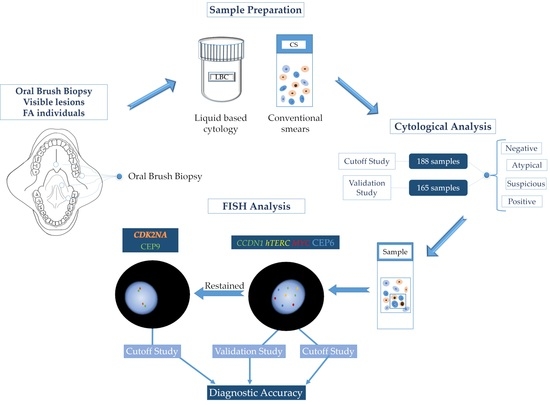
A New Multi-Color FISH Assay for Brush Biopsy-Based Detection of Chromosomal Aneuploidy in Oral (Pre)Cancer in Patients with Fanconi Anemia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
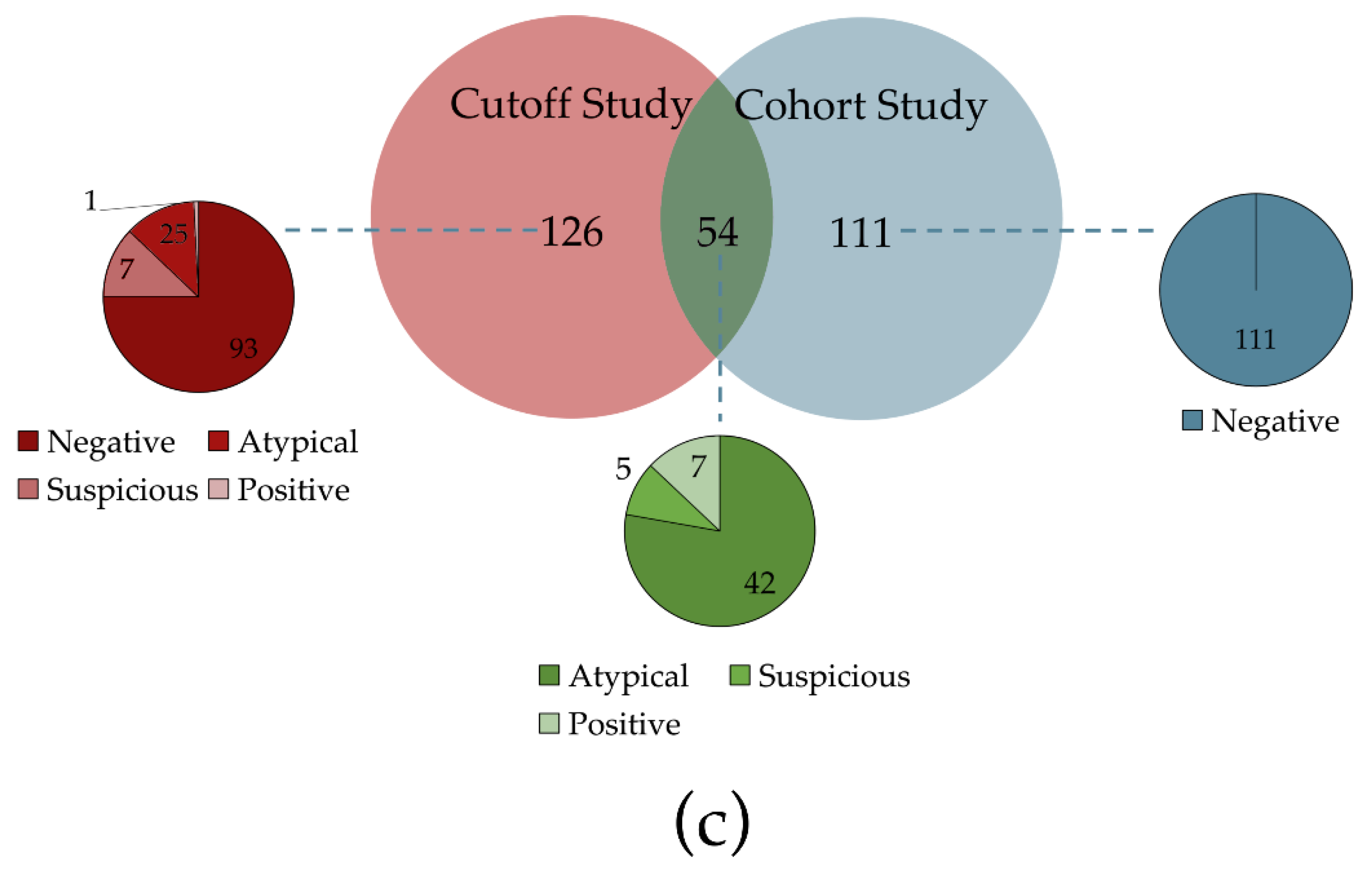
2.1. Study Design and Samples
2.1.1. Cutoff Study
2.1.2. Validation Study
2.2. Re-Evaluation of Cytological Samples
2.3. FISH
2.3.1. Hybridization
2.3.2. Re-Hybridization
2.3.3. FISH Analysis
2.3.4. FISH Evaluation Protocol
- At least 3 FISH probes showed a gain (>2 signals) (definition 1 (D1));
- All 4 probes showed a gain (>2 signals) (definition 2 (D2));
- All 4 probes showed a gain (>2 signals) or 3 probes showed a gain (>2 signals) if one of these probes was MYC with a gain of ≥4 Signals (definition 3 (D3));
- All 4 probes showed a gain (>2 signals) or 3 probes showed a gain (>2 signals) if one of these probes was MYC with a gain of ≥5 Signals (definition 4 (D4)).
2.4. Reference Standard
2.5. Statistical Analysis
3. Results
3.1. Patients and Samples
3.2. Multi-Color FISH Assay
3.3. 9p21 FISH Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Definition | Cutoff-Point | Sensitivity | Specificity | Youden Index | Definition | Cutoff-Point | Sensitivity | Specificity | Youden Index |
|---|---|---|---|---|---|---|---|---|---|
| D1 | 1 | 0.974 | 0.438 | 0.412 | D2 | 1 | 0.949 | 0.579 | 0.527 |
| 2 | 0.974 | 0.587 | 0.561 | 2 | 0.923 | 0.686 | 0.609 | ||
| 3 | 0.949 | 0.653 | 0.602 | 3 | 0.923 | 0.752 | 0.675 | ||
| 4 | 0.923 | 0.686 | 0.609 | 4 | 0.872 | 0.810 | 0.682 | ||
| 5 | 0.923 | 0.711 | 0.634 | 5 | 0.821 | 0.843 | 0.663 | ||
| 6 | 0.897 | 0.727 | 0.625 | 6 | 0.795 | 0.860 | 0.654 | ||
| 7 | 0.897 | 0.760 | 0.658 | 7 | 0.769 | 0.868 | 0.637 | ||
| 8 | 0.897 | 0.769 | 0.666 | 8 | 0.692 | 0.884 | 0.577 | ||
| 9 | 0.872 | 0.785 | 0.657 | 9 | 0.641 | 0.893 | 0.534 | ||
| 10 | 0.846 | 0.785 | 0.631 | 10 | 0.641 | 0.901 | 0.542 | ||
| 11 | 0.846 | 0.793 | 0.640 | 11 | 0.590 | 0.909 | 0.499 | ||
| 12 | 0.795 | 0.802 | 0.597 | 12 | 0.564 | 0.917 | 0.481 | ||
| 14 | 0.744 | 0.802 | 0.545 | 13 | 0.513 | 0.934 | 0.447 | ||
| 15 | 0.718 | 0.818 | 0.536 | 15 | 0.487 | 0.950 | 0.438 | ||
| 16 | 0.718 | 0.835 | 0.553 | 17 | 0.462 | 0.950 | 0.412 | ||
| 18 | 0.718 | 0.843 | 0.561 | 20 | 0.436 | 0.950 | 0.386 | ||
| 19 | 0.692 | 0.843 | 0.535 | 21 | 0.410 | 0.967 | 0.377 | ||
| 20 | 0.667 | 0.860 | 0.526 | 23 | 0.333 | 0.967 | 0.300 | ||
| 21 | 0.641 | 0.860 | 0.501 | 26 | 0.308 | 0.967 | 0.275 | ||
| D3 | 1 | 0.974 | 0.529 | 0.503 | D4 | 1 | 0.949 | 0.570 | 0.519 |
| 2 | 0.949 | 0.636 | 0.585 | 2 | 0.949 | 0.686 | 0.635 | ||
| 3 | 0.949 | 0.711 | 0.659 | 3 | 0.949 | 0.736 | 0.684 | ||
| 4 | 0.923 | 0.752 | 0.675 | 4 | 0.897 | 0.785 | 0.683 | ||
| 5 | 0.872 | 0.802 | 0.673 | 5 | 0.872 | 0.826 | 0.698 | ||
| 6 | 0.872 | 0.818 | 0.690 | 6 | 0.872 | 0.835 | 0.707 | ||
| 7 | 0.872 | 0.826 | 0.698 | 7 | 0.846 | 0.843 | 0.689 | ||
| 8 | 0.846 | 0.826 | 0.673 | 8 | 0.744 | 0.843 | 0.587 | ||
| 9 | 0.821 | 0.835 | 0.655 | 9 | 0.718 | 0.860 | 0.577 | ||
| 10 | 0.769 | 0.835 | 0.604 | 10 | 0.692 | 0.876 | 0.568 | ||
| 11 | 0.718 | 0.843 | 0.561 | 11 | 0.615 | 0.884 | 0.500 | ||
| 12 | 0.667 | 0.843 | 0.510 | 12 | 0.590 | 0.901 | 0.491 | ||
| 13 | 0.641 | 0.860 | 0.501 | 13 | 0.590 | 0.926 | 0.515 | ||
| 14 | 0.641 | 0.868 | 0.509 | 14 | 0.538 | 0.926 | 0.464 | ||
| 15 | 0.641 | 0.901 | 0.542 | 15 | 0.538 | 0.934 | 0.472 | ||
| 16 | 0.590 | 0.909 | 0.499 | 16 | 0.538 | 0.950 | 0.489 | ||
| 17 | 0.590 | 0.934 | 0.524 | 18 | 0.513 | 0.950 | 0.463 | ||
| 19 | 0.564 | 0.942 | 0.506 | 21 | 0.487 | 0.967 | 0.454 |
| Definition | Cutoff-Point | Sensitivity | Specificity | Youden Index | Definition | Cutoff-Point | Sensitivity | Specificity | Youden Index |
|---|---|---|---|---|---|---|---|---|---|
| D1 | 1 | 1.000 | 0.025 | 0.025 | D2 | 1 | 0.974 | 0.150 | 0.124 |
| 3 | 0.974 | 0.075 | 0.049 | 2 | 0.947 | 0.250 | 0.197 | ||
| 4 | 0.947 | 0.150 | 0.097 | 3 | 0.947 | 0.350 | 0.297 | ||
| 5 | 0.947 | 0.175 | 0.122 | 4 | 0.895 | 0.500 | 0.395 | ||
| 6 | 0.921 | 0.225 | 0.146 | 5 | 0.842 | 0.550 | 0.392 | ||
| 7 | 0.921 | 0.300 | 0.221 | 6 | 0.816 | 0.600 | 0.416 | ||
| 8 | 0.921 | 0.325 | 0.246 | 7 | 0.789 | 0.625 | 0.414 | ||
| 9 | 0.895 | 0.375 | 0.270 | 8 | 0.711 | 0.675 | 0.386 | ||
| 10 | 0.868 | 0.375 | 0.243 | 9 | 0.658 | 0.700 | 0.358 | ||
| 11 | 0.868 | 0.400 | 0.268 | 11 | 0.605 | 0.725 | 0.330 | ||
| 12 | 0.816 | 0.425 | 0.241 | 12 | 0.579 | 0.750 | 0.329 | ||
| 14 | 0.763 | 0.425 | 0.188 | 13 | 0.526 | 0.800 | 0.326 | ||
| 15 | 0.737 | 0.475 | 0.212 | 15 | 0.500 | 0.850 | 0.350 | ||
| 16 | 0.737 | 0.500 | 0.237 | 17 | 0.474 | 0.850 | 0.324 | ||
| 18 | 0.737 | 0.525 | 0.262 | 20 | 0.447 | 0.850 | 0.297 | ||
| 19 | 0.711 | 0.525 | 0.236 | 21 | 0.421 | 0.900 | 0.321 | ||
| 20 | 0.684 | 0.575 | 0.259 | 23 | 0.342 | 0.900 | 0.242 | ||
| 21 | 0.658 | 0.575 | 0.233 | 26 | 0.316 | 0.900 | 0.216 | ||
| 23 | 0.658 | 0.625 | 0.283 | 28 | 0.289 | 0.950 | 0.239 | ||
| 24 | 0.658 | 0.675 | 0.333 | 30 | 0.263 | 0.950 | 0.213 | ||
| 25 | 0.658 | 0.700 | 0.358 | 32 | 0.211 | 0.975 | 0.186 | ||
| 26 | 0.632 | 0.750 | 0.382 | 34 | 0.184 | 0.975 | 0.159 | ||
| D3 | 1 | 1.000 | 0.075 | 0.075 | D4 | 1 | 0.974 | 0.125 | 0.099 |
| 2 | 0.974 | 0.125 | 0.099 | 2 | 0.974 | 0.250 | 0.224 | ||
| 3 | 0.974 | 0.250 | 0.224 | 3 | 0.974 | 0.325 | 0.299 | ||
| 4 | 0.947 | 0.350 | 0.297 | 4 | 0.921 | 0.450 | 0.371 | ||
| 5 | 0.895 | 0.425 | 0.320 | 5 | 0.895 | 0.500 | 0.395 | ||
| 6 | 0.895 | 0.475 | 0.370 | 6 | 0.895 | 0.525 | 0.420 | ||
| 7 | 0.895 | 0.500 | 0.395 | 7 | 0.868 | 0.550 | 0.418 | ||
| 8 | 0.868 | 0.500 | 0.368 | 8 | 0.763 | 0.550 | 0.313 | ||
| 9 | 0.842 | 0.525 | 0.367 | 9 | 0.737 | 0.600 | 0.337 | ||
| 10 | 0.789 | 0.525 | 0.314 | 10 | 0.711 | 0.650 | 0.361 | ||
| 11 | 0.737 | 0.550 | 0.287 | 11 | 0.632 | 0.650 | 0.282 | ||
| 12 | 0.684 | 0.550 | 0.234 | 12 | 0.605 | 0.700 | 0.305 | ||
| 13 | 0.658 | 0.575 | 0.233 | 13 | 0.605 | 0.775 | 0.380 | ||
| 14 | 0.658 | 0.600 | 0.258 | 14 | 0.553 | 0.775 | 0.328 | ||
| 15 | 0.658 | 0.700 | 0.358 | 15 | 0.553 | 0.800 | 0.353 | ||
| 16 | 0.605 | 0.725 | 0.330 | 16 | 0.553 | 0.850 | 0.403 | ||
| 17 | 0.605 | 0.800 | 0.405 | 18 | 0.526 | 0.850 | 0.376 | ||
| 19 | 0.579 | 0.825 | 0.404 | 21 | 0.500 | 0.900 | 0.400 |
Appendix B
| Enhanced Follow-Up (Range 2–5 Years) without HG OED or SCC | ||
| FP FISH | Histology | |
| Oral Region | Result | Time Interval * |
| Gingiva | --- | --- |
| Gingiva | --- | --- |
| Gingiva | --- | --- |
| Gingiva | --- | --- |
| Gingiva | --- | --- |
| Cheek | --- | --- |
| Tongue | --- | --- |
| Tongue | --- | --- |
| Tongue | --- | --- |
| Gingiva | --- | --- |
| HG OED or SCC Somewhere in the Oral Cavity but not in the Vicinity of the Brushed Lesion | ||
| FP FISH | Histology | |
| Oral Region | Result | Time Interval |
| Gingiva | SCC | NR |
| Gingiva | SCC, | <1 year |
| HG OED | <1 year | |
| SCC, | 2 Years | |
| Tongue | HG OED | NR |
| Gingiva | SCC | NR |
| Tongue | SCC | NR |
| HG OED or SCC in the vicinity of oral brushing or at the same place in the clinical course | ||
| FP FISH | Histology | |
| Oral Region | Result | Time Interval |
| Gingiva | SCC, | <1 year |
| Gingiva | HG OED | NR |
| Gingiva | HG OED | NR |
| Gingiva | SCC | <1 year |
| Cheek | SCC | 2 Years |
| Tongue | HG OED | 1 Year |
| Lip | SCC | NR |
| Cheek | SCC | 2 Years |
| Gingiva | SCC | NR |
References
- Nalepa, G.; Clapp, D.W. Fanconi Anaemia and Cancer: An Intricate Relationship. Nat. Rev. Cancer 2018, 18, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Knies, K.; Inano, S.; Ramírez, M.J.; Ishiai, M.; Surrallés, J.; Takata, M.; Schindler, D. Biallelic Mutations in the Ubiquitin Ligase RFWD3 Cause Fanconi Anemia. J. Clin. Invest. 2017, 127, 3013–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, D.M.; Mitchell, D.K.; Abdul-Sater, Z.; Chan, K.-K.; Sun, Z.; Sheth, A.; He, Y.; Jiang, L.; Yuan, J.; Sharma, R.; et al. Mitotic Errors Promote Genomic Instability and Leukemia in a Novel Mouse Model of Fanconi Anemia. Front. Oncol. 2021, 11, 4498. [Google Scholar] [CrossRef]
- Michl, J.; Zimmer, J.; Tarsounas, M. Interplay between Fanconi Anemia and Homologous Recombination Pathways in Genome Integrity. EMBO J. 2016, 35, 909–923. [Google Scholar] [CrossRef]
- Cheung, R.S.; Taniguchi, T. Recent Insights into the Molecular Basis of Fanconi Anemia: Genes, Modifiers, and Drivers. Int. J. Hematol. 2017, 106, 335–344. [Google Scholar] [CrossRef] [Green Version]
- Dufour, C. How I Manage Patients with Fanconi Anaemia. Br. J. Haematol. 2017, 178, 32–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutler, D.I.; Patel, K.R.; Auerbach, A.D.; Kennedy, J.; Lach, F.P.; Sanborn, E.; Cohen, M.A.; Kuhel, W.I.; Smogorzewska, A. Natural History and Management of Fanconi Anemia Patients with Head and Neck Cancer: A 10-Year Follow-Up. Laryngoscope 2016, 126, 870–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutler, D.I.; Singh, B.; Satagopan, J.; Batish, S.D.; Berwick, M.; Giampietro, P.F.; Hanenberg, H.; Auerbach, A.D. A 20-Year Perspective on the International Fanconi Anemia Registry (IFAR). Blood 2003, 101, 1249–1256. [Google Scholar] [CrossRef]
- Singh, B.; Greenberger, J.; Kutler, D. Head and Neck Cancer in Patients with Fanconi Anemia. In Fanconi Anemia Clinical Care Guidelines; Frohnmayer, L., Ravenhorst, S.V., Wirkkula, L., Eds.; Fanconi Anemia Research Fund: Eugene, OR, USA, 2020; pp. 89–101. [Google Scholar]
- Velleuer, E.; Dietrich, R.; Pomjanski, N.; de Santana Almeida Araujo, I.K.; Silva de Araujo, B.E.; Sroka, I.; Biesterfeld, S.; Böcking, A.; Schramm, M. Diagnostic Accuracy of Brush Biopsy–Based Cytology for the Early Detection of Oral Cancer and Precursors in Fanconi Anemia. Cancer Cytopathol. 2020, 128, 403–413. [Google Scholar] [CrossRef]
- Savic, S.; Bubendorf, L. Common Fluorescence in Situ Hybridization Applications in Cytology. Arch. Pathol. Lab. Med. 2016, 140, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Schramm, M.; Wrobel, C.; Born, I.; Kazimirek, M.; Pomjanski, N.; William, M.; Kappes, R.; Gerharz, C.D.; Biesterfeld, S.; Böcking, A. Equivocal Cytology in Lung Cancer Diagnosis: Improvement of Diagnostic Accuracy Using Adjuvant Multicolor FISH, DNA-image Cytometry, and Quantitative Promoter Hypermethylation Analysis. Cancer Cytopathol. 2011, 119, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Silva de Araujo, B.E.; Velleuer, E.; Dietrich, R.; Pomjanski, N.; de Santana Almeida Araujo, I.K.; Schlensog, M.; Wells, S.I.; Dorsman, J.C.; Schramm, M. Detection of Cytogenetic Changes and Chromosomal Aneuploidy with Fluorescent in Situ Hybridization in Cytological Specimens of Oral Cancers in Fanconi Anemia—Proof of Concept. Clin. Exp. Dent. Res. 2022, 8, 108–116. [Google Scholar] [CrossRef] [PubMed]
- de Miranda Onofre, F.B.; Onofre, A.S.C.; Pomjanski, N.; Buckstegge, B.; Grote, H.J.; Böcking, A. 9p21 Deletion in the Diagnosis of Malignant Mesothelioma in Serous Effusions Additional to Immunocytochemistry, DNA-ICM, and AgNOR Analysis. Cancer Cytopathol. 2008, 114, 204–215. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, A.K.; Chan, J.K.; Grandis, J.R.; Slootweg, P.J. WHO Classification of Head and Neck Tumours, 4th ed.; IARC WHO Classification of Tumours; World Health Organization (WHO): Lyon, France, 2017; Volume 9. [Google Scholar]
- Youden, W.J. Index for Rating Diagnostic Tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Schwarz, S.; Bier, J.; Driemel, O.; Reichert, T.E.; Hauke, S.; Hartmann, A.; Brockhoff, G. Losses of 3p14 and 9p21 as Shown by Fluorescence in Situ Hybridization Are Early Events in Tumorigenesis of Oral Squamous Cell Carcinoma and Already Occur in Simple Keratosis. Cytom. Part J. Int. Soc. Anal. Cytol. 2008, 73, 305–311. [Google Scholar] [CrossRef]
- van Harten, A.M.; Poell, J.B.; Buijze, M.; Brink, A.; Wells, S.I.; Leemans, C.R.; Wolthuis, R.M.; Brakenhoff, R.H. Characterization of a Head and Neck Cancer-Derived Cell Line Panel Confirms the Distinct TP53-Proficient Copy Number-Silent Subclass. Oral Oncol. 2019, 98, 53–61. [Google Scholar] [CrossRef]
- Kipp, B.R.; Barr Fritcher, E.G.; Pettengill, J.E.; Halling, K.C.; Clayton, A.C. Improving the Accuracy of Pancreatobiliary Tract Cytology with Fluorescence in Situ Hybridization: A Molecular Test with Proven Clinical Success. Cancer Cytopathol. 2013, 121, 610–619. [Google Scholar] [CrossRef]
- Furquim, C.P.; Pivovar, A.; Amenábar, J.M.; Bonfim, C.; Torres-Pereira, C.C. Oral Cancer in Fanconi Anemia: Review of 121 Cases. Crit. Rev. Oncol. Hematol. 2018, 125, 35–40. [Google Scholar] [CrossRef]
- Biesterfeld, S.; Gerres, K.; Fischer-Wein, G.; Böcking, A. Polyploidy in Non-Neoplastic Tissues. J. Clin. Pathol. 1994, 47, 38–42. [Google Scholar] [CrossRef] [Green Version]
- Segura, S.; Rozas-Muñoz, E.; Toll, A.; Martín-Ezquerra, G.; Masferrer, E.; Espinet, B.; Rodriguez, M.; Baro, T.; Barranco, C.; Pujol, R. Evaluation of MYC Status in Oral Lichen Planus in Patients with Progression to Oral Squamous Cell Carcinoma. Br. J. Dermatol. 2013, 169, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Martin-Ezquerra, G.; Salgado, R.; Toll, A.; Gilaberte, M.; Baro, T.; Alameda Quitllet, F.; Yebenes, M.; Sole, F.; Garcia-Muret, M.; Espinet, B. Multiple Genetic Copy Number Alterations in Oral Squamous Cell Carcinoma: Study of MYC, TP53, CCDN1, EGFR and ERBB2 Status in Primary and Metastatic Tumours. Br. J. Dermatol. 2010, 163, 1028–1035. [Google Scholar] [CrossRef]
- Sokolova, I.A.; Halling, K.C.; Jenkins, R.B.; Burkhardt, H.M.; Meyer, R.G.; Seelig, S.A.; King, W. The Development of a Multitarget, Multicolor Fluorescence in Situ Hybridization Assay for the Detection of Urothelial Carcinoma in Urine. J. Mol. Diagn. 2000, 2, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Nagai, T.; Naiki, T.; Etani, T.; Iida, K.; Noda, Y.; Shimizu, N.; Isobe, T.; Nozaki, S.; Okamura, T.; Ando, R. UroVysion Fluorescence in Situ Hybridization in Urothelial Carcinoma: A Narrative Review and Future Perspectives. Transl. Androl. Urol. 2021, 10, 1908. [Google Scholar] [CrossRef]
- Lochhead, P.; Chan, A.T.; Nishihara, R.; Fuchs, C.S.; Beck, A.H.; Giovannucci, E.; Ogino, S. Etiologic Field Effect: Reappraisal of the Field Effect Concept in Cancer Predisposition and Progression. Mod. Pathol. 2015, 28, 14–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, H.; Brown, R.E. Field Effect in Cancer–an Update. Ann. Clin. Lab. Sci. 2009, 39, 331–337. [Google Scholar]
- Smetsers, S.E.; Velleuer, E.; Dietrich, R.; Wu, T.; Brink, A.; Buijze, M.; Deeg, D.J.; Soulier, J.; Leemans, C.R.; Braakhuis, B.J. Noninvasive Molecular Screening for Oral Precancer in Fanconi Anemia Patients. Cancer Prev. Res. 2015, 8, 1102–1111. [Google Scholar] [CrossRef] [Green Version]
- Velleuer, E.; Dietrich, R. Fanconi Anemia: Young Patients at High Risk for Squamous Cell Carcinoma. Mol. Cell. Pediatr. 2014, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Ha, P.K.; Chang, S.S.; Glazer, C.A.; Califano, J.A.; Sidransky, D. Molecular Techniques and Genetic Alterations in Head and Neck Cancer. Oral Oncol. 2009, 45, 335–339. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Mu, Q.; Thiviyanathan, V.; Annapragada, A.; Vigneswaran, N. Cancer Stem Cells Are Enriched in Fanconi Anemia Head and Neck Squamous Cell Carcinomas. Int. J. Oncol. 2014, 45, 2365–2372. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.Y.; Oh, S.Y.; Kim, H.; Hwang, T.S. DNA Degradation in Liquid-based Cytology and Its Comparison with Conventional Smear. Diagn. Cytopathol. 2016, 44, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.L.H.; Sanders, M.A.; Patel, K.; Dietrich, R.; Noonan, R.J.; Lach, F.P.; White, R.R.; Goldfarb, A.; Hadi, K.; Edwards, M.M.; et al. Fanconi Anemia Pathway Deficiency Drives Copy Number Variation in Squamous Cell Carcinomas. bioRxiv 2021. [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinomas. Nature 2015, 517, 576. [Google Scholar] [CrossRef] [Green Version]
- Leemans, C.R.; Snijders, P.J.; Brakenhoff, R.H. The Molecular Landscape of Head and Neck Cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]



| Cutoff Study | ||||||
| Characteristics | All | No HSCT | HSCT | |||
| Negative FUP | Positive FUP | Negative FUP | Positive FUP | Negative FUP | Positive FUP | |
| Total No. | 121 | 39 | 38 | 15 | 83 | 24 |
| Sex, F/M | 56/65 | 13/26 | 20/18 | 11/4 | 36/47 | 2/22 |
| Median age at brush cytology (Range), Y | 26 (10–46) | 30 (9–40) | 33 (10–46) | 33 (9–40) | 25 (10–41) | 29 (16–36) |
| Median follow-up time (Range), Mo | 45 (9–86) | 31 (0–101) | 35 (9–66) | 9 (0–55) | 48 (14–86) | 58 (7–101) |
| Validation Study | ||||||
| Characteristics | All | no HSCT | HSCT | |||
| Negative FUP | Positive FUP | Negative FUP | Positive FUP | Negative FUP | Positive FUP | |
| Total No. | 129 | 23 | 39 | 9 | 90 | 14 |
| Sex, F/M | 60/69 | 6/17 | 21/18 | 4/5 | 39/51 | 2/12 |
| Median age at brush cytology (Range), Y | 27 (10–46) | 30 (16–36) | 32 (10–46) | 30 (16–35) | 26 (10–41) | 22 (16–36) |
| Median follow-up time (Range), Mo | 33 (7–99) | 16 (7–58) | 32 (9–59) | 9 (7–31) | 36 (7–99) | 20 (7–58) |
| Definition | Sensitivity | Specificity | Youden Index | AUC | AUC p-Value | AUC 5% Confidence Interval | ||
|---|---|---|---|---|---|---|---|---|
| Cutoff Study | All specimens | D1 | 89.7% (35/39) | 72.7% (88/121) | 0.625 | 0.812 | <0.001 | 0.738–0.886 |
| D2 | 79.5% (31/39) | 86.0% (104/121) | 0.654 | 0.827 | <0.001 | 0.745–0.909 | ||
| D3 | 87.2% (34/39) | 81.8% (99/121) | 0.690 | 0.845 | <0.001 | 0.772–0.918 | ||
| D4 | 87.2% (34/39) | 83.5% (101/121) | 0.707 | 0.853 | <0.001 | 0.781–0.925 | ||
| Atypical, suspicious and positive cytology only | D1 | 92.1% (35/38) | 22.5% (9/40) | 0.146 | 0.573 | 0.267 * | 0.446–0.700 | |
| D2 | 81.6% (31/38) | 60.0% (24/40) | 0.416 | 0.708 | 0.002 | 0.591–0.825 | ||
| D3 | 89.5% (34/38) | 47.5% (19/40) | 0.370 | 0.685 | 0.005 | 0.566–0.804 | ||
| D4 | 89.5% (34/38) | 52.5% (21/40) | 0.420 | 0.710 | 0.001 | 0.593–0.826 | ||
| Validation Study | All specimens | D2 | 82.6% (19/23) | 84.5% (109/129) | 0.671 | 0.836 | <0.001 | 0.739–0.932 |
| D4 | 87.0% (20/23) | 82.9% (107/129) | 0.699 | 0.850 | <0.001 | 0.761–0.938 | ||
| Atypical, suspicious and positive cytology only | D2 | 81.8% (18/22) | 55.2% (16/29) | 0.370 | 0.685 | 0.025 | 0.537–0.833 | |
| D4 | 86.4% (19/22) | 51.7% (15/29) | 0.381 | 0.690 | 0.021 | 0.544–0.837 | ||
| Cutoff Study | ||||||
| FISH | Cytology | HSCT | ||||
| Negative | Atypical | Suspicious | Positive | No | Yes | |
| FN | 1 | 7 | 0 | 0 | 2 | 6 |
| TN | 80 | 22 | 2 | 0 | 34 | 70 |
| FP | 1 | 16 | 0 | 0 | 4 | 13 |
| TP | 0 | 15 | 8 | 8 | 13 | 18 |
| Validation Study | ||||||
| FISH | Cytology | HSCT | ||||
| Negative | Atypical | Suspicious | Positive | No | Yes | |
| FN | 0 | 4 | 0 | 0 | 2 | 2 |
| TN | 93 | 15 | 1 | 0 | 33 | 77 |
| FP | 7 | 13 | 0 | 0 | 6 | 14 |
| TP | 1 | 7 | 4 | 7 | 7 | 12 |
| Cutoff | Sensitivity | Specificity | AUC | p-Value | AUC 5% Confidence Interval | ||
|---|---|---|---|---|---|---|---|
| Cutoff Study | All specimens | 16 | 77.3% | 63.9% | 0.752 | <0.001 | 0.662–0.842 |
| Atypical, suspicious and positive cytology only | 16 | 55.3% | 65.7% | 0.587 | 0.200 * | 0.455–0.719 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva de Araujo, B.E.; Markgraf, M.; de Santana Almeida Araujo, I.K.; Velleuer, E.; Dietrich, R.; Pomjanski, N.; Schramm, M. A New Multi-Color FISH Assay for Brush Biopsy-Based Detection of Chromosomal Aneuploidy in Oral (Pre)Cancer in Patients with Fanconi Anemia. Cancers 2022, 14, 3468. https://doi.org/10.3390/cancers14143468
Silva de Araujo BE, Markgraf M, de Santana Almeida Araujo IK, Velleuer E, Dietrich R, Pomjanski N, Schramm M. A New Multi-Color FISH Assay for Brush Biopsy-Based Detection of Chromosomal Aneuploidy in Oral (Pre)Cancer in Patients with Fanconi Anemia. Cancers. 2022; 14(14):3468. https://doi.org/10.3390/cancers14143468
Chicago/Turabian StyleSilva de Araujo, Bruno Eduardo, Mona Markgraf, Isabela Karoline de Santana Almeida Araujo, Eunike Velleuer, Ralf Dietrich, Natalia Pomjanski, and Martin Schramm. 2022. "A New Multi-Color FISH Assay for Brush Biopsy-Based Detection of Chromosomal Aneuploidy in Oral (Pre)Cancer in Patients with Fanconi Anemia" Cancers 14, no. 14: 3468. https://doi.org/10.3390/cancers14143468