Metabolism-Related Gene Expression in Circulating Tumor Cells from Patients with Early Stage Non-Small Cell Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
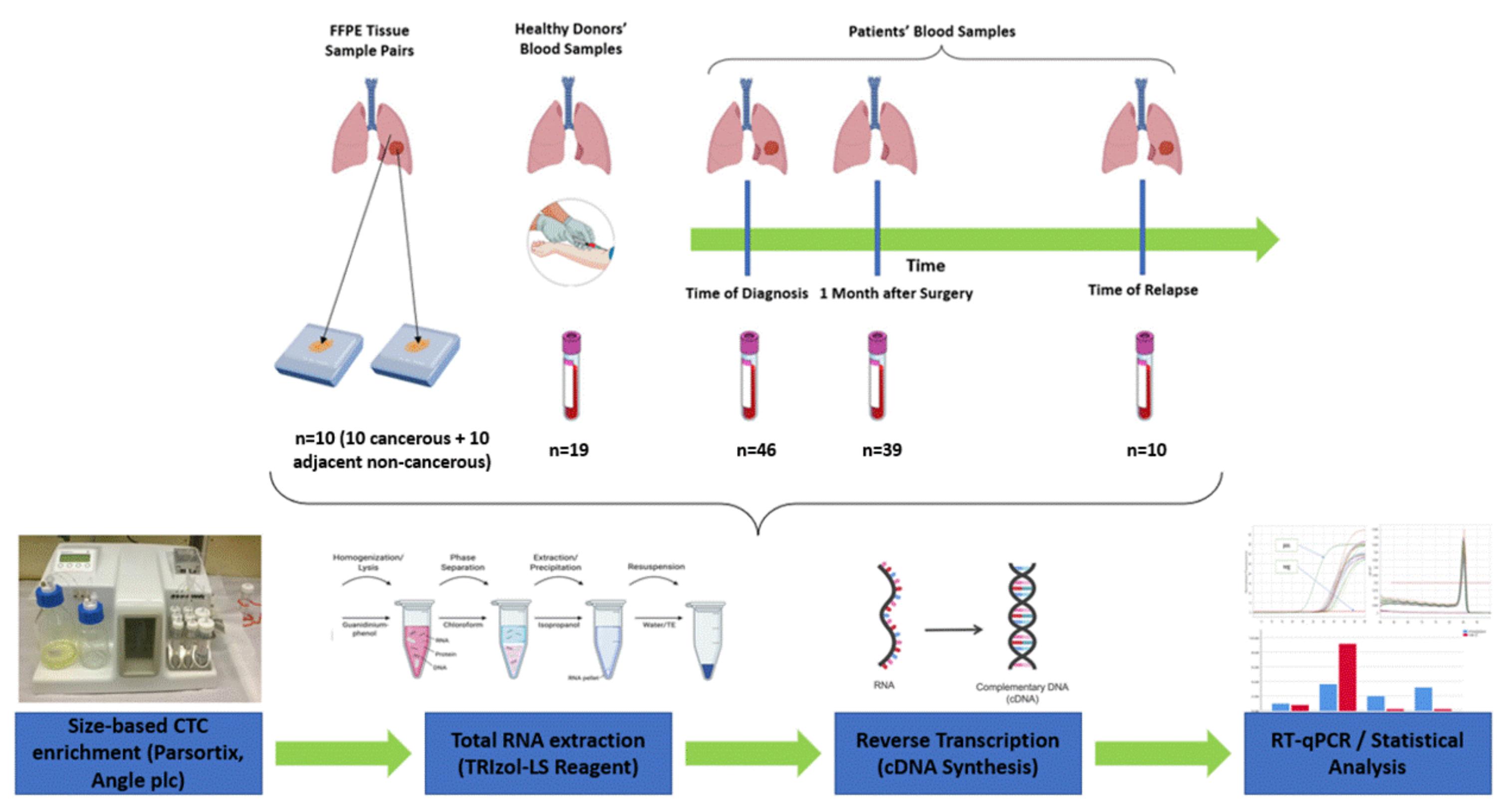
2.1. Clinical Samples
2.2. CTC Enrichment Using the Parsortix Size-Based Microfluidic Device
2.3. RNA Extraction from Fresh Frozen Tissues
2.4. RT-qPCR
2.5. Optimization of Experimental Conditions
2.6. Statistical Analysis
3. Results
3.1. TCGA Analysis
3.2. MRG Expression in NSCLC Paired Tissues
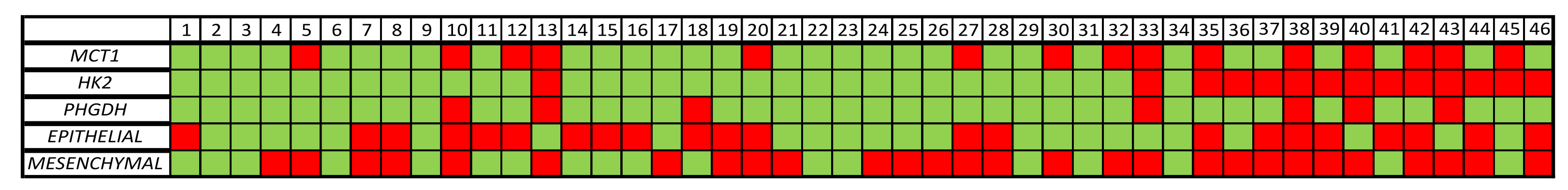
3.3. MRG Expression in the CTC Fraction of HD and NSCLC Patients
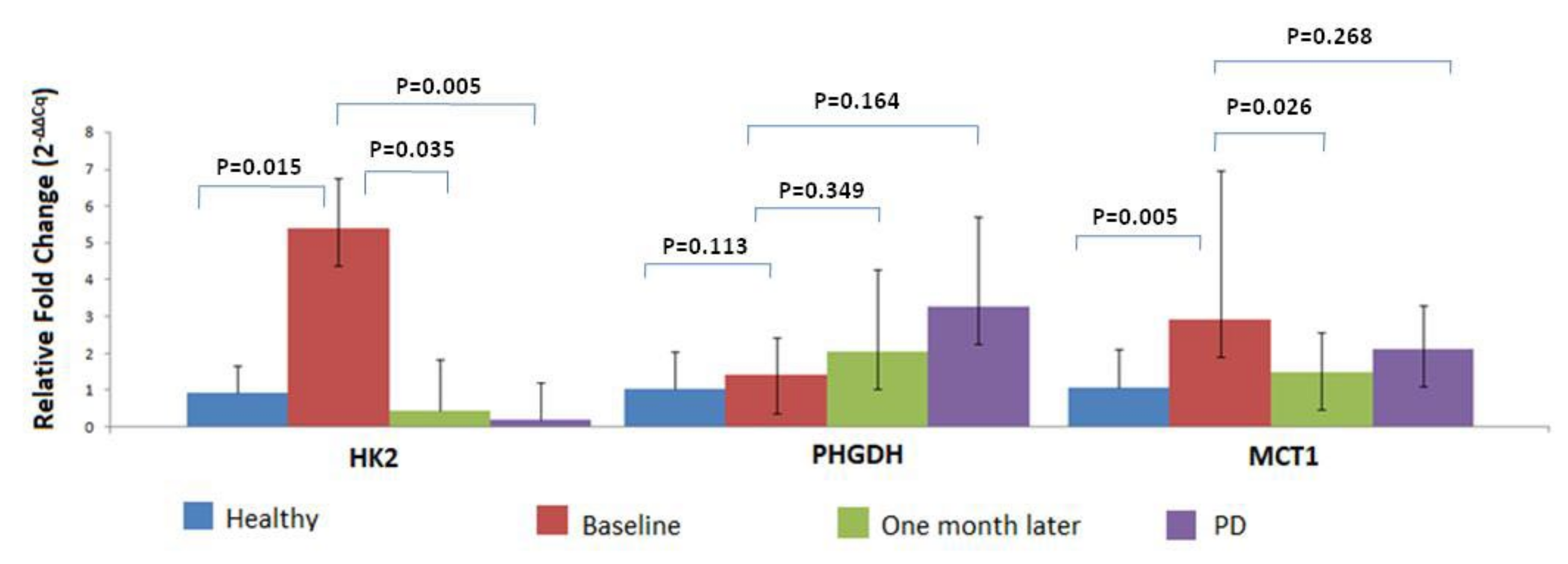
3.4. MRG Overexpression in CTC Fraction at Different Timepoints
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer, J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Tsuboi, M.; John, T.; Grohe, C.; Majern, M.; Goldman, G.J.; Laktionov, K.; Kim, S.-W.; Kato, T.; Vu, H.-V.; et al. Osimertinib in Resected EGFR -Mutated Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wang, C.; Wan, S.; Mu, Z.; Zhang, Z.; Abu-Khalaf, M.M.; Fellin, F.M.; Silver, D.P.; Neupane, M.; Jaslow, R.J.; et al. Association of clinical outcomes in metastatic breast cancer patients with circulating tumour cell and circulating cell-free DNA. Eur. J. Cancer 2019, 106, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Pierga, J.-Y.; Reuben, J.; Rademaker, A.; Davis, A.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. 2019, 134, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Hou, J.-M.; Sloane, R.; Lancashire, L.; Priest, L.; Nonaka, D.; Ward, T.H.; Backen, A.; Clack, G.; Hughes, A.; et al. Analysis of Circulating Tumor Cells in Patients with Non-small Cell Lung Cancer Using Epithelial Marker-Dependent and -Independent Approaches. J. Thorac. Oncol. 2012, 7, 306–315. [Google Scholar] [CrossRef] [Green Version]
- Lianidou, E.S.; Markou, A.; Strati, A. The Role of CTCs as Tumor Biomarkers. Adv. Exp. Med. Biol. 2015, 867, 341–367. [Google Scholar] [CrossRef]
- Cohen, E.N.; Jayachandran, G.; Gao, H.; Qiao, W.; Liu, S.; He, J.; Qiao, Y.; Yao, L.; Lin, S.H.; Reuben, J.M. Enumeration and molecular characterization of circulating tumor cells enriched by microcavity array from stage III non-small cell lung cancer patients. Transl. Lung Cancer Res. 2020, 9, 1974–1985. [Google Scholar] [CrossRef]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E. RNA-Based CTC Analysis Provides Prognostic Information in Metastatic Breast Cancer. Diagnostics 2021, 11, 513. [Google Scholar] [CrossRef]
- Ntzifa, A.; Strati, A.; Kallergi, G.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. Gene expression in circulating tumor cells reveals a dynamic role of EMT and PD-L1 during osimertinib treatment in NSCLC patients. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Markou, A.; Tzanikou, E.; Strati, A.; Zavridou, M.; Mastoraki, S.; Bournakis, E.; Lianidou, E. PIM-1 Is Overexpressed at a High Frequency in Circulating Tumor Cells from Metastatic Castration-Resistant Prostate Cancer Patients. Cancers 2020, 12, 1188. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [Green Version]
- Levine, A.J.; Puzio-Kuter, A.M. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, L.; Zhang, Y.; Wang, J.; Deng, Y.; Lin, D. Inhibition of glycolytic enzyme hexokinase II (HK2) suppresses lung tumor growth. Cancer Cell Int. 2016, 16, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Smith, C.P.; Wilbur, R.R.; Cain, C.P.; Kallu, S.R.; Valasapalli, S.; Sahoo, A.; Guda, M.R.; Tsung, A.J.; Velpula, K.K. An overview of MCT1 and MCT4 in GBM: Small molecule transporters with large implications. Am. J. Cancer Res. 2018, 8, 1967–1976. [Google Scholar]
- Pinheiro, C.; Miranda-Gonçalves, V.; Longatto-Filho, A.; Vicente, A.L.S.A.; Berardinelli, G.N.; Scapulatempo-Neto, C.; Costa, R.F.A.; Viana, C.R.; Reis, R.M.; Baltazar, F.; et al. The metabolic microenvironment of melanomas: Prognostic value of MCT1 and MCT4. Cell Cycle 2016, 15, 1462–1470. [Google Scholar] [CrossRef] [Green Version]
- Payen, V.L.; Hsu, M.Y.; Rädecke, K.S.; Wyart, E.; Vazeille, T.; Bouzin, C.; Porporato, P.E.; Sonveaux, P. Monocarboxylate Transporter MCT1 Promotes Tumor Metastasis Independently of Its Activity as a Lactate Transporter. Cancer Res. 2017, 77, 5591–5601. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zheng, A.; Hydbring, P.; Ambroise, G.; Ouchida, A.T.; Goiny, M.; Vakifahmetoglu-Norberg, H.; Norberg, E. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep. 2017, 19, 2289–2303. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Ma, J.; Wang, X.; Ma, T.; Zhang, S.; Wang, W.; Zhou, X.; Shi, J. High Expression of PHGDH Predicts Poor Prognosis in Non–Small Cell Lung Cancer. Transl. Oncol. 2016, 9, 592–599. [Google Scholar] [CrossRef]
- Polański, R.; Hodgkinson, C.L.; Fusi, A.; Nonaka, D.; Priest, L.; Kelly, P.; Trapani, F.; Bishop, P.W.; White, A.; Critchlow, S.E.; et al. Activity of the Monocarboxylate Transporter 1 Inhibitor AZD3965 in Small Cell Lung Cancer. Clin. Cancer Res. 2014, 20, 926–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markou, A.; Tzanikou, E.; Kallergi, G.; Pantazaka, E.; Georgoulias, V.; Kotsakis, A.; Lianidou, E. Evaluation of Monocarboxylate Transporter 4 (MCT4) Expression and Its Prognostic Significance in Circulating Tumor Cells From Patients With Early Stage Non-Small-Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 641978. [Google Scholar] [CrossRef]
- Yang, L.; Yan, X.; Chen, J.; Zhan, Q.; Hua, Y.; Xu, S.; Li, Z.; Wang, Z.; Dong, Y.; Zuo, D.; et al. Hexokinase 2 discerns a novel circulating tumor cell population associated with poor prognosis in lung cancer patients. Proc. Natl. Acad. Sci. USA 2021, 118, e2012228118. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Z.; Tang, Y.; Lu, X.; Chen, J.; Dong, Y.; Wu, B.; Wang, C.; Yang, L.; Guo, Z.; et al. Liquid biopsy-based single-cell metabolic phenotyping of lung cancer patients for informative diagnostics. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turetta, M.; Bulfoni, M.; Brisotto, G.; Fasola, G.; Zanello, A.; Biscontin, E.; Mariuzzi, L.; Steffan, A.; Di Loreto, C.; Cesselli, D.; et al. Assessment of the Mutational Status of NSCLC Using Hypermetabolic Circulating Tumor Cells. Cancers 2018, 10, 270. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Li, N.; Lin, L.; Li, H.; Lin, J.-M. Metabolism-Based Capture and Analysis of Circulating Tumor Cells in an Open Space. Anal. Chem. 2021, 93, 6955–6960. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Z.; Li, Z.; Kim, J.; Deng, Y.; Li, Y.; Heath, J.R.; Wei, W.; Lu, S.; Shi, Q. High-throughput screening of rare metabolically active tumor cells in pleural effusion and peripheral blood of lung cancer patients. Proc. Natl. Acad. Sci. USA 2017, 114, 2544–2549. [Google Scholar] [CrossRef] [Green Version]
- Mongersun, A.; Smeenk, I.; Pratx, G.; Asuri, P.; Abbyad, P. Droplet Microfluidic Platform for the Determination of Single-Cell Lactate Release. Anal. Chem. 2016, 88, 3257–3263. [Google Scholar] [CrossRef] [Green Version]
- Chudziak, J.; Burt, D.J.; Mohan, S.; Rothwell, D.G.; Mesquita, B.; Antonello, J.; Dalby, S.; Ayub, M.; Priest, L.; Carter, L.; et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst 2016, 141, 669–678. [Google Scholar] [CrossRef]
- Porras, T.B.; Kaur, P.; Ring, A.; Schechter, N.; Lang, J.E. Challenges in using liquid biopsies for gene expression profiling. Oncotarget 2018, 9, 7036–7053. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vanhove, K.; Graulus, G.-J.; Mesotten, L.; Thomeer, M.; Derveaux, E.; Noben, J.-P.; Guedens, W.; Adriaensens, P. The Metabolic Landscape of Lung Cancer: New Insights in a Disturbed Glucose Metabolism. Front. Oncol. 2019, 9, 1215. [Google Scholar] [CrossRef] [Green Version]
- Bose, S.; Zhang, C.; Le, A. Glucose Metabolism in Cancer: The Warburg Effect and Beyond. Adv. Exp. Med. Biol. 2021, 1311, 3–15. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Cayrefourcq, L.; Mazard, T.; Maudelonde, T.; Assenat, E.; Assou, S. Molecular Portrait of Metastasis-Competent Circulating Tumor Cells in Colon Cancer Reveals the Crucial Role of Genes Regulating Energy Metabolism and DNA Repair. Clin. Chem. 2017, 63, 700–713. [Google Scholar] [CrossRef]
- DE LA Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. 2019, 9, 1143. [Google Scholar] [CrossRef] [Green Version]
- Gallamini, A.; Zwarthoed, C.; Borra, A. Positron Emission Tomography (PET) in Oncology. Cancers 2014, 6, 1821–1889. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.; Marayati, R.; Moffitt, R.; Yeh, J.J. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget 2017, 8, 56081–56094. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Gao, F.; Ma, X.; Wang, R.; Dong, X.; Wang, W. Deguelin inhibits non-small cell lung cancer via down-regulating Hexokinases II-mediated glycolysis. Oncotarget 2017, 8, 32586–32599. [Google Scholar] [CrossRef] [Green Version]
- Zavridou, M.; Mastoraki, S.; Strati, A.; Koutsodontis, G.; Klinakis, A.; Psyrri, A.; Lianidou, E. Direct comparison of size-dependent versus EpCAM-dependent CTC enrichment at the gene expression and DNA methylation level in head and neck squamous cell carcinoma. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, K.; Torres-Gérica, P.; Jiménez-Segovia, A.; Ramos-Muñoz, E.; Crespo-Toro, L.; Fuentes, P.; Toribio, M.L.; Callejas-Hernández, F.; Carrato, A.; Bermejo, M.L.G.; et al. Cyclooxygenase 2 Effector Genes as Potential Inflammation-Related Biomarkers for Colorectal Cancer Circulating Tumor Cells Detection by Liquid Biopsy. Front. Pharmacol. 2022, 12, 806395. [Google Scholar] [CrossRef]
- Obermayr, E.; Agreiter, C.; Schuster, E.; Fabikan, H.; Weinlinger, C.; Baluchova, K.; Hamilton, G.; Hochmair, M.; Zeillinger, R. Molecular Characterization of Circulating Tumor Cells Enriched by A Microfluidic Platform in Patients with Small-Cell Lung Cancer. Cells 2019, 8, 880. [Google Scholar] [CrossRef] [Green Version]
- Obermayr, E.; Koppensteiner, N.; Heinzl, N.; Schuster, E.; Holzer, B.; Fabikan, H.; Weinlinger, C.; Illini, O.; Hochmair, M.; Zeillinger, R. Cancer Stem Cell-Like Circulating Tumor Cells Are Prognostic in Non-Small Cell Lung Cancer. J. Pers. Med. 2021, 11, 1225. [Google Scholar] [CrossRef]
- Obermayr, E.; Maritschnegg, E.; Agreiter, C.; Pecha, N.; Speiser, P.; Helmy-Bader, S.; Danzinger, S.; Krainer, M.; Singer, C.; Zeillinger, R. Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget 2017, 9, 812–823. [Google Scholar] [CrossRef] [Green Version]
- Faubert, B.; Li, K.Y.; Cai, L.; Hensley, C.T.; Kim, J.; Zacharias, L.G.; Yang, C.; Do, Q.N.; Doucette, S.; Burguete, D.; et al. Lactate Metabolism in Human Lung Tumors. Cell 2017, 171, 358–371.e9. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Li, M.; Yu, X.; Gao, F.; Li, W. Repression of Hexokinases II-Mediated Glycolysis Contributes to Piperlongumine-Induced Tumor Suppression in Non-Small Cell Lung Cancer Cells. Int. J. Biol. Sci. 2019, 15, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Dhup, S.; Dadhich, R.K.; Copetti, T.; Sonveaux, P. Anticancer Targets in the Glycolytic Metabolism of Tumors: A Comprehensive Review. Front. Pharmacol. 2011, 2, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Su, J.; Sun, M.; Song, J.; Sun, H.; Fan, J.; Chen, G.; Shan, C.; Qi, Q.; Zhang, S. Octamer transcription factor-1 induces the Warburg effect via up-regulation of hexokinase 2 in non-small cell lung cancer. Mol. Cell. Biochem. 2021, 476, 3423–3431. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.-H.; Hu, X.-P.; Xiang, X.-P.; Fang, L. High expression of monocarboxylate transporter 4 (MCT 4), but not MCT 1, predicts poor prognosis in patients with non-small cell lung cancer. Transl. Cancer Res. 2021, 10, 1336–1345. [Google Scholar] [CrossRef]
- Kershaw, S.; Cummings, J.; Morris, K.; Tugwood, J.; Dive, C. Optimisation of immunofluorescence methods to determine MCT1 and MCT4 expression in circulating tumour cells. BMC Cancer 2015, 15, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Cruz, M.D.; Ledbetter, S.; Chowdhury, S.; Tiwari, A.K.; Momi, N.; Wali, R.K.; Bliss, C.; Huang, C.; Lichtenstein, D.; Bhattacharya, S.; et al. Metabolic reprogramming of the premalignant colonic mucosa is an early event in carcinogenesis. Oncotarget 2017, 8, 20543–20557. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Semenza, G.L.; Simons, J.W.; De Marzo, A.M. Up-regulation of hypoxia-inducible factor 1α is an early event in prostate carcinogenesis. Cancer Detect. Prev. 2004, 28, 88–93. [Google Scholar] [CrossRef]
- Domingo-Vidal, M.; Whitaker-Menezes, D.; Martos-Rus, C.; Tassone, P.; Snyder, C.M.; Tuluc, M.; Philp, N.; Curry, J.; Martinez-Outschoorn, U. Cigarette Smoke Induces Metabolic Reprogramming of the Tumor Stroma in Head and Neck Squamous Cell Carcinoma. Mol. Cancer Res. 2019, 17, 1893–1909. [Google Scholar] [CrossRef] [Green Version]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [Green Version]
- Estrella, V.; Chen, T.; Lloyd, M.; Wojtkowiak, J.; Cornnell, H.H.; Ibrahim-Hashim, A.; Bailey, K.; Balagurunathan, Y.; Rothberg, J.M.; Sloane, B.F.; et al. Acidity Generated by the Tumor Microenvironment Drives Local Invasion. Cancer Res. 2013, 73, 1524–1535. [Google Scholar] [CrossRef] [Green Version]
- Rivello, F.; Matuła, K.; Piruska, A.; Smits, M.; Mehra, N.; Huck, W.T.S. Probing single-cell metabolism reveals prognostic value of highly metabolically active circulating stromal cells in prostate cancer. Sci. Adv. 2020, 6, 3849. Available online: https://www.science.org (accessed on 24 May 2020). [CrossRef]

| HK2 a | p-Value | PHGDH a | p-Value | MCT1 a | p-Value | |||
|---|---|---|---|---|---|---|---|---|
| Gender | Male | 30 | 9 | 0.593 | 5 | 0.535 | 8 | 0.105 |
| Female | 16 | 5 | 2 | 8 | ||||
| Age | ≤65 | 26 | 8 | 0.758 | 5 | 11 | 0.061 | |
| >65 | 19 | 5 | 2 | 3 | ||||
| n.a. | 1 | 1 | 1 | 1 | ||||
| Stage | I | 22 | 6 | 0.379 | 2 | 0.040 | 8 | 0.438 |
| II | 10 | 4 | 3 | 3 | ||||
| III | 13 | 3 | 1 | 3 | ||||
| n.a. | 1 | 1 | 1 | 1 | ||||
| Size | ≤5 | 33 | 10 | 0.976 | 6 | 0.378 | 12 | 0.392 |
| >5 | 13 | 4 | 1 | 3 | ||||
| Type | SCC | 24 | 7 | 0.838 | 5 | 0.148 | 8 | 0.904 |
| ADENO | 19 | 5 | 1 | 6 | ||||
| n.a. | 3 | 1 | 2 | 1 | ||||
| Smoking status | No | 4 | 2 | 0.333 | 0 | 0.322 | 0 | 0.017 |
| Ex | 17 | 3 | 1 | 2 | ||||
| Yes | 20 | 7 | 4 | 10 | ||||
| n.a. | 5 | 3 | 1 | 1 | ||||
| Lymph Nodes | Yes | 15 | 3 | 0.290 | 2 | 0.807 | 3 | 0.427 |
| No | 31 | 11 | 5 | 11 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafeiriadou, A.; Kollias, I.; Londra, T.; Tsaroucha, E.; Georgoulias, V.; Kotsakis, A.; Lianidou, E.; Markou, A. Metabolism-Related Gene Expression in Circulating Tumor Cells from Patients with Early Stage Non-Small Cell Lung Cancer. Cancers 2022, 14, 3237. https://doi.org/10.3390/cancers14133237
Zafeiriadou A, Kollias I, Londra T, Tsaroucha E, Georgoulias V, Kotsakis A, Lianidou E, Markou A. Metabolism-Related Gene Expression in Circulating Tumor Cells from Patients with Early Stage Non-Small Cell Lung Cancer. Cancers. 2022; 14(13):3237. https://doi.org/10.3390/cancers14133237
Chicago/Turabian StyleZafeiriadou, A., I. Kollias, T. Londra, E. Tsaroucha, V. Georgoulias, A. Kotsakis, E. Lianidou, and A. Markou. 2022. "Metabolism-Related Gene Expression in Circulating Tumor Cells from Patients with Early Stage Non-Small Cell Lung Cancer" Cancers 14, no. 13: 3237. https://doi.org/10.3390/cancers14133237
APA StyleZafeiriadou, A., Kollias, I., Londra, T., Tsaroucha, E., Georgoulias, V., Kotsakis, A., Lianidou, E., & Markou, A. (2022). Metabolism-Related Gene Expression in Circulating Tumor Cells from Patients with Early Stage Non-Small Cell Lung Cancer. Cancers, 14(13), 3237. https://doi.org/10.3390/cancers14133237








