 75 | 0 | 14 (22.6%) | 25 (37.3%) | 36 (50%) | - | - | - |
| Cirrhosis | - | - | - | - | - | 245 b | NA | NA |
| Negative | 106 | 44 (100%) | 62 (100%) | 0 | 0 | - | - | - |
| Positive | 139 | 0 | 0 | 67 (100%) | 72 (100%) | -- | - | - |
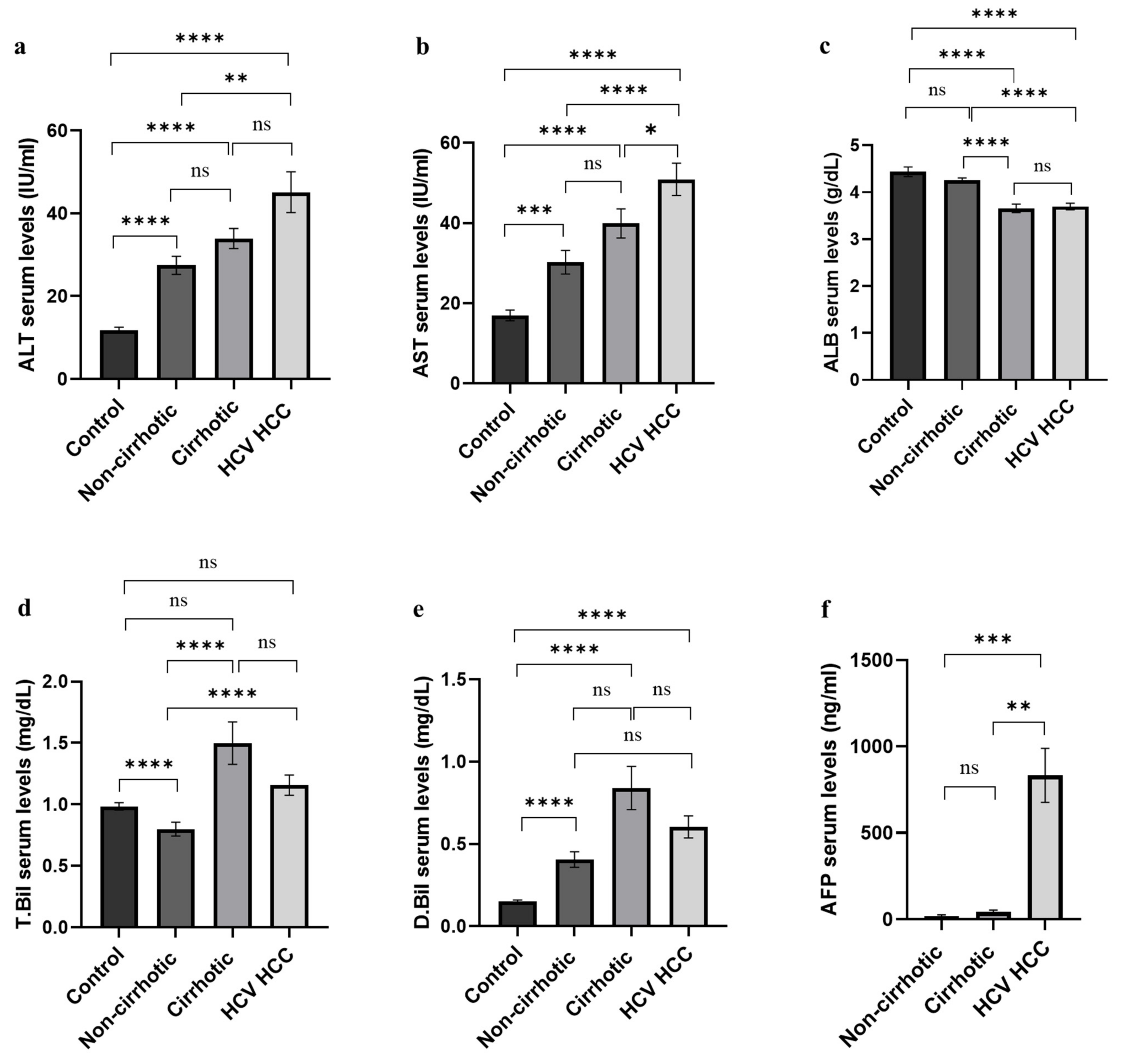
| ALT | 12 ± 0.7 | 27 ± 2.2 | 34 ± 2.4 | 45 ± 5 | 90.3 a | <0.0001 | 0.179 d (N.S) | |
| ≤40 IU/L | 187 | 42 (100%) | 51 (83.6%) | 50 (77%) | 44 (63%) | 22.75 b | NA | - |
| >40 IU/L | 51 | 0 | 10 (16.4%) | 15 (23%) | 26 (37%) | - | - | |
| Missing | 7 | 2 | 1 | 2 | 2 | - | - | - |
| AST | - | 17 ± 1.3 | 30 ± 3 | 40 ± 3.6 | 51 ± 4 | 70.1 a | <0.0001 | 0.046 |
| ≤40 IU/L | 167 | 41 (97.6%) | 51 (83.6%) | 39 (60%) | 36 (51.4%) | 35.34 b | <0.0001 | - |
| >40 IU/L | 71 | 1 (2.4%) | 10 (16.4%) | 26 (40%) | 34 (48.6%) | - | - | - |
| Missing | 7 | 2 | 1 | 2 | 2 | - | - | - |
| ALB | 4.4 ± 0.1 | 4.3 ± 0.5 | 3.7 ± 0.1 | 3.7 ± 0.1 | 23.82 c | <0.0001 | 0.981 e (N.S) | |
| >4 g/dL | 122 | 28 (66.7%) | 51 (83.6%) | 22 (33.8%) | 21 (30.4%) | 49.41 b | <0.0001 | - |
| ≤4 g/dL | 115 | 14 (33.3%) | 10 (16.4%) | 43 (66.2%) | 48 (69.6%) | - | - | - |
| Missing | 8 | 2 | 1 | 2 | 3 | - | - | - |
| T. Bil | 1 ± 0.03 | 0.8 ± 0.06 | 1.2 ± 0.2 | 1 ± 0.1 | 33.12 a | <0.0001 | 0.774 d (N.S) | |
| ≤1.25 mg/dL | 191 | 41 (97.6%) | 57 (93.4%) | 44 (67.7%) | 49 (71%) | 25.19 b | <0.0001 | - |
| >1.25 mg/dL | 46 | 1 (2.4%) | 4 (6.6%) | 21 (32.3%) | 20 (29%) | - | - | - |
| Missing | 8 | 2 | 1 | 2 | 3 | - | - | - |
| D. Bil | 0.1 ± 0.01 | 0.4 ± 0.05 | 0.8 ± 0.13 | 0.6 ± 0.1 | 71.23 a | <0.0001 | 0.749 d (N.S) | |
| ≤0.35 mg/dL | 134 | 42 (100%) | 33 (54.1%) | 27 (41.5%) | 32 (46.4) | 41.29 b | NA | - |
| >0.35 mg/dL | 103 | 0 | 28 (45.9%) | 38 (58.5%) | 37 (53.6%) | - | - | - |
| Missing | 8 | 2 | 1 | 2 | 3 | - | - | - |
| AFP | - | - | 18.6 ± 7.4 | 41.8 ± 11.8 | 834.5 ± 156.4 | 19.24 a | <0.0001 | <0.0001 |
| <20 ng/mL | 153 | NA | 56 (91.8%) | 55 (83.33%) | 42 (60.9%) | 19.71 b | <0.0001 | - |
| 20–400 ng/mL | 16 | NA | 5 (8.2%) | 10 (15.15%) | 1 (1.4%) | 8.45 b | 0.015 | - |
| >400 ng/mL | 27 | NA | 0 | 1 (1.52%) | 26 (37.7%) | 51.2 b | NA | - |
| Missing | 5 | NA | 1 | 1 | 3 | - | - | - |
| Hemoglobin (g/dL) | - | 13.4 ± 0.3 | 12.9 ± 0.2 | 11.5 ± 0.22 | 12.8 ± 0.2 | 12.37 c | <0.0001 | <0.0001 e |
| WBCs (×109/L) | - | 7.63 ± 0.42 | 4.75 ± 0.19 | 4.19 ± 0.09 | 4.614 ± 0.1 | 61.1 a | <0.0001 | 0.015 d |
| RBCs (×103/mm3) | - | 4.77 ± 0.274 | 12.9 ± 0.226 | 11.491 ± 0.22 | 12.79 ± 0.23 | 106.9 a | <0.0001 | <0.0001 d |
| Platelets (×109/L) | - | 301 ± 12.4 | 220 ± 13.7 | 130 ± 7.6 | 141 ± 9.1 | 84.6 a | <0.0001 | 0.481 d (N.S) |
| Groups | Statistics | |||||
|---|---|---|---|---|---|---|
| Parameter | No. of Participants | HCV Non-Cirrhotic | HCV Cirrhotic | HCV-HCC | χ2 (a) | p-Value |
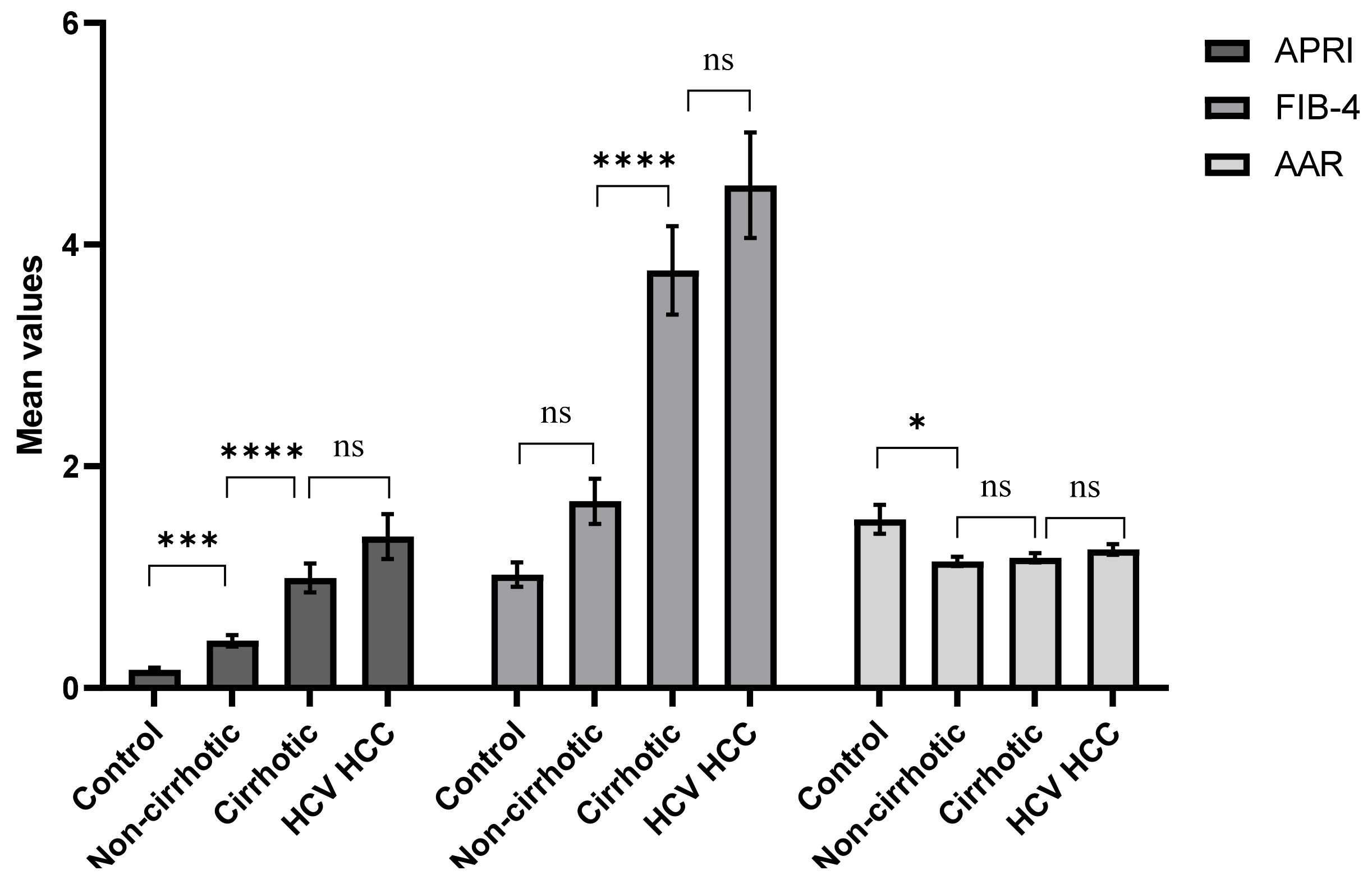
| APRI score | 213 | 0.424 ± 0.051 | 0.992 ± 0.13 | 01.365 ± 0.202 | 102.65 | <0.0001 |
| <0.5 | 113 | 36 (76.6%) | 20 (33.9%) | 16 (24.6%) | 73.747 | <0.0001 |
| 0.5–1.5 | 74 | 10 (21.3%) | 29 (49.2%) | 34 (52.3%) | 37.409 | <0.0001 |
| >1.5 | 26 | 1 (2.1%) | 10 (16.9%) | 15 (23.1%) | 20.165 | <0.0001 |
| Missing | 32 | 15 | 8 | 7 | - | - |
| FIB-4 score | 192 | 1.684 ± 0.204 | 3.767 ± 0.399 | 4.534 ± 0.476 | 86.193 | <0.0001 |
| <1.45 | 67 | 23 (62.2%) | 3 (6%) | 7 (11.1%) | 85.387 | <0.0001 |
| 1.45–3.27 | 71 | 10 (27%) | 29 (58%) | 25 (39.7%) | 18.686 | <0.0001 |
| >3.27 | 54 | 4 (10.8%) | 18 (36%) | 31 (49.2%) | 34.641 | <0.0001 |
| Missing | 53 | 25 | 17 | 9 | - | - |
| AAR | 238 | 1.14 ± 0.041 | 1.17 ± 0.043 | 1.25 ± 0.047 | 9.73 | 0.001 |
| ≤1 | 54 | 15 (24.6%) | 17 (26.2%) | 11 (15.7%) | 2.805 | 0.423 |
| >1 | 184 | 46 (75.4%) | 48 (73.8%) | 59 (84.3%) | - | - |
| Missing | 7 | 1 | 2 | 2 | - | - |
| Parameter | No. of Participants | HCV Cirrhotic | HCV-HCC | |
|---|---|---|---|---|
| CTP score | A | 100 | 46 (69.7%) | 54 (78.3%) |
| B | 30 | 16 (24.2%) | 14 (20.3%) | |
| C | 5 | 4 (6.1%) | 1 (1.4%) | |
| Missing | 4 | 1 | 3 | |
| Ascites | Absent | 117 | 20 (29.8%) | 35 (48.6%) |
| Present | 31 | 15 (22.4%) | 16 (22.2%) | |
| Unknown | 53 | 32 (47.8%) | 21 (29.2%) | |
| Focal lesions number | Single | - | - | 46 (63.9%) |
| Multiple | - | - | 26 (36.1%) | |
| Focal lesions size (Measured by CT) | Tumor ≤ 3 cm | - | - | 22 (40.7%) |
| Tumor > 3 cm | - | - | 32 (59.2%) | |
| Missing | - | - | 18 | |
| Performance status (PS) | PS = 0 | - | - | 35 (48.7%) |
| PS = 1–2 | - | - | 31 (43%) | |
| PS > 2 | - | - | 6 (8.3%) | |
| BCLC staging system | 0 | - | - | 10 (17.9%) |
| A | - | - | 36 (64.2%) | |
| B | - | - | 10 (17.9%) | |
| Missing | - | - | 16 | |
| Target | Groups | Statistics | |||||
|---|---|---|---|---|---|---|---|
| Control Mean ∆Ct (n = 41) | HCV Non- cirrhotic Mean ∆Ct (n = 44) | HCV Cirrhotic Mean ∆Ct (n = 51) | HCV-HCC Mean ∆Ct (n = 52) | Test Statistics (a) | p-Value (Cont. vs. HCV and HCC) (b) | p-value (HCV vs. HCC) (b) | |
| miR-142 | 3.847 ± 0.42 | −0.816 ± 0.47 | −1.58 ± 0.5 | −2.25 ± 0.32 | 74.022 | <0.0001 | 0.625 (N.S.) |
| miR-150 | −2.402 ± 0.55 | −7.42 ± 0.35 | −7.37 ± 0.35 | −8.49 ± 0.33 | 64.834 | <0.0001 | 0.109 (N.S.) |
| miR-183 | 4.21 ± 0.401 | −0.254 ± 0.29 | −1.24 ± 0.303 | −1.22 ± 0.32 | 80.109 | <0.0001 | 0.771 (N.S.) |
| miR-199a | 2.13 ± 0.49 | −2.62 ± 0.35 | −3.63 ± 0.32 | −4.213 ± 0.26 | 81.468 | <0.0001 | 0.429 (N.S.) |
| miR-215 | 4.81 ± 0.45 | 0.217 ± 0.29 | 0.148 ± 0.27 | −0.374 ± 0.21 | 76.745 | <0.0001 | 0.264 (N.S.) |
| mir-217 | 4.684 ± 0.48 | −0.54 ± 0.395 | −0.759 ± 0.33 | −0.358 ± 0.29 | 72.422 | <0.0001 | 0.521 (N.S.) |
| miR-224 | 3.469 ± 0.46 | −0.012 ± 0.41 | −0.737 ± 0.3 | −1.294 ± 0.25 | 60.309 | <0.0001 | 0.34 (N.S.) |
| miR-424 | 2.401 ± 0.45 | −2.79 ± 0.37 | −3.663 ± 0.36 | −4.796 ± 0.303 | 90.225 | <0.0001 | 0.05 |
| miR-3607 | 3.358 ± 0.4 | 0.77 ± 0.25 | 0.93 ± 0.17 | 0.317 ± 0.21 | 46.934 | <0.0001 | 0.479 (N.S.) |
| Target | Groups | Statistics | ||||
|---|---|---|---|---|---|---|
| Control Fold Change Mean Rank (n = 41) | HCV Non-Cirrhotic Fold Change Mean Rank (n = 44) | HCV Cirrhotic Fold Change Mean Rank (n = 51) | HCV-HCC Fold Change Mean Rank (n = 52) | Test Statistics (a) | p-Value | |
| miR-142 | 31.634 | 98.659 | 115.098 | 120.346 | 74.0224 | <0.0001 |
| miR-150 | 35.976 | 104.636 | 104.235 | 122.519 | 64.3728 | <0.0001 |
| miR-183 | 28.879 | 99.705 | 119.971 | 116.856 | 80.1087 | <0.0001 |
| miR-199a | 30.39 | 92.886 | 116.431 | 124.904 | 81.4677 | <0.0001 |
| miR-215 | 31.488 | 104.989 | 109.069 | 121.019 | 72.812 | <0.0001 |
| mir-217 | 31.39 | 111.875 | 119.039 | 105.49 | 72.4221 | <0.0001 |
| miR-224 | 38.146 | 98.091 | 110.235 | 120.462 | 60.2673 | <0.0001 |
| miR-424 | 27.049 | 95.568 | 111.549 | 130.058 | 90.2253 | <0.0001 |
| miR-3607 | 44.634 | 108.92 | 99.4314 | 116.779 | 46.9342 | <0.0001 |
| Factor | Statistics | miR-142 2−∆∆Ct | miR-150 2−∆∆Ct | miR-183 2−∆∆Ct | miR-199a 2−∆∆Ct | miR-215 2−∆∆Ct | miR-217 2−∆∆Ct | miR-224 2−∆∆Ct | miR-424 2−∆∆Ct | miR-3607 2−∆∆Ct |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-142 2−∆∆Ct | rho | 1.000 | 0.619 ** | 0.403 ** | 0.624 ** | 0.371 ** | 0.350 ** | 0.579 ** | 0.651 ** | 0.359 ** |
| p-value | - | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| miR-150 2−∆∆Ct | rho | 0.619 ** | 1.000 | 0.378 ** | 0.656 ** | 0.409 ** | 0.315 ** | 0.767 ** | 0.774 ** | 0.420 ** |
| p-value | <0.0001 | - | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| miR-183 2−∆∆Ct | rho | 0.403 ** | 0.378 ** | 1.000 | 0.617 ** | 0.707 ** | 0.725 ** | 0.439 ** | 0.495 ** | 0.497 ** |
| p-value | <0.0001 | <0.0001 | - | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| miR-199a 2−∆∆Ct | rho | 0.624 ** | 0.656 ** | 0.617 ** | 1.000 | 0.659 ** | 0.541 ** | 0.727 ** | 0.828 ** | 0.581 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | - | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| miR-215 2−∆∆Ct | rho | 0.371 ** | 0.409 ** | 0.707 ** | 0.659 ** | 1.000 | 0.681 ** | 0.527 ** | 0.582 ** | 0.621 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | - | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| miR-217 2−∆∆Ct | rho | 0.350 ** | 0.315 ** | 0.725 ** | 0.541 ** | 0.681 ** | 1.000 | 0.339 ** | 0.441 ** | 0.503 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | - | <0.0001 | <0.0001 | <0.0001 | |
| miR-224 2−∆∆Ct | rho | 0.579 ** | 0.767 ** | 0.439 ** | 0.727 ** | 0.527 ** | 0.339 ** | 1.000 | 0.769 ** | 0.569 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | - | <0.0001 | <0.0001 | |
| miR-424 2−∆∆Ct | rho | 0.651 ** | 0.774 ** | 0.495 ** | 0.828 ** | 0.582 ** | 0.441 ** | 0.769 ** | 1.000 | 0.545 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | - | <0.0001 | |
| miR-3607 2−∆∆Ct | rho | 0.359 ** | 0.420 ** | 0.497 ** | 0.581 ** | 0.621 ** | 0.503 ** | 0.569 ** | 0.545 ** | 1.000 |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Age | rho | 0.021 | 0.049 | 0.206 ** | 0.160 * | 0.170 * | 0.081 | 0.082 | 0.127 | −0.047 |
| p-value | 0.781 | 0.527 | 0.007 | 0.037 | 0.026 | 0.291 | 0.285 | 0.098 | 0.538 | |
| Gender | rho | 0.153 * | 0.103 | 0.168 * | 0.074 | 0.084 | 0.251 ** | 0.046 | 0.056 | 0.055 |
| p-value | 0.036 | 0.159 | 0.021 | 0.315 | 0.252 | 0.001 | 0.530 | 0.442 | 0.450 | |
| Cirrhosis | rho | 0.472 ** | 0.385 ** | 0.485 ** | 0.532 ** | 0.418 ** | 0.360 ** | 0.424 ** | 0.535 ** | 0.278 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.000 | <0.0001 | |
| ALT | rho | 0.452 ** | 0.402 ** | 0.404 ** | 0.481 ** | 0.422 ** | 0.322 ** | 0.483 ** | 0.511 ** | 0.268 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| AST | rho | 0.379 ** | 0.299 ** | 0.323 ** | 0.402 ** | 0.356 ** | 0.244 ** | 0.387 ** | 0.416 ** | 0.202 ** |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| AFP | rho | 0.131 | −0.067 | 0.081 | 0.170 * | 0.082 | −0.046 | 0.082 | 0.127 | 0.090 |
| p-value | 0.117 | 0.420 | 0.332 | 0.040 | 0.324 | 0.583 | 0.328 | 0.128 | 0.281 | |
| T. Bil | rho | −0.064 | −0.219 ** | −0.008 | −0.006 | −0.005 | −0.139 | −0.057 | −0.059 | −0.139 |
| p-value | 0.392 | 0.003 | 0.915 | 0.930 | 0.945 | 0.060 | 0.443 | 0.427 | 0.059 | |
| D. Bil | rho | 0.293 ** | 0.179 * | 0.423 ** | 0.364 ** | 0.423 ** | 0.389 ** | 0.312 ** | 0.353 ** | 0.284 ** |
| p-value | <0.0001 | 0.015 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| ALB | rho | −0.211 ** | −0.071 | −0.229 ** | −0.258 ** | −0.266 ** | −0.159 * | −0.195 ** | −0.265 ** | −0.132 |
| p-value | 0.004 | 0.339 | 0.002 | <0.0001 | <0.0001 | 0.031 | 0.008 | <0.0001 | 0.074 | |
| CTP score | rho | −0.037 | −0.259 ** | 0.154 | 0.055 | 0.123 | 0.231 * | −0.057 | 0.045 | 0.214 * |
| p-value | 0.715 | 0.009 | 0.124 | 0.587 | 0.219 | 0.020 | 0.570 | 0.658 | 0.032 | |
| BCLC | rho | −0.295 | −0.110 | −0.035 | 0.136 | 0.168 | −0.042 | −0.210 | −0.115 | −0.056 |
| p-value | 0.065 | 0.500 | 0.829 | 0.404 | 0.301 | 0.798 | 0.194 | 0.481 | 0.730 |
| miRNA | AUC | 95% CI | Cut-Off | Sensitivity | Specificity | PPV | NPV | Accuracy | SE | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| HCC vs. Control | ||||||||||
| miR-424 | 0.993 | 0.98–1 | 9.05 | 100 | 90.24 | 92.86 | 100 | 95.7 | 0.005 | <0.0001 |
| miR-199a | 0.968 | 0.93–1 | 18.22 | 92.31 | 95.12 | 96 | 90.7 | 93.55 | 0.02 | <0.0001 |
| miR-142 | 0.972 | 0.95–0.99 | 10.80 | 92.31 | 90.24 | 92.31 | 90.24 | 91.4 | 0.014 | <0.0001 |
| miR-215 | 0.958 | 0.92–0.994 | 13.89 | 92.31 | 90.24 | 92.31 | 90.24 | 91.4 | 0.019 | <0.0001 |
| miR-224 | 0.921 | 0.87–0.97 | 9.60 | 80.77 | 87.8 | 89.36 | 78.26 | 83.87 | 0.027 | <0.0001 |
| miR-150 | 0.928 | 0.88–0.98 | 10.36 | 88.46 | 82.93 | 86.8 | 85 | 86.02 | 0.024 | <0.0001 |
| miR-3607 | 0.868 | 0.79–0.95 | 3.96 | 82.7 | 80.49 | 84.31 | 78.57 | 81.72 | 0.041 | <0.0001 |
| miR-183 | 0.957 | 0.92–0.998 | 9.14 | 94.23 | 90.24 | 92.45 | 92.5 | 92.47 | 0.021 | <0.0001 |
| miR-217 | 0.933 | 0.88–0.98 | 9.54 | 86.54 | 85.37 | 88.24 | 83.33 | 86.02 | 0.026 | <0.0001 |
| Combined panel a | 0.969 | 0.928–1 | 100 | 95.12 | 96.3 | 100 | 97.85 | 0.021 | <0.0001 | |
| HCV vs. Control | ||||||||||
| miR-424 | 0.940 | 0.9–0.98 | 7.173 | 85.26 | 85.37 | 93.1 | 71.43 | 85.29 | 0.019 | <0.0001 |
| miR-199a | 0.919 | 0.86–0.97 | 8.418 | 84.21 | 85.37 | 93.02 | 70 | 84.56 | 0.028 | <0.0001 |
| miR-142 | 0.903 | 0.85–0.96 | 7.165 | 86.32 | 85.37 | 93.18 | 72.92 | 86.03 | 0.026 | <0.0001 |
| miR-215 | 0.913 | 0.86–0.96 | 6.196 | 86.32 | 80.49 | 91.11 | 71.74 | 84.56 | 0.06 | <0.0001 |
| miR-224 | 0.863 | 0.8–0.93 | 5.729 | 80 | 78.05 | 89.41 | 62.74 | 79.41 | 0.034 | <0.0001 |
| miR-150 | 0.882 | 0.83–0.94 | 6.966 | 82.11 | 73.17 | 87.64 | 63.83 | 79.41 | 0.029 | <0.0001 |
| miR-3607 | 0.824 | 0.74–0.91 | 3.271 | 81.05 | 75.61 | 88.5 | 63.27 | 79.41 | 0.044 | <0.0001 |
| miR-183 | 0.941 | 0.9–0.98 | 6.266 | 91.58 | 87.8 | 94.57 | 81.82 | 90.44 | 0.022 | <0.0001 |
| miR-217 | 0.927 | 0.88–0.97 | 8.496 | 88.42 | 85.37 | 93.33 | 76.1 | 87.5 | 0.023 | <0.0001 |
| Combined panel a | 0.871 | 0.8–0.941 | 90.53 | 85.37 | 93.48 | 79.55 | 88.97 | 0.036 | <0.0001 | |
| HCC vs. non-HCC | ||||||||||
| miR-424 | 0.761 | 0.7–0.83 | 27.943 | 80.77 | 60.29 | 43.75 | 89.13 | 65.96 | 0.035 | <0.0001 |
| miR-199a | 0.724 | 0.65–0.8 | 28.769 | 78.85 | 58.09 | 41.84 | 87.78 | 63.83 | 0.037 | <0.0001 |
| miR-142 | 0.69 | 0.61–0.77 | 21.811 | 76.92 | 58.09 | 41.24 | 86.81 | 63.3 | 0.039 | <0.0001 |
| miR-215 | 0.695 | 0.62–0.77 | 22.138 | 73.08 | 55.15 | 38.38 | 84.27 | 60.11 | 0.039 | <0.0001 |
| miR-224 | 0.691 | 0.61–0.77 | 10.321 | 73.08 | 54.41 | 38 | 84.1 | 59.57 | 0.04 | <0.0001 |
| miR-150 | 0.706 | 0.63–0.79 | 23.534 | 71.15 | 54.41 | 37.38 | 83.15 | 59.04 | 0.041 | <0.0001 |
| miR-3607 | 0.664 | 0.58–0.75 | 5.797 | 71.15 | 52.94 | 36.63 | 82.76 | 57.98 | 0.042 | 0.001 |
| miR-183 | 0.664 | 0.59–0.74 | 18.872 | 61.54 | 59.56 | 36.78 | 80.2 | 60.11 | 0.041 | <0.0001 |
| Combined panel b | 0.68 | 0.593–0.767 | 80.77 | 61.03 | 44.21 | 89.25 | 66.49 | 0.044 | <0.0001 | |
| HCC vs. non-HCC (SVR group) | ||||||||||
| miR-424 | 0.8 | 0.72–0.89 | 24.165 | 79.17 | 62.5 | 32.76 | 92.86 | 65.63 | 0.044 | <0.0001 |
| miR-199a | 0.74 | 0.65–0.83 | 28.769 | 83.33 | 64.42 | 35.1 | 94.37 | 67.97 | 0.045 | <0.0001 |
| miR-142 | 0.794 | 0.71–0.88 | 38.561 | 75 | 70.19 | 36.73 | 92.41 | 71.1 | 0.042 | <0.0001 |
| miR-215 | 0.742 | 0.65–0.83 | 24.654 | 75 | 64.42 | 32.73 | 91.78 | 66.41 | 0.045 | <0.0001 |
| miR-224 | 0.703 | 0.6–0.8 | 11.042 | 70.83 | 61.54 | 29.82 | 90.14 | 63.28 | 0.051 | 0.002 |
| miR-150 | 0.735 | 0.63–0.84 | 23.534 | 75 | 59.62 | 30 | 91.18 | 62.5 | 0.053 | <0.0001 |
| miR-3607 | 0.763 | 0.67–0.86 | 8.695 | 75 | 66.35 | 33.96 | 92 | 67.97 | 0.05 | <0.0001 |
| miR-183 | 0.728 | 0.63–0.82 | 18.872 | 62.5 | 65.38 | 29.41 | 88.31 | 64.84 | 0.048 | 0.001 |
| miR-217 | 0.673 | 0.57–0.78 | 24.745 | 75 | 57.7 | 29.03 | 90.91 | 60.94 | 0.052 | 0.008 |
| Combined panel a | 0.683 | 0.562–0.804 | 83.33 | 63.73 | 35.09 | 94.2 | 67.46 | 0.062 | 0.005 | |
| HCC vs. non-HCC (Naïve group) | ||||||||||
| miR-424 | 0.835 | 0.76–0.91 | 29.098 | 85.71 | 75.34 | 57.14 | 93.22 | 78.22 | 0.039 | <0.0001 |
| miR-199a | 0.821 | 0.74–0.9 | 22.67 | 89.29 | 68.5 | 52.1 | 94.34 | 74.26 | 0.041 | <0.0001 |
| miR-142 | 0.737 | 0.64–0.83 | 12.933 | 85.71 | 64.38 | 48 | 92.16 | 70.3 | 0.049 | <0.0001 |
| miR-215 | 0.772 | 0.68–0.86 | 17.087 | 85.71 | 63.01 | 47.06 | 92 | 69.31 | 0.046 | <0.0001 |
| miR-224 | 0.78 | 0.7–0.87 | 9.76 | 82.14 | 61.64 | 45.1 | 90 | 67.33 | 0.046 | <0.0001 |
| miR-150 | 0.791 | 0.7–0.88 | 11.152 | 82.14 | 60.27 | 44.23 | 89.8 | 66.34 | 0.046 | <0.0001 |
| miR-3607 | 0.695 | 0.59–0.8 | 5.0867 | 71.43 | 61.64 | 41.67 | 84.91 | 64.36 | 0.052 | 0.002 |
| miR-183 | 0.748 | 0.65–0.84 | 14.496 | 85.71 | 64.38 | 48 | 92.16 | 70.3 | 0.049 | <0.0001 |
| miR-217 | 0.677 | 0.58–0.78 | 13.518 | 75 | 60.27 | 42 | 86.27 | 64.36 | 0.052 | 0.006 |
| Combined panel c | 0.809 | 0.718–0.901 | 89.29 | 72.6 | 55.56 | 94.64 | 77.23 | 0.047 | <0.0001 | |
| HCC vs. HCV (cirrhotic and non-cirrhotic) | ||||||||||
| miR-424 | 0.662 | 0.57–0.75 | 79.387 | 63.46 | 57.9 | 45.21 | 74.32 | 59.86 | 0.046 | 0.001 |
| miR-199a | 0.618 | 0.53–0.71 | 38.725 | 63.46 | 50.52 | 41.25 | 71.64 | 55.1 | 0.047 | 0.018 |
| miR-150 | 0.61 | 0.51–0.71 | 45.166 | 55.77 | 50.53 | 38.16 | 67.61 | 52.38 | 0.05 | 0.028 |
| AFP | 0.688 | 0.60–0.77 | 6.25 | 62.32 | 64.57 | 48.86 | 75.93 | 63.78 | 0.043 | <0.0001 |
| Combined panel d | 0.567 | 0.469–0.644 | 61.54 | 56.84 | 43.84 | 72.97 | 58.5 | 0.05 | 0.182 (ns) | |
| Combined panel d + AFP | 0.61 | 0.512–0.707 | 62.75 | 56.84 | 43.84 | 73.97 | 58.9 | 0.05 | 0.03 | |
| HCC vs. HCV (cirrhotic and non-cirrhotic) (SVR) | ||||||||||
| miR-424 | 0.679 | 0.55–0.81 | 79.387 | 66.67 | 60.32 | 39.02 | 82.61 | 62.07 | 0.065 | 0.01 |
| miR-142 | 0.665 | 0.54–0.79 | 112.42 | 66.67 | 69.84 | 45.71 | 84.62 | 68.97 | 0.063 | 0.018 |
| miR-3607 | 0.672 | 0.54–0.8 | 9.325 | 70.83 | 53.97 | 36.96 | 82.93 | 58.62 | 0.067 | 0.014 |
| AFP | 0.615 | 0.5–0.73 | 6.05 | 51.43 | 60.67 | 33.96 | 76.06 | 58.065 | 0.059 | 0.047 |
| Combined panel d | 0.653 | 0.514–0.792 | 70.83 | 61.9 | 41.46 | 84.78 | 64.37 | 0.071 | 0.028 | |
| Combined panel d + AFP | 0.676 | 0.542–0.809 | 70.83 | 73.02 | 50 | 86.79 | 72.41 | 0.068 | 0.012 | |
| miRNA | Normalized LR Weights | Cut-off | AUC | Specificity | Sensitivity | Accuracy |
|---|---|---|---|---|---|---|
| HCC vs. Control | ||||||
| miR-150 | 0.205523 | - | - | - | - | - |
| miR-199a | 0.145376 | - | - | - | - | - |
| miR-215 | 0.145271 | - | - | - | - | - |
| miR-424 | 0.138455 | - | - | - | - | - |
| miR-224 | 0.127198 | - | - | - | - | - |
| miR-217 | 0.097305 | - | - | - | - | - |
| miR-183 | 0.074394 | - | - | - | - | - |
| miR-142 | 0.064225 | - | - | - | - | - |
| miR-3607 | 0.002254 | - | - | - | - | - |
| Combined panel | 0.121 | 0.95 | 95.12 | 94.23 | 94.62 | |
| HCV vs. Control | ||||||
| miR-424 | 0.182482 | - | - | - | - | - |
| miR-142 | 0.155125 | - | - | - | - | - |
| miR-215 | 0.142391 | - | - | - | - | - |
| miR-224 | 0.142301 | - | - | - | - | - |
| miR-150 | 0.132367 | - | - | - | - | - |
| miR-183 | 0.074106 | - | - | - | - | |
| miR-199a | 0.070252 | - | - | - | - | - |
| miR-217 | 0.062827 | - | - | - | - | - |
| miR-3607 | −0.03815 | - | - | - | - | - |
| Combined panel | 0.101 | 0.91 | 90.24 | 91.58 | 91.18 | |
| HCC vs. non-HCC | ||||||
| miR-150 | 0.280744 | - | - | - | - | - |
| miR-424 | 0.164503 | - | - | - | - | - |
| miR-199a | 0.124309 | - | - | - | - | - |
| miR-142 | 0.10468 | - | - | - | - | - |
| miR-183 | 0.094862 | - | - | - | - | - |
| miR-215 | 0.088024 | - | - | - | - | - |
| miR-224 | 0.058672 | - | - | - | - | - |
| miR-3607 | 0.048511 | - | - | - | - | - |
| miR-217 | −0.03569 | - | - | - | - | - |
| Combined panel | 0.111 | 0.65 | 65.44 | 65.38 | 65.43 | |
| HCC vs. HCV | ||||||
| miR-150 | 0.281315 | - | - | - | - | |
| miR-424 | 0.190728 | - | - | - | - | - |
| miR-3607 | 0.122029 | - | - | - | - | - |
| miR-183 | 0.098993 | - | - | - | - | - |
| miR-224 | 0.092565 | - | - | - | - | |
| miR-199a | 0.065576 | - | - | - | - | - |
| miR-215 | 0.043295 | - | - | - | - | - |
| miR-142 | 0.033518 | - | - | - | - | - |
| miR-217 | −0.07198 | - | - | - | - | - |
| Combined panel | 0.182 | 0.62 | 62.11 | 61.54 | 61.9 | |
| miRNA | Related Pathways |
|---|---|
| miR-142 | Cell migration, neoplasm metastasis, colony formation, cell growth, neoplasms, apoptosis, cell cycle, cell proliferation |
| miR-150 | cell migration, neoplasm metastasis, colony formation, vascularization, breast cancer, cell growth, neoplasms, apoptosis, cell cycle |
| miR-183 | Breast cancer related |
| miR-199a | Melanoma, breast cancer related, liver cancer, cell migration, renal cell carcinoma, neoplasm metastasis, cell proliferation |
| miR-215 | Liver cancer, cell migration, cell proliferation, cell cycle arrest, G1/S transition checkpoint, colorectal cancer, hepatocellular carcinoma |
| miR-217 | Cell migration, renal cell carcinoma, neoplasm metastasis, cell proliferation, hypertrophy, senescence, apoptosis, colony formation, lumen formation |
| miR-224 | Breast cancer related |
| miR-3607 | Liver cancer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabbish, A.M.; Abdelzaher, H.M.; Abohawya, M.; Shamma, S.; Mahmoud, Y.H.; Maged, A.; Manaa, M.; Hassany, M.; Kobeissy, F.; Bazgir, O.; et al. Prognostic MicroRNA Panel for HCV-Associated HCC: Integrating Computational Biology and Clinical Validation. Cancers 2022, 14, 3036. https://doi.org/10.3390/cancers14133036
Dabbish AM, Abdelzaher HM, Abohawya M, Shamma S, Mahmoud YH, Maged A, Manaa M, Hassany M, Kobeissy F, Bazgir O, et al. Prognostic MicroRNA Panel for HCV-Associated HCC: Integrating Computational Biology and Clinical Validation. Cancers. 2022; 14(13):3036. https://doi.org/10.3390/cancers14133036
Chicago/Turabian StyleDabbish, Areeg M., Hana M. Abdelzaher, Moustafa Abohawya, Samir Shamma, Yosra H. Mahmoud, Amr Maged, Mohamed Manaa, Mohamed Hassany, Firas Kobeissy, Omid Bazgir, and et al. 2022. "Prognostic MicroRNA Panel for HCV-Associated HCC: Integrating Computational Biology and Clinical Validation" Cancers 14, no. 13: 3036. https://doi.org/10.3390/cancers14133036
APA StyleDabbish, A. M., Abdelzaher, H. M., Abohawya, M., Shamma, S., Mahmoud, Y. H., Maged, A., Manaa, M., Hassany, M., Kobeissy, F., Bazgir, O., El-Fawal, H., Azzazy, H. M. E., & Abdelnaser, A. (2022). Prognostic MicroRNA Panel for HCV-Associated HCC: Integrating Computational Biology and Clinical Validation. Cancers, 14(13), 3036. https://doi.org/10.3390/cancers14133036








