Combined Inhibition of FOSL-1 and YAP Using siRNA-Lipoplexes Reduces the Growth of Pancreatic Tumor
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Liposomes
2.2. Preparation of Lipoplexes
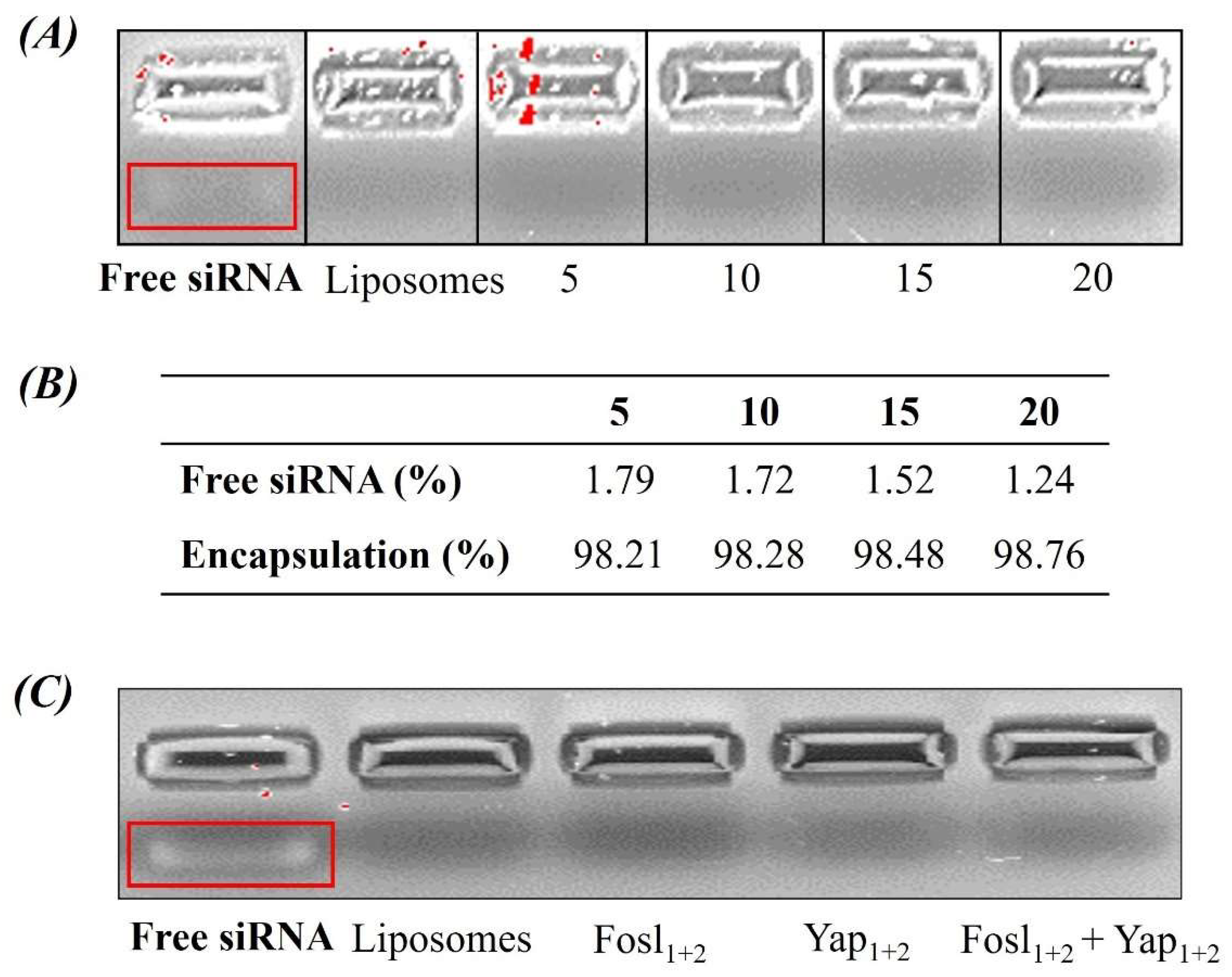
2.3. Determination of siRNA Encapsulation
2.3.1. Agarose Gel Electrophoresis
2.3.2. Quant-it™ RiboGreen RNA Assay Kit
2.4. Physical-Chemical Characterization
2.5. Sterility
2.6. In Vitro Studies
2.6.1. Cell Lines
2.6.2. Cellular Uptake
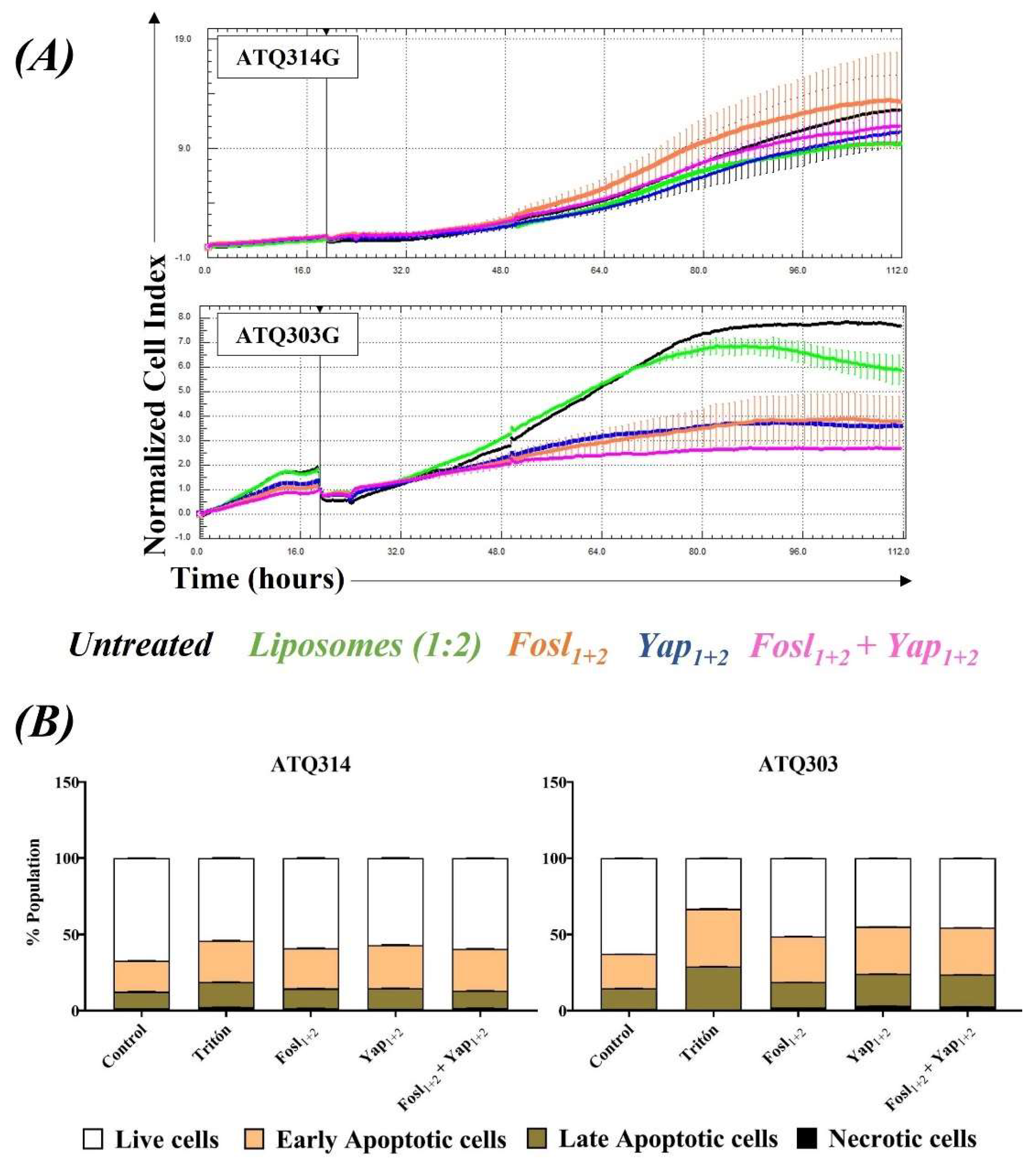
2.6.3. Cell Viability in the Presence of Liposomes and Lipoplexes
2.6.4. Activation of the Complement Cascade
2.6.5. Hemolysis
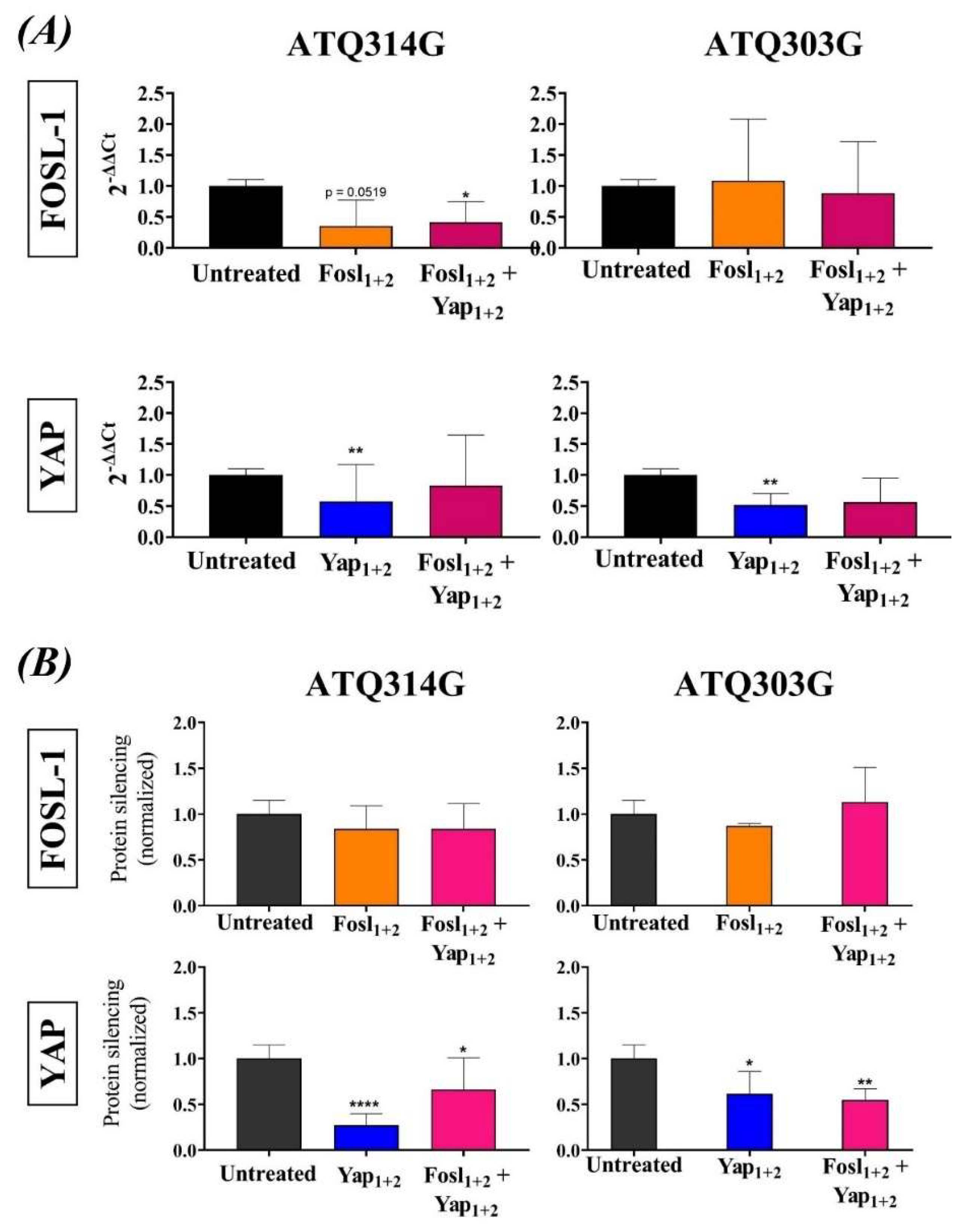
2.6.6. FOSL-1 and YAP Silencing
2.7. In Vivo Studies
2.7.1. qRT-PCR to Analyze the Silencing of FOSL-1 and YAP in the Tumors
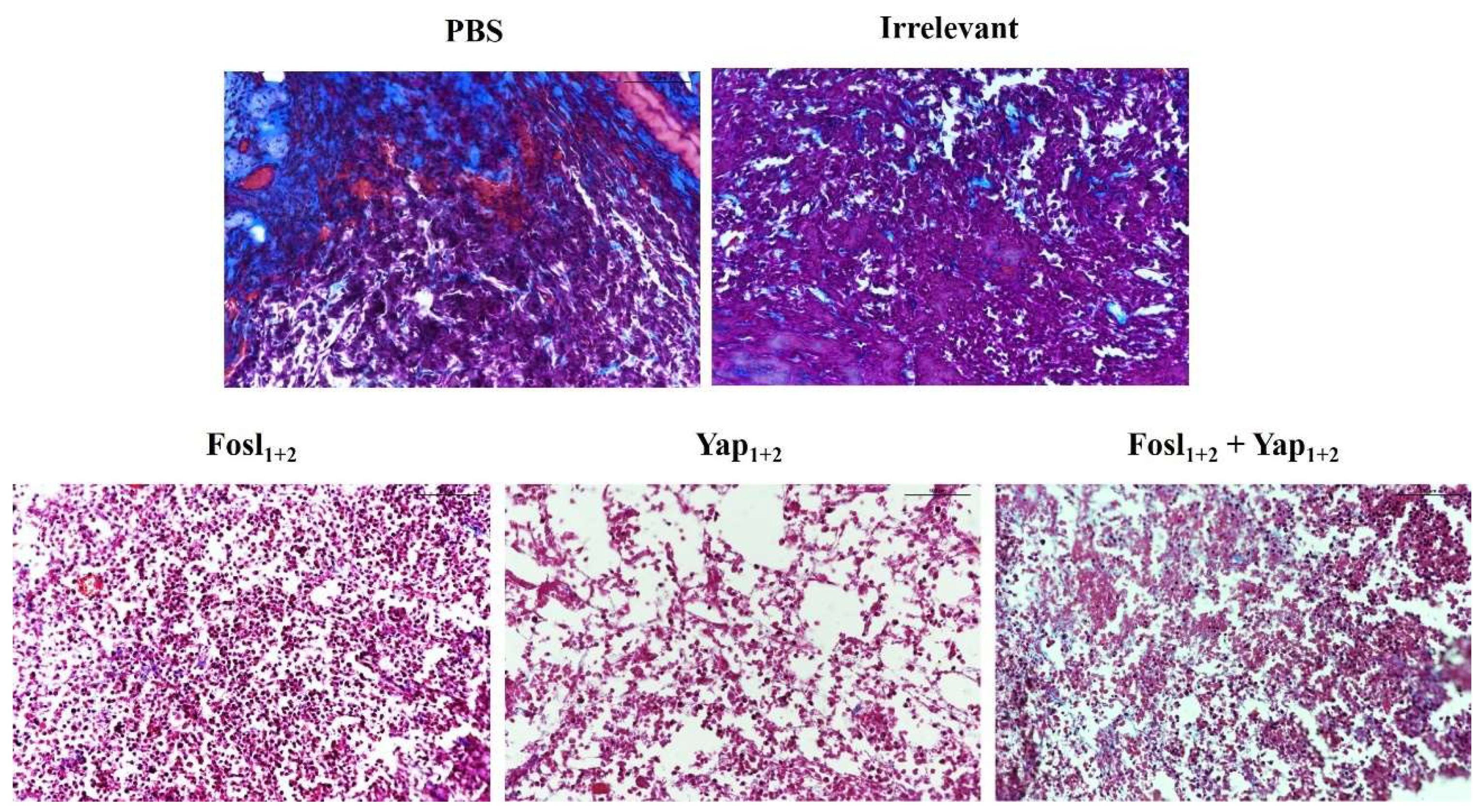
2.7.2. Histological Studies
2.7.3. Statistical Analysis
3. Results and Discussion
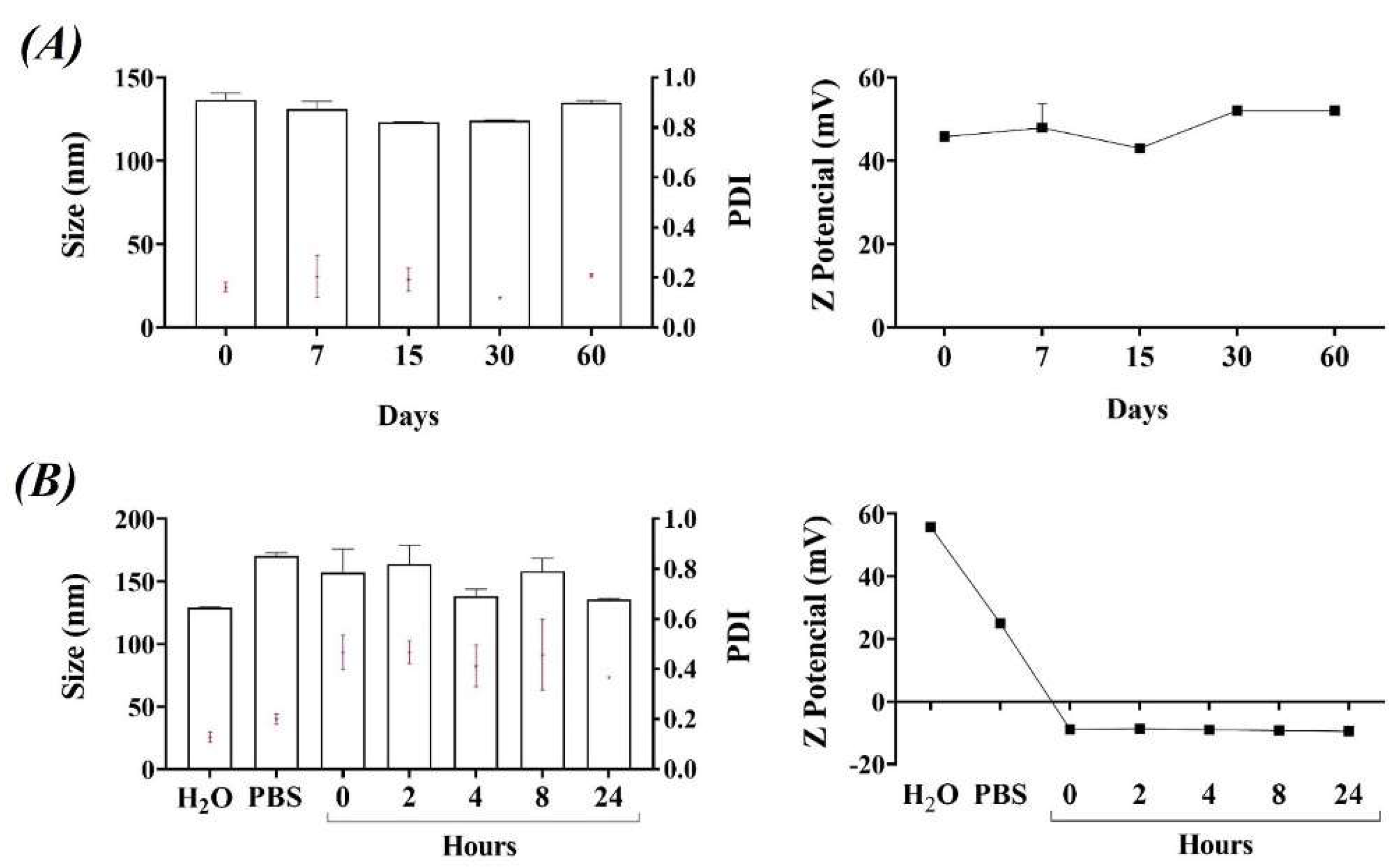
3.1. Physicochemical Characterization and Stability of Liposomes and Lipoplexes
3.2. Cellular Uptake of Liposomes
3.3. FOSL-1 and YAP mRNA and Protein Silencing
3.4. Cytocompatibility of Liposomes and Therapeutic Effect of Lipoplexes
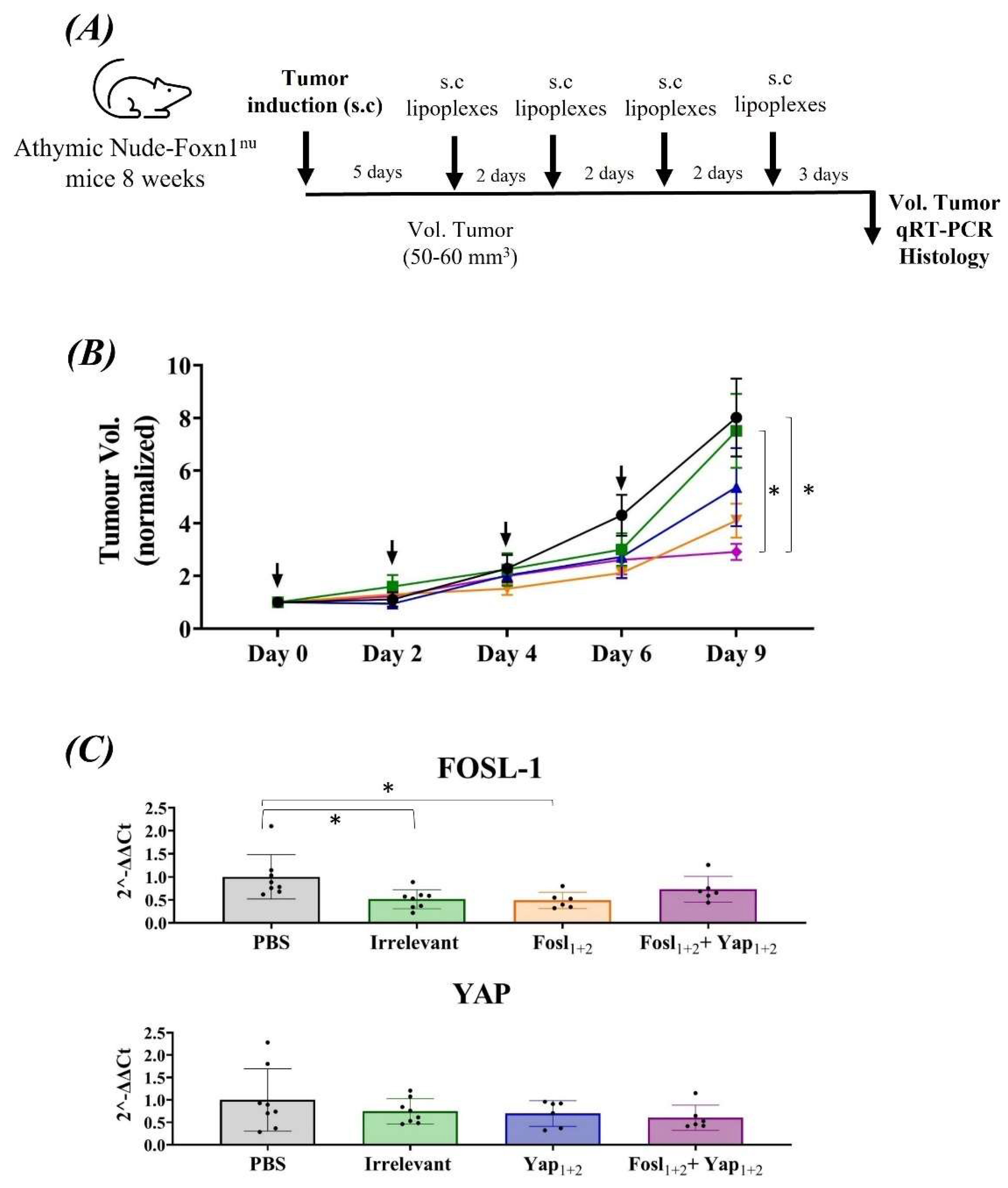
3.5. Anti-Tumor Efficacy in Pancreatic Allografts in Immunosuppressed and Immunocompetent Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schizas, D.; Charalampakis, N.; Kole, C.; Economopoulou, P.; Koustas, E.; Gkotsis, E.; Ziogas, D.; Psyrri, A.; Karamouzis, M.V. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat. Rev. 2020, 86, 102016. [Google Scholar] [CrossRef] [PubMed]
- Hessmann, E.; Schneider, G. New Insights Into Pancreatic Cancer: Notes from a Virtual Meeting. Gastroenterology 2021, 161, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, C.; Xie, K.-P. Therapeutic resistance of pancreatic cancer: Roadmap to its reversal. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188461. [Google Scholar] [CrossRef]
- Wu, J.; Cai, J. Dilemma and Challenge of Immunotherapy for Pancreatic Cancer. Dig. Dis. Sci. 2021, 66, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Gillson, J.; Ramaswamy, Y.; Singh, G.; Gorfe, A.A.; Pavlakis, N.; Samra, J.; Mittal, A.; Sahni, S. Small Molecule KRAS Inhibitors: The Future for Targeted Pancreatic Cancer Therapy? Cancers 2020, 12, 1341. [Google Scholar] [CrossRef]
- Rozengurt, E.; Sinnett-Smith, J.; Eibl, G. Yes-associated protein (YAP) in pancreatic cancer: At the epicenter of a targetable signaling network associated with patient survival. Signal Transduct. Target. Ther. 2018, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Eisenbarth, D.; Choi, W.; Kim, H.; Choi, C.; Lee, D.; Lim, D.S. YAP and AP-1 Cooperate to Initiate Pancreatic Cancer Development from Ductal Cells in Mice. Cancer Res. 2020, 80, 4768–4779. [Google Scholar] [CrossRef]
- Pocaterra, A.; Romani, P.; Dupont, S. YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 2020, 133, jcs230425. [Google Scholar]
- Kawamoto, R.; Nakano, N.; Ishikawa, H.; Tashiro, E.; Nagano, W.; Sano, K.; Irie, M.; Ikuta, M.; Kishi, F.; Nakane, T.; et al. Narciclasine is a novel YAP inhibitor that disturbs interaction between YAP and TEAD4. BBA Adv. 2021, 1, 100008. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhu, J.S.; Gao, C.P.; Li, L.P.; Zhou, C.; Wang, H.; Liu, X.G. siRNA targeting YAP gene inhibits gastric carcinoma growth and tumor metastasis in SCID mice. Oncol. Lett. 2016, 11, 2806–2814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Sun, Y.; Wan, G.; Sun, J.; Sun, J.; Pan, C. Knockdown of YAP inhibits growth in Hep-2 laryngeal cancer cells via epithelial-mesenchymal transition and the Wnt/β-catenin pathway. BMC Cancer 2019, 19, 654. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Yuan, Z.; Du, K.Y.; Fang, L.; Lyu, J.; Zhang, C.; He, A.; Eshaghi, E.; Zeng, K.; Ma, J.; et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019, 26, 2758–2773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Chen, R. Mechanisms of Hippo pathway in the pancreatic cancer. J. Pancreatol. 2020, 3, 21–28. [Google Scholar] [CrossRef]
- Kapoor, A.; Yao, W.; Ying, H.; Hua, S.; Liewen, A.; Wang, Q.; Zhong, Y.; Wu, C.J.; Sadanandam, A.; Hu, B.; et al. Yap1 Activation Enables Bypass of Oncogenic Kras Addiction in Pancreatic Cancer. Cell 2014, 158, 185–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhillon, A.S.; Tulchinsky, E. FRA-1 as a driver of tumour heterogeneity: A nexus between oncogenes and embryonic signalling pathways in cancer. Oncogene 2015, 34, 4421–4428. [Google Scholar]
- Jiang, X.; Xie, H.; Dou, Y.; Yuan, J.; Zeng, D.; Xiao, S. Expression and function of FRA1 protein in tumors. Mol. Biol. Rep. 2020, 47, 737–752. [Google Scholar] [CrossRef]
- Vallejo, A.; Perurena, N.; Guruceaga, E.; Mazur, P.K.; Martinez-Canarias, S.; Zandueta, C.; Valencia, K.; Arricibita, A.; Gwinn, D.; Sayles, L.C.; et al. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat. Commun. 2017, 8, 14294. [Google Scholar] [CrossRef] [Green Version]
- Keshamouni, V.G. Excavation of FOSL1 in the Ruins of KRAS-Driven Lung Cancer. Am. J. Respir. Cell Mol. Biol. 2018, 58, 551–552. [Google Scholar] [CrossRef]
- Yang, W.; Meng, L.; Chen, K.; Tian, C.; Peng, B.; Zhong, L.; Zhang, C.; Yang, X.; Zou, J.; Yang, S.; et al. Preclinical pharmacodynamic evaluation of a new Src/FOSL1 inhibitor, LY-1816, in pancreatic ductal adenocarcinoma. Cancer Sci. 2019, 110, 1408–1419. [Google Scholar] [CrossRef]
- Subhan, M.A.; Torchilin, V.P. Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl. Res. 2019, 214, 62–91. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. 2017, 13, 48–57. [Google Scholar]
- Sousa, A.R.; Oliveira, A.V.; Oliveira, M.J.; Sarmento, B. Nanotechnology-based siRNA delivery strategies for metastatic colorectal cancer therapy. Int. J. Pharm. 2019, 568, 118530. [Google Scholar] [CrossRef] [PubMed]
- Gomes-da-Silva, L.C.; Fonseca, N.A.; Moura, V.; de Lima, M.C.P.; Simões, S.; Moreira, J.N. Lipid-based nanoparticles for siRNA delivery in cancer therapy: Paradigms and challenges. Acc. Chem. Res. 2012, 45, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.C.N.; Fernandes, J.; Gonçalves, A.; Gomes, A.C.; Oliveira, M.E.C.D.R. Lipid-based Nanocarriers for siRNA Delivery: Challenges, Strategies and the Lessons Learned from the DODAX: MO Liposomal System. Curr. Drug Targets 2019, 20, 29–50. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [Green Version]
- Nogueira, E.; Freitas, J.; Loureiro, A.; Nogueira, P.; Gomes, A.C.; Preto, A.; Carmo, A.M.; Moreira, A.; Cavaco-Paulo, A. Neutral PEGylated liposomal formulation for efficient folate-mediated delivery of MCL1 siRNA to activated macrophages. Colloids Surf. B Biointerfaces 2017, 155, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Hart, S.L.; Du, Z. Assembly strategy of liposome and polymer systems for siRNA delivery. Int. J. Pharm. 2021, 592, 120033. [Google Scholar] [CrossRef]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef]
- Shim, M.S.; Kwon, Y.J. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010, 277, 4814–4827. [Google Scholar] [CrossRef]
- Barba, A.A.; Cascone, S.; Caccavo, D.; Lamberti, G.; Chiarappa, G.; Abrami, M.; Grassi, G.; Grassi, M.; Tomaiuolo, G.; Guido, S.; et al. Engineering approaches in siRNA delivery. Int. J. Pharm. 2017, 525, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.P.N.; Coutinho, P.J.G.; Oliveira, M.E.C.D.R. Characterization of monoolein-based lipoplexes using fluorescence spectroscopy. J. Fluoresc. 2008, 18, 555–562. [Google Scholar] [CrossRef] [Green Version]
- Real-Oliveira, M.E.; Silva, J.P.; Coutinho, P.J.; Coutinho, O.M.; Gomes, A.F.; Casal, M.P. Use of Monoolein as a New Auxiliary Lipid in Lipofection. International Patent WO2010/020935, 2010. [Google Scholar]
- Oliveira, A.C.; Martens, T.F.; Raemdonck, K.; Adati, R.D.; Feitosa, E.; Botelho, C.; Gomes, A.C.; Braeckmans, K.; Real Oliveira, M.E.C.D. Dioctadecyldimethylammonium:Monoolein Nanocarriers for Efficient in Vitro Gene Silencing. ACS Appl. Mater. Interfaces 2014, 6, 6977–6989. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, A.C.; Raemdonck, K.; Martens, T.; Rombouts, K.; Simón-Vázquez, R.; Botelho, C.; Lopes, I.; Lúcio, M.; González-Fernández, Á.; Real Oliveira, M.E.; et al. Stealth monoolein-based nanocarriers for delivery of siRNA to cancer cells. Acta Biomater. 2015, 25, 216–229. [Google Scholar] [CrossRef] [Green Version]
- Lopes, I.; CN Oliveira, A.; PSárria, M.; PNeves Silva, J.; Gonçalves, O.; Gomes, A.C.; Real Oliveira, M.E. Monoolein-based nanocarriers for enhanced folate receptor-mediated RNA delivery to cancer cells. J. Liposome Res. 2016, 26, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Guerra, C.; Collado, M.; Navas, C.; Schuhmacher, A.J.; Hernández-Porras, I.; Cañamero, M.; Rodriguez-Justo, M.; Serrano, M.; Barbacid, M. Pancreatitis-Induced Inflammation Contributes to Pancreatic Cancer by Inhibiting Oncogene-Induced Senescence. Cancer Cell 2011, 19, 728–739. [Google Scholar] [CrossRef] [Green Version]
- Diego-González, L.; Crecente-Campo, J.; Paul, M.J.; Singh, M.; Reljic, R.; Alonso, M.J.; González-Fernández, Á.; Simón-Vázquez, R. Design of Polymeric Nanocapsules for Intranasal Vaccination against Mycobacterium Tuberculosis: Influence of the Polymeric Shell and Antigen Positioning. Pharmaceutics 2020, 12, 489. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; McNeil, S.E. Understanding the correlation between in vitro and in vivo immunotoxicity tests for nanomedicines. J. Control. Release 2013, 172, 456–466. [Google Scholar] [CrossRef] [Green Version]
- Nogueira, E.; Gomes, A.C.; Preto, A.; Cavaco-Paulo, A. Design of liposomal formulations for cell targeting. Colloids Surf. B Biointerfaces 2015, 136, 514–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, A.C.N.; Sárria, M.; Moreira, P.P.; Fernandes, J.; Castro, L.; Lopes, I.; Côrte-Real, M.; Cavaco-Paulo, A.; Oliveira, M.E.C.D.R.; Gomes, A.C. Counter ions and constituents combination affect DODAX: MO nanocarriers toxicity in vitro and in vivo. Toxicol. Res. 2016, 5, 1244–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Wang, X.; Cheng, H.; Fang, M.; Ning, P.; Zhou, Y.; Chen, W.; Song, H. A polycation coated liposome as efficient siRNA carrier to overcome multidrug resistance. Colloids Surf. B Biointerfaces 2017, 159, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ahn, H.J. PEGylated DC-Chol/DOPE cationic liposomes containing KSP siRNA as a systemic siRNA delivery Carrier for ovarian cancer therapy. Biochem. Biophys. Res. Commun. 2018, 503, 1716–1722. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Neupane, Y.R.; Shafi, S.; Mangla, B.; Kohli, K. PEGylated liposomes as an emerging therapeutic platform for oral nanomedicine in cancer therapy: In vitro and in vivo assessment. J. Mol. Liq. 2020, 303, 112649. [Google Scholar] [CrossRef]
- Silva, J.P.N.; Oliveira, A.C.N.; Gomes, A.C.; Oliveir, M.E.C.D.R. Development of Dioctadecyldimethylammonium Bromide/Monoolein Liposomes for Gene Delivery. In Cell Interaction; Chapter: 10; Intechopen: London, UK, 2012; pp. 245–272. [Google Scholar]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef] [Green Version]
- Roumenina, L.T.; Daugan, M.V.; Petitprez, F.; Sautès-Fridman, C.; Fridman, W.H. Context-dependent roles of complement in cancer. Nat. Rev. Cancer 2019, 19, 698–715. [Google Scholar] [CrossRef] [Green Version]
- Kemp, S.B.; Steele, N.G.; Carpenter, E.S.; Donahue, K.L.; Bushnell, G.G.; Morris, A.H.; The, S.; Orbach, S.M.; Sirihorachai, V.R.; Nwosu, Z.C.; et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci. Alliance 2021, 4, e202000935. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Q.; Li, T.; Liao, Q.; Zhao, Y. Role of the complement system in the tumor microenvironment. Cancer Cell Int. 2019, 19, 300. [Google Scholar] [CrossRef]
- Bettac, L.; Denk, S.; Seufferlein, T.; Huber-Lang, M. Complement in Pancreatic Disease—Perpetrator or Savior? Front. Immunol. 2017, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Revel, M.; Daugan, M.; Sautés-Fridman, C.; Fridman, W.; Roumenina, L. Complement System: Promoter or Suppressor of Cancer Progression? Antibodies 2020, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.J. YAP/TAZ: Drivers of Tumor Growth, Metastasis, and Resistance to Therapy. BioEssays 2020, 42, 1900162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talotta, F.; Casalino, L.; Verde, P. The nuclear oncoprotein Fra-1: A transcription factor knocking on therapeutic applications’ door. Oncogene 2020, 39, 4491–4506. [Google Scholar] [CrossRef]
- Pham, T.H.; Hagenbeek, T.J.; Lee, H.J.; Li, J.; Rose, C.M.; Lin, E.; Yu, M.; Martin, S.E.; Piskol, R.; Lacap, J.A.; et al. Machine-Learning and Chemicogenomics Approach Defines and Predicts Cross-Talk of Hippo and MAPK Pathways. Cancer Discov. 2021, 11, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Abylkassov, R.; Xie, Y. Role of Yes-associated protein in cancer: An update. Oncol. Lett. 2016, 12, 2277–2282. [Google Scholar] [CrossRef] [Green Version]
- Meng, Z.; Moroishi, T.; Guan, K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldker, N.; Ferrazi, F.; Schuhwerk, H.; Widholz, S.A.; Guenther, K.; Frisch, I.; Jakob, K.; Kleemann, J.; Riegel, D.; Bönisch, U.; et al. Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. EMBO J. 2020, 39, e103209. [Google Scholar] [CrossRef]
- Cobbaut, M.; Karagil, S.; Bruno, L.; de la Loza, M.C.D.; Mackenzie, F.E.; Stolonski, M.; Elbediwy, A. Dysfunctional Mechanotransduction through the YAP/TAZ/Hippo Pathway as a Feature of Chronic Disease. Cells 2020, 9, 151. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Lu, X.; Dey, P.; Deng, P.; Wu, C.C.; Jiang, S.; Fang, Z.; Zhao, K.; Konaparthi, R.; Hua, S.; et al. Targeting YAP-Dependent MDSC Infiltration Impairs Tumor Progression. Cancer Discov. 2016, 6, 80–95. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Wang, X.; Weng, Y.-H.; Jin, X.; Ji, J.-L.; Guo, L.; Hu, B.; Liu, N.; Cheng, Q.; Zhang, J.; et al. siRNA Knockdown of RRM2 Effectively Suppressed Pancreatic Tumor Growth Alone or Synergistically with Doxorubicin. Mol. Ther. Nucleic Acids 2018, 12, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, L.; Chen, H.; Kong, R.; Pan, S.; Hu, J.; Wang, Y.; Li, Y.; Sun, B. Silencing IGFBP-2 decreases pancreatic cancer metastasis and enhances chemotherapeutic sensitivity. Oncotarget 2017, 8, 61674–61686. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| siRNA | 5′–3′ Strand | 3′–5′ Strand |
|---|---|---|
| Irrelevant | GCAAACCACCAAUCUAACA | CGUUUGGUGGUUAGAUUGU |
| Fosl1 | GGGCAGCUGCUAUUUAUUUUU | UUCCCGUCGACGAUAAAUAAA |
| Fosl2 | GGUGCCCUUUGACUAGCCUTT | TTCCACGGGAAACUGAUCGGA |
| Yap1 | GCUUUCUCACGUGGUUAUAUU | UUCGAAAGAGUGCACCAAUAU |
| Yap2 | CCAAGCUAGAUAAAGAAAGTT | GTGGUUCGAUCUAUUUCUUUC |
| Primers | Forward (Fw) | Reverse (Rv) |
| GAPDH | 5′-CCTCACCACCATGGAGGAGGC-3′ | 5′-GGCATGGACTGTGGTCATGAG-3′ |
| FOSL-1 | 5′-ATGTACCGAGACTACGGGGAA-3′ | 5′-CTGCTGCTGTCGATGCTTG-3′ |
| YAP | 5′-GGATGTCTCAGGAATTGAGAACA-3′ | 5′-ATGCTGTAGCTGCTCATGCTGA-3′ |
| Size (nm) | PDI | ζ-Potential (mV) | |
|---|---|---|---|
| Liposomes | 139.0 ± 17.12 | 0.170 ± 0.03 | (+) 50.7 ± 5.70 |
| Lipoplexes (Fosl1+2) | 129.6 ± 12.50 | 0.200 ± 0.02 | (+) 54.0 ± 0.28 |
| Lipoplexes (Yap1+2) | 136.0 ± 13.20 | 0.100 ± 0.05 | (+) 41.3 ± 0.85 |
| Lipoplexes (Fosl1+2 + Yap1+2) | 136.7 ± 13.60 | 0.160 ± 0.02 | (+) 39.6 ± 12.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diego-González, L.; Fernández-Carrera, A.; Igea, A.; Martínez-Pérez, A.; Real Oliveira, M.E.C.D.; Gomes, A.C.; Guerra, C.; Barbacid, M.; González-Fernández, Á.; Simón-Vázquez, R. Combined Inhibition of FOSL-1 and YAP Using siRNA-Lipoplexes Reduces the Growth of Pancreatic Tumor. Cancers 2022, 14, 3102. https://doi.org/10.3390/cancers14133102
Diego-González L, Fernández-Carrera A, Igea A, Martínez-Pérez A, Real Oliveira MECD, Gomes AC, Guerra C, Barbacid M, González-Fernández Á, Simón-Vázquez R. Combined Inhibition of FOSL-1 and YAP Using siRNA-Lipoplexes Reduces the Growth of Pancreatic Tumor. Cancers. 2022; 14(13):3102. https://doi.org/10.3390/cancers14133102
Chicago/Turabian StyleDiego-González, Lara, Andrea Fernández-Carrera, Ana Igea, Amparo Martínez-Pérez, M. Elisabete C. D. Real Oliveira, Andreia C. Gomes, Carmen Guerra, Mariano Barbacid, África González-Fernández, and Rosana Simón-Vázquez. 2022. "Combined Inhibition of FOSL-1 and YAP Using siRNA-Lipoplexes Reduces the Growth of Pancreatic Tumor" Cancers 14, no. 13: 3102. https://doi.org/10.3390/cancers14133102
APA StyleDiego-González, L., Fernández-Carrera, A., Igea, A., Martínez-Pérez, A., Real Oliveira, M. E. C. D., Gomes, A. C., Guerra, C., Barbacid, M., González-Fernández, Á., & Simón-Vázquez, R. (2022). Combined Inhibition of FOSL-1 and YAP Using siRNA-Lipoplexes Reduces the Growth of Pancreatic Tumor. Cancers, 14(13), 3102. https://doi.org/10.3390/cancers14133102









