Deubiquitinases in Cancers: Aspects of Proliferation, Metastasis, and Apoptosis
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Ubiquitination
1.2. Deubiquitination
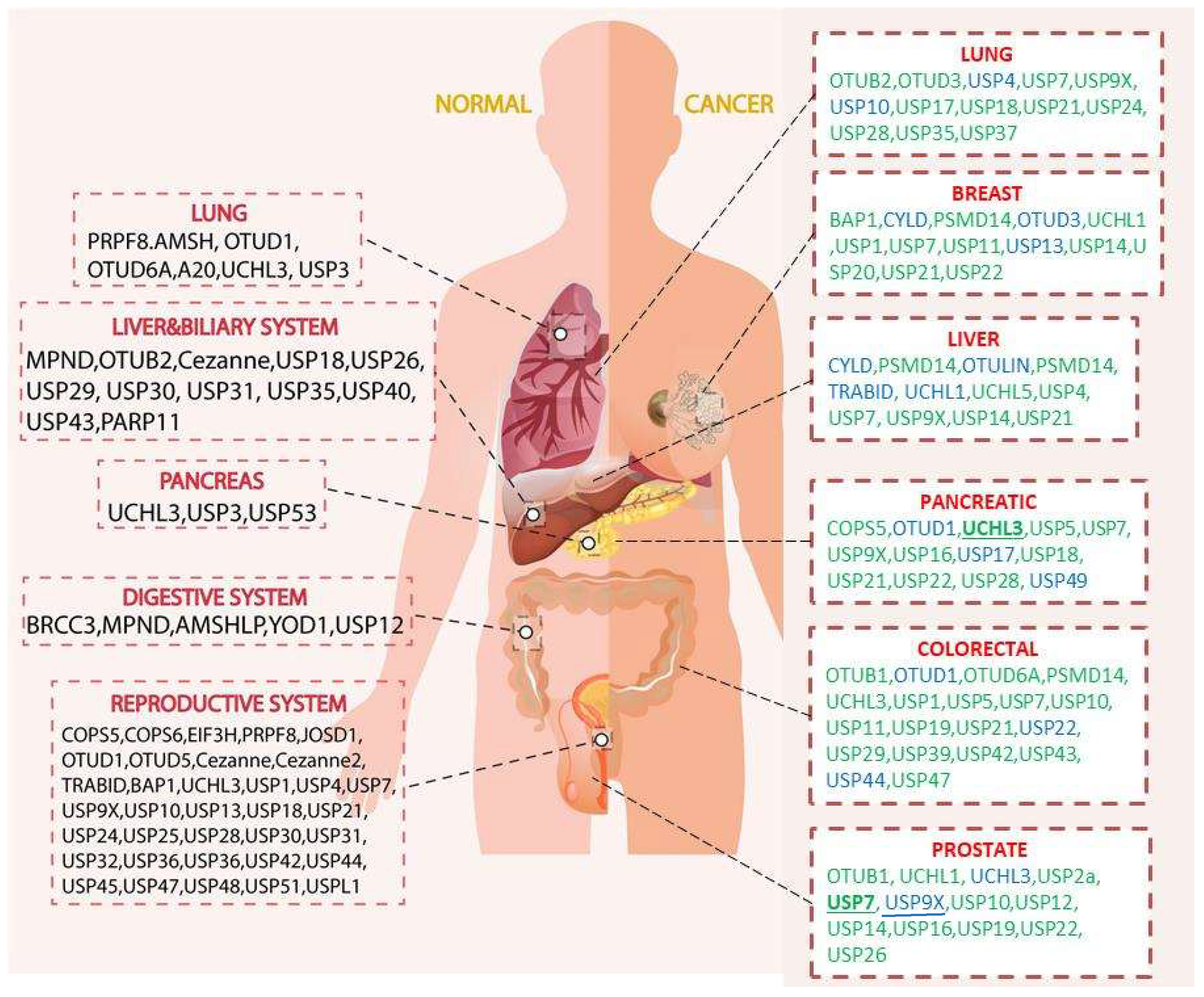
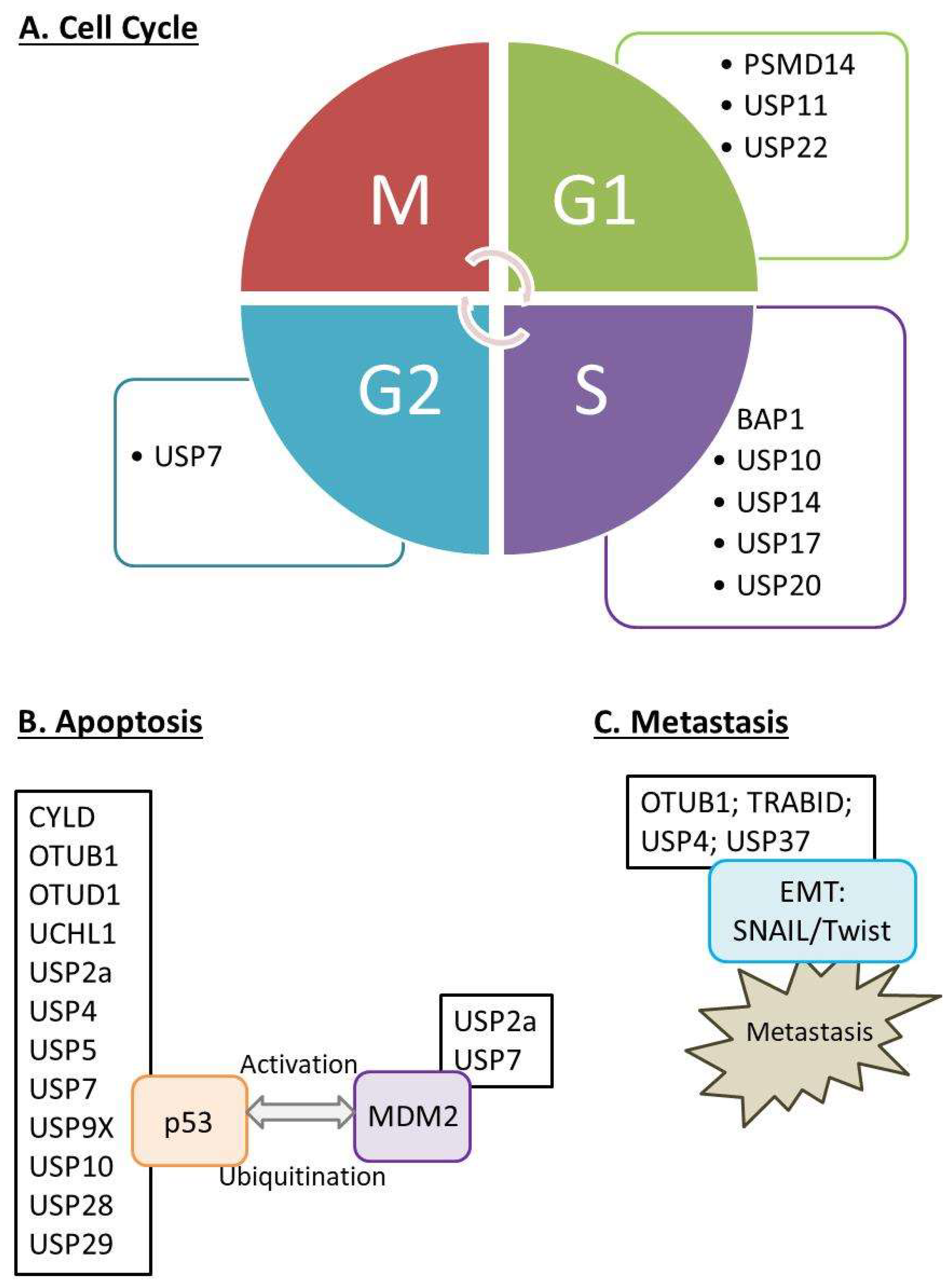
1.3. Biological Functions of DUBs and Their Expressions in Selected Organs
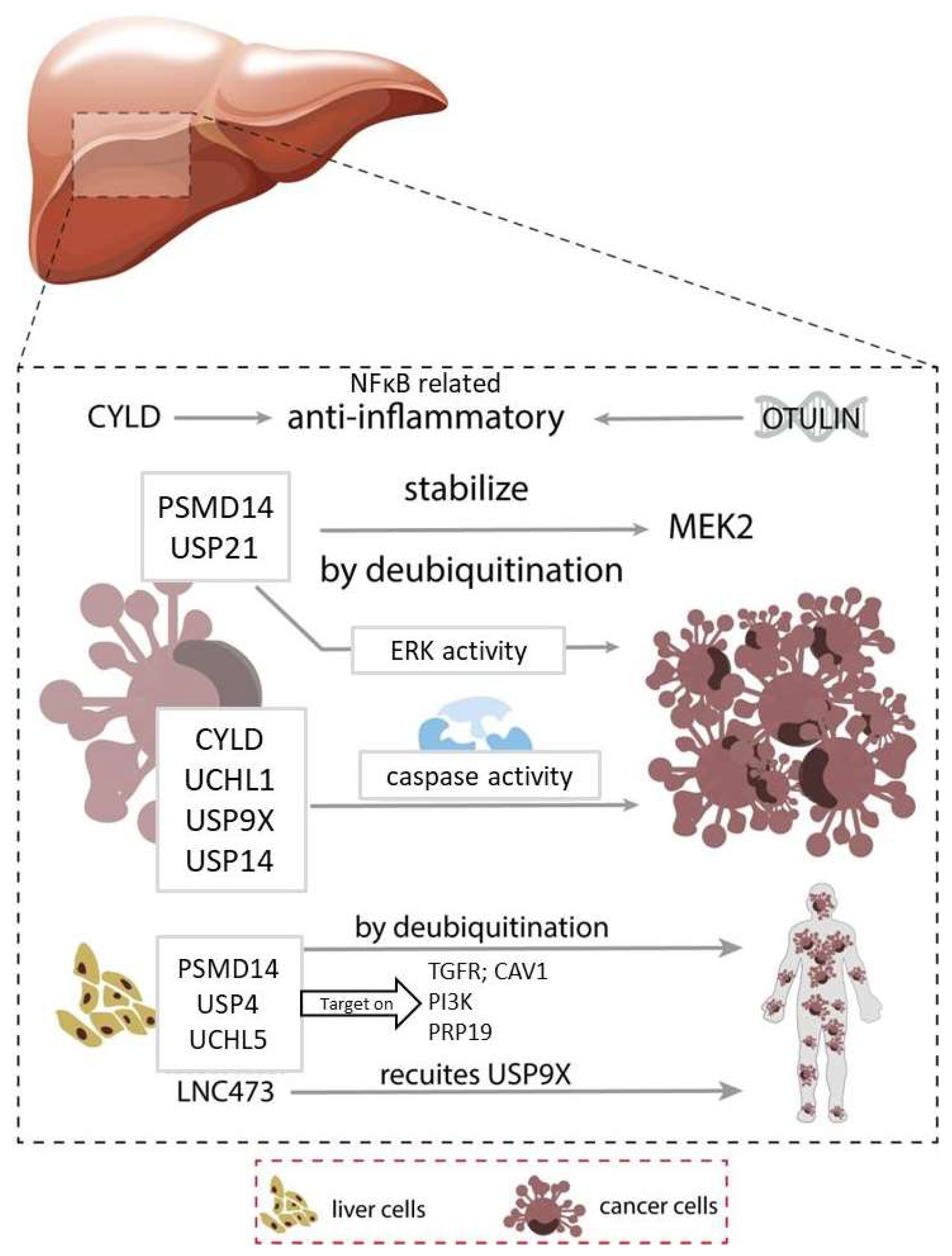
2. Liver Cancer
2.1. Inflammation
2.2. Cell Proliferation
2.3. Migration, Invasion & Metastasis
2.4. Apoptosis
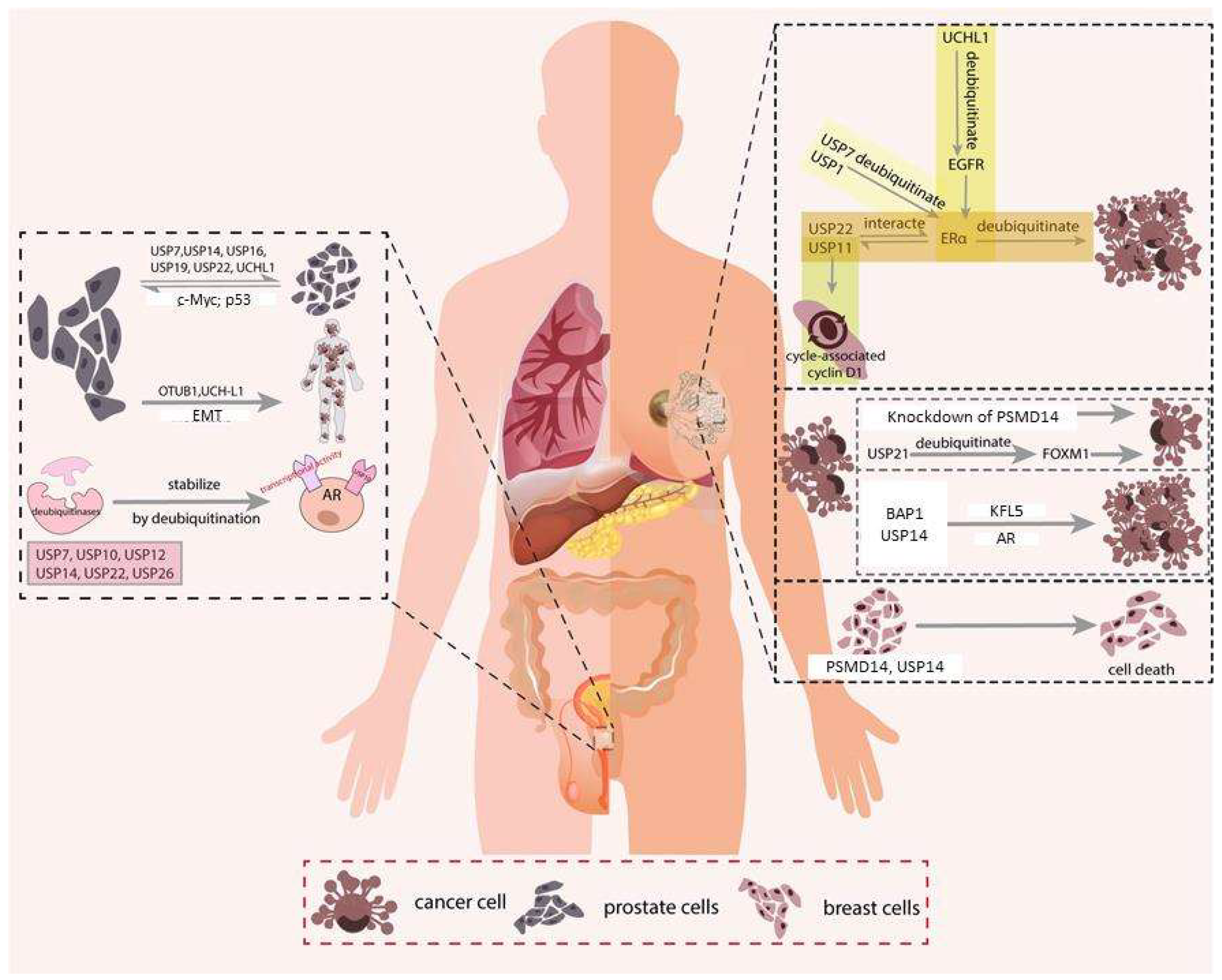
3. Breast Cancer
3.1. ERα Signaling
3.2. Cell Proliferation
3.3. Migration, Invasion & Metastasis
3.4. Apoptosis
4. Prostate Cancer
4.1. Androgen Receptor (AR)
4.2. Cell Proliferation
4.3. Migration, Invasion & Metastasis
4.4. Apoptosis
5. Colorectal Cancer
5.1. Wnt Signaling
5.2. Cell Proliferation
5.3. Migration, Invasion & Metastasis
5.4. Apoptosis
6. Pancreatic Cancer
6.1. Akt Signaling
6.2. Cell Proliferation
6.3. Migration, Invasion & Metastasis
6.4. Apoptosis
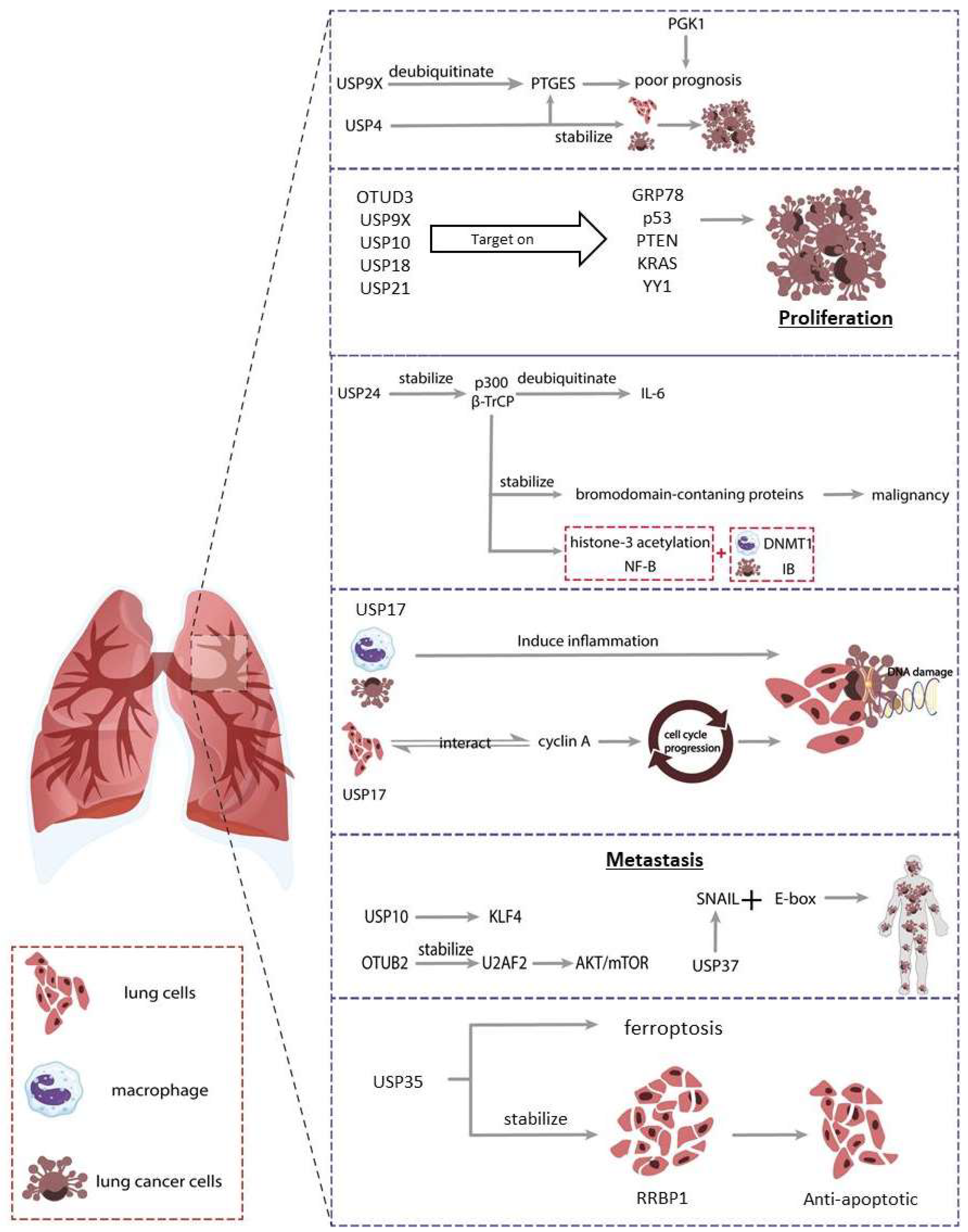
7. Lung Cancer
7.1. Proliferation
7.2. Migration, Invasion & Metastasis
7.3. Apoptosis
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pickart, C.M. Ubiquitin Enters the New Millennium. Mol. Cell 2001, 8, 499–504. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, R.C.; Wendland, B. Ubiquitin: Not just for proteasomes anymore. Curr. Opin. Cell Biol. 2003, 15, 184–190. [Google Scholar] [CrossRef]
- Petroski, M.D. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008, 9 (Suppl. 1), S7. [Google Scholar] [CrossRef] [Green Version]
- Nijman, S.M.B.; Luna-Vargas, M.P.A.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.G.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [Green Version]
- Nijman, S.M.; Huang, T.T.; Dirac, A.M.; Brummelkamp, T.R.; Kerkhoven, R.M.; D’Andrea, A.D.; Bernards, R. The Deubiquitinating Enzyme USP1 Regulates the Fanconi Anemia Pathway. Mol. Cell 2005, 17, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from Genes to Organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Rehman, S.A.A.; Kristariyanto, Y.A.; Choi, S.-Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [Green Version]
- Hermanns, T.; Pichlo, C.; Woiwode, I.; Klopffleisch, K.; Witting, K.F.; Ovaa, H.; Baumann, U.; Hofmann, K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat. Commun. 2018, 9, 799. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Saad, Y.; Lei, T.; Wang, J.; Qi, D.; Yang, Q.; Kolattukudy, P.E.; Fu, M. Mcp-induced protein 1 deubiquitinates traf proteins and negatively regulates jnk and nf-kappab signaling. J. Exp. Med. 2010, 207, 2959–2973. [Google Scholar] [CrossRef] [Green Version]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Eletr, Z.M.; Wilkinson, K.D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 2013, 1843, 114–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta—Mol. Cell Res. 2004, 1695, 189–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, K.R.; Catic, A.; Schlieker, C.; Ploegh, H.L. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat. Chem. Biol. 2007, 3, 697–705. [Google Scholar] [CrossRef]
- Song, L.; Rape, M. Reverse the curse—The role of deubiquitination in cell cycle control. Curr. Opin. Cell Biol. 2008, 20, 156–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, K.P.; Chen, J.; Tse, W.K.F. Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. Int. J. Mol. Sci. 2020, 21, 2548. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Venuto, S.; Merla, G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.; Zhang, Y.; Galardy, P.J. DUBs and cancer: The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 2009, 8, 1688–1697. [Google Scholar] [CrossRef]
- Vodermaier, H.C. APC/C and SCF: Controlling Each Other and the Cell Cycle. Curr. Biol. 2004, 14, R787–R796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernald, K.; Kurokawa, M. Evading apoptosis in cancer. Trends Cell Biol. 2013, 23, 620–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Zhou, Z.; Wu, G.; Chen, Q.; Wan, Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacol. Ther. 2017, 177, 96–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bednash, J.S.; Mallampalli, R.K. Targeting deubiquitinases in cancer. Methods Mol. Biol. 2018, 1731, 295–305. [Google Scholar] [PubMed]
- Levine, A.J. p53, the Cellular Gatekeeper for Growth and Division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Brooks, C.L.; Li, M.; Hu, M.; Shi, Y.; Gu, W. The p53--mdm2--hausp complex is involved in p53 stabilization by hausp. Oncogene 2007, 26, 7262–7266. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, L.F.; Sparks, A.; Allende-Vega, N.; Xirodimas, D.; Lane, D.; Saville, M.K. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007, 26, 976–986. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Liu, R.; Zhang, W.; Qian, S.; Wang, J.-H. MicroRNA-205 regulates ubiquitin specific peptidase 7 protein expression in hepatocellular carcinoma cells. Mol. Med. Rep. 2015, 12, 4652–4656. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, A.; Suresh, B.; Sarodaya, N.; Ko, N.-R.; Oh, S.-J.; Kim, K.-S.; Ramakrishna, S. Ubiquitin Specific Protease 29 Functions as an Oncogene Promoting Tumorigenesis in Colorectal Carcinoma. Cancers 2021, 13, 2706. [Google Scholar] [CrossRef]
- Ouyang, S.W.; Liu, T.T.; Liu, X.S.; Zhu, F.X.; Zhu, F.M.; Liu, X.N.; Peng, Z.H. Usp10 regulates musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett. 2019, 593, 406–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potu, H.; Peterson, L.F.; Pal, A.; Verhaegen, M.; Cao, J.; Talpaz, M.; Donato, N.J. Usp5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget 2014, 5, 5559–5569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, B.; Vallés, A.M.; Edme, N. Induction and regulation of epithelial-mesenchymal transitions. Biochem. Pharmacol. 2000, 60, 1091–1099. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Li, M.; Wang, X.; Li, L.; Li, Q.; Hou, Z.; Jia, H.; Liu, S. USP37 Promotes Lung Cancer Cell Migration by Stabilizing Snail Protein via Deubiquitination. Front. Genet. 2019, 10, 1324. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, Y.; Zhu, R.; Ding, F.; Cao, X.; Lin, D.; Liu, Z. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene 2018, 37, 3356–3368. [Google Scholar] [CrossRef]
- Shen, G.; Lin, Y.; Yang, X.; Zhang, J.; Xu, Z.; Jia, H. MicroRNA-26b inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X. BMC Cancer 2014, 14, 393. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, S.; Wu, H.; Lu, J.; Lu, Y.; Wang, F.; Zhao, W.; Zhan, P.; Lu, J.; Fang, Q.; et al. Deubiquitinase PSMD14 enhances hepatocellular carcinoma growth and metastasis by stabilizing GRB2. Cancer Lett. 2020, 469, 22–34. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Mahjoubin-Tehran, M.; De Vincentis, A.; Mikhailidis, D.P.; Atkin, S.L.; Mantzoros, C.S.; Jamialahmadi, T.; Sahebkar, A. Non-alcoholic fatty liver disease and steatohepatitis: State of the art on effective therapeutics based on the gold standard method for diagnosis. Mol. Metab. 2021, 50, 101049. [Google Scholar] [CrossRef]
- Nowarski, R.; Gagliani, N.; Huber, S.; Flavell, R.A. Innate Immune Cells in Inflammation and Cancer. Cancer Immunol. Res. 2013, 1, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Akaike, T. Nitric oxide and oxygen radicals in infection, inflammation, and cancer. Biochem. Biokhimiia. 1998, 63, 854–865. [Google Scholar]
- Capece, D.; Fischietti, M.; Verzella, D.; Gaggiano, A.; Cicciarelli, G.; Tessitore, A.; Zazzeroni, F.; Alesse, E. The inflammatory microenvironment in hepatocellular carcinoma: A pivotal role for tumor-associated macrophages. BioMed. Res. Int. 2013, 2013, 187204. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Maeda, S. Inflammation- and stress-related signaling pathways in hepatocarcinogenesis. World J. Gastroenterol. 2012, 18, 4071–4081. [Google Scholar] [CrossRef]
- Pannem, R.R.; Dorn, C.; Ahlqvist, K.; Bosserhoff, A.K.; Hellerbrand, C.; Massoumi, R. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis 2014, 35, 461–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellerbrand, C.; Bumes, E.; Bataille, F.; Dietmaier, W.; Massoumi, R.; Bosserhoff, A. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 2007, 28, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Urbanik, T.; Boger, R.J.; Longerich, T.; Becker, K.; Ehrenberg, K.R.; Hövelmeyer, N.; Hahn, M.; Schuchmann, M.; Jäger, D.; Waisman, A.; et al. Liver specific deletion of cyldexon7/8 induces severe biliary damage, fibrosis and increases hepatocarcinogenesis in mice. J. Hepatol. 2012, 57, 995–1003. [Google Scholar] [CrossRef]
- Kovalenko, A.; Chable-Bessia, C.; Cantarella, G.; Israel, A.; Wallach, D.; Courtois, G. The tumour suppressor cyld negatively regulates nf-kappa b signalling by deubiquitination. Nature 2003, 424, 801–805. [Google Scholar] [CrossRef]
- Trompouki, E.; Hatzivassiliou, E.; Tsichritzis, T.; Farmer, H.; Ashworth, A.; Mosialos, G. Cyld is a deubiquitinating enzyme that negatively regulates nf-kappa b activation by tnfr family members. Nature 2003, 424, 793–796. [Google Scholar] [CrossRef]
- Xiao-Jing, Z.; Huang, Z.; Yan, Z.; Wang, X.; Zhao, L.-P.; Wang, P.-X.; Zhang, X.-J.; Alves-Bezerra, M.; Cai, L.; Zhang, P.; et al. The deubiquitinating enzyme cylindromatosis mitigates nonalcoholic steatohepatitis. Nat. Med. 2018, 24, 213–223. [Google Scholar] [CrossRef]
- Verboom, L.; Martens, A.; Priem, D.; Hoste, E.; Sze, M.; Vikkula, H.; Van Hove, L.; Voet, S.; Roels, J.; Maelfait, J.; et al. OTULIN Prevents Liver Inflammation and Hepatocellular Carcinoma by Inhibiting FADD- and RIPK1 Kinase-Mediated Hepatocyte Apoptosis. Cell Rep. 2020, 30, 2237–2247.e2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damgaard, R.B.; Jolin, H.E.; Allison, M.E.D.; Davies, S.E.; Titheradge, H.L.; McKenzie, A.N.J.; Komander, D. OTULIN protects the liver against cell death, inflammation, fibrosis, and cancer. Cell Death Differ. 2020, 27, 1457–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Cui, K.; Prochownik, E.V.; Li, Y. The deubiquitinase USP21 stabilizes MEK2 to promote tumor growth. Cell Death Dis. 2018, 9, 482. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, J.; Jin, W.; Cao, J.; Fu, T.; Ma, D.; Zhang, Y. Lidocaine inhibits the proliferation and invasion of hepatocellular carcinoma by downregulating USP14 induced PI3K/Akt pathway. Pathol.—Res. Pract. 2020, 216, 152963. [Google Scholar] [CrossRef]
- Huang, G.; Li, L.M.; Zhou, W.P. USP14 activation promotes tumor progression in hepatocellular carcinoma. Oncol. Rep. 2015, 34, 2917–2924. [Google Scholar] [CrossRef]
- Yu, J.; Tao, Q.; Cheung, K.F.; Jin, H.; Poon, F.F.; Wang, X.; Li, H.; Cheng, Y.Y.; Röcken, C.; Ebert, M.P.A.; et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology 2008, 48, 508–518. [Google Scholar] [CrossRef]
- Li, M.; Brooks, C.L.; Kon, N.; Gu, W. A Dynamic Role of HAUSP in the p53-Mdm2 Pathway. Mol. Cell 2004, 13, 879–886. [Google Scholar] [CrossRef]
- Li, M.; Chen, D.; Shiloh, A.; Luo, J.; Nikolaev, A.Y.; Qin, J.; Gu, W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 2002, 416, 648–653. [Google Scholar] [CrossRef]
- Cummins, J.M.; Vogelstein, B. HAUSP is Required for p53 Destabilization. Cell Cycle 2004, 3, 689–692. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cao, M.; Dong, J.; Li, C.; Xu, W.; Zhan, Y.; Wang, X.; Yu, M.; Ge, C.; Ge, Z.; et al. ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat. Commun. 2014, 5, 5059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Fu, D.; Tang, W.; Cai, Y.; Ma, D.; Wang, H.; Xue, R.; Liu, T.; Huang, X.; Dong, L.; et al. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim. Biophys. Acta 2013, 1833, 559–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, C.; Liu, Y.; Mei, Y.; Zou, M.; Zhao, Z.; Ye, M.; Wu, X. Ubiquitin-specific protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-β signaling-induced epithelial-mesenchymal transition. Aging 2018, 10, 2783–2799. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, F.; Li, X.; Gong, Z.; Wang, L.-W. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2018, 499, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Giubellino, A.; Burke, T.R., Jr.; Bottaro, D.P. Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets 2008, 12, 1021–1033. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Xu, X.; Yang, Z.; Zhang, L.; Liu, Y.; Ma, A.; Xu, G.; Tang, M.; Jing, T.; Wu, L.; et al. POH1 contributes to hyperactivation of TGF-β signaling and facilitates hepatocellular carcinoma metastasis through deubiquitinating TGF-β receptors and caveolin-1. eBioMedicine 2019, 41, 320–332. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Qu, C.; Hong, X.; Jia, Y.; Lin, M.; Luo, Y.; Lin, F.; Xie, X.; Xie, X.; Huang, J.; et al. Trabid inhibits hepatocellular carcinoma growth and metastasis by cleaving RNF8-induced K63 ubiquitination of Twist1. Cell Death Differ. 2019, 26, 306–320. [Google Scholar] [CrossRef] [Green Version]
- Urbanik, T.; Köhler, B.C.; Boger, R.J.; Wörns, M.A.; Heeger, S.; Otto, G.; Hövelmeyer, N.; Galle, P.R.; Schuchmann, M.; Waisman, A.; et al. Down-regulation of cyld as a trigger for nf-κb activation and a mechanism of apoptotic resistance in hepatocellular carcinoma cells. Int. J. Oncol. 2011, 38, 121–131. [Google Scholar]
- Liu, H.; Chen, W.; Liang, C.; Chen, B.W.; Zhi, X.; Zhang, S.; Zheng, X.; Bai, X.; Liang, T. WP1130 increases doxorubicin sensitivity in hepatocellular carcinoma cells through usp9x-dependent p53 degradation. Cancer Lett. 2015, 361, 218–225. [Google Scholar] [CrossRef]
- Nagai, H.; Noguchi, T.; Homma, K.; Katagiri, K.; Takeda, K.; Matsuzawa, A.; Ichijo, H. Ubiquitin-like Sequence in ASK1 Plays Critical Roles in the Recognition and Stabilization by USP9X and Oxidative Stress-Induced Cell Death. Mol. Cell 2009, 36, 805–818. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, L.; Dou, Y.; Song, D.; Deng, H. Glycogen synthase kinase-3β antagonizes ROS-induced hepatocellular carcinoma cell death through suppression of the apoptosis signal-regulating kinase 1. Med Oncol. 2016, 33, 60. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Lumachi, F.; Luisetto, G.; Basso, S.M.; Basso, U.; Brunello, A.; Camozzi, V. Endocrine therapy of breast cancer. Curr. Med. Chem. 2011, 18, 513–522. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, H.; Yao, J. ERα, A Key Target for Cancer Therapy: A Review. OncoTargets Ther. 2020, 13, 2183–2191. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhong, X.; Wang, C.; Luo, H.; Lin, L.; Sun, H.; Sun, G.; Zeng, K.; Zou, R.; Liu, W.; et al. USP22 positively modulates ERα action via its deubiquitinase activity in breast cancer. Cell Death Differ. 2020, 27, 3131–3145. [Google Scholar] [CrossRef]
- Niu, Z.; Li, X.; Feng, S.; Huang, Q.; Zhuang, T.; Yan, C.; Qian, H.; Ding, Y.; Zhu, J.; Xu, W. The deubiquitinating enzyme USP1 modulates ERα and modulates breast cancer progression. J. Cancer 2020, 11, 6992–7000. [Google Scholar] [CrossRef]
- Xia, X.; Liao, Y.; Huang, C.; Liu, Y.; He, J.; Shao, Z.; Jiang, L.; Dou, Q.P.; Liu, J.; Huang, H. Deubiquitination and stabilization of estrogen receptor α by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019, 465, 118–128. [Google Scholar] [CrossRef]
- Dwane, L.; O’Connor, A.E.; Das, S.; Moran, B.; Mulrane, L.; Pinto-Fernandez, A.; Ward, E.; Blümel, A.M.; Cavanagh, B.L.; Mooney, B.; et al. A Functional Genomic Screen Identifies the Deubiquitinase USP11 as a Novel Transcriptional Regulator of ERα in Breast Cancer. Cancer Res. 2020, 80, 5076–5088. [Google Scholar] [CrossRef]
- Chen, X.-S.; Wang, K.-S.; Guo, W.; Li, L.-Y.; Yu, P.; Sun, X.-Y.; Wang, H.-Y.; Guan, Y.-D.; Tao, Y.-G.; Ding, B.-N.; et al. UCH-L1-mediated Down-regulation of Estrogen Receptor α Contributes to Insensitivity to Endocrine Therapy for Breast Cancer. Theranostics 2020, 10, 1833–1848. [Google Scholar] [CrossRef]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Tong, D.; Czerwenka, K.; Heinze, G.; Ryffel, M.; Schuster, E.; Witt, A.; Leodolter, S.; Zeillinger, R. Expression of KLF5 is a Prognostic Factor for Disease-Free Survival and Overall Survival in Patients with Breast Cancer. Clin. Cancer Res. 2006, 12, 2442–2448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, K.; Miki, Y.; Onodera, Y.; Nakamura, Y.; Ishida, T.; Watanabe, M.; Inoue, S.; Sasano, H.; Suzuki, T. Krüppel-like factor 5 in human breast carcinoma: A potent prognostic factor induced by androgens. Endocr.-Relat. Cancer 2012, 19, 741–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Zhou, Z.; Chen, W.; Wang, C.; Zhang, H.; Ge, G.; Shao, M.; You, D.; Fan, Z.; Xia, H.; et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat. Commun. 2015, 6, 8471. [Google Scholar] [CrossRef]
- Peters, A.A.; Buchanan, G.; Ricciardelli, C.; Bianco-Miotto, T.; Centenera, M.M.; Harris, J.M.; Jindal, S.; Segara, D.; Jia, L.; Moore, N.L.; et al. Androgen Receptor Inhibits Estrogen Receptor-α Activity and Is Prognostic in Breast Cancer. Cancer Res. 2009, 69, 6131–6140. [Google Scholar] [CrossRef] [Green Version]
- Cochrane, D.R.; Bernales, S.; Jacobsen, B.M.; Cittelly, D.M.; Howe, E.N.; D’Amato, N.C.; Spoelstra, N.S.; Edgerton, S.M.; Jean, A.; Guerrero, J.; et al. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res. 2014, 16, R7. [Google Scholar] [CrossRef] [Green Version]
- Doane, A.S.; Danso, M.; Lal, P.; Donaton, M.; Zhang, L.; Hudis, C.; Gerald, W.L. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 2006, 25, 3994–4008. [Google Scholar] [CrossRef] [Green Version]
- Ni, M.; Chen, Y.; Lim, E.; Wimberly, H.; Bailey, S.T.; Imai, Y.; Rimm, D.L.; Liu, X.S.; Brown, M. Targeting Androgen Receptor in Estrogen Receptor-Negative Breast Cancer. Cancer Cell 2011, 20, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Xia, X.; Liu, N.; Cai, J.; Guo, Z.; Li, Y.; Jiang, L.; Dou, Q.P.; Tang, D.; Huang, H.; et al. Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene 2018, 37, 1896–1910. [Google Scholar] [CrossRef]
- Luo, G.; Hu, N.; Xia, X.; Zhou, J.; Ye, C. RPN11 deubiquitinase promotes proliferation and migration of breast cancer cells. Mol. Med. Rep. 2017, 16, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Laoukili, J.; Kooistra, M.R.H.; Brás, A.; Kauw, J.; Kerkhoven, R.M.; Morrison, A.; Clevers, H.; Medema, R. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005, 7, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Hu, Y.; Chen, D.; Linghu, R.; Wang, Y.; Kou, X.; Yang, J.; Jiao, S. Ubiquitin specific protease 21 upregulation in breast cancer promotes cell tumorigenic capability and is associated with the NOD-like receptor signaling pathway. Oncol. Lett. 2016, 6, 4531–4537. [Google Scholar] [CrossRef] [PubMed]
- Arceci, A.; Bonacci, T.; Wang, X.; Stewart, K.; Damrauer, J.S.; Hoadley, K.; Emanuele, M.J. FOXM1 Deubiquitination by USP21 Regulates Cell Cycle Progression and Paclitaxel Sensitivity in Basal-like Breast Cancer. Cell Rep. 2019, 26, 3076–3086.e3076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; González-Prieto, R.; Zhang, M.; Geurink, P.P.; Kooij, R.; Iyengar, P.V.; van Dinther, M.; Bos, E.; Zhang, X.; Le Dévédec, S.E.; et al. Deubiquitinase activity profiling identifies uchl1 as a candidate oncoprotein that promotes tgfβ-induced breast cancer metastasis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 1460–1473. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Shen, M.; Jiang, Y.-Z.; Zhang, R.; Zheng, H.; Wei, Y.; Shao, Z.-M.; Kang, Y. Deubiquitinase USP20 promotes breast cancer metastasis by stabilizing SNAI2. Genes Dev. 2020, 34, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Ma, A.; Zhang, L.; Jin, W.-L.; Qian, Y.; Xu, G.; Qiu, B.; Yang, Z.; Liu, Y.; Xia, Q.; et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 2015, 6, 8704. [Google Scholar] [CrossRef]
- Song, Y.; Li, S.; Ray, A.; Das, D.S.; Qi, J.; Samur, M.K.; Tai, Y.-T.; Munshi, N.; Carrasco, R.D.; Chauhan, D.; et al. Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene 2017, 36, 5631–5638. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-H.; Lu, S.-X.; Liu, L.-L.; Li, Y.; Yang, X.; He, Y.-F.; Chen, S.-L.; Cai, S.-H.; Wang, H.; Yun, J.-P. POH1 Knockdown Induces Cancer Cell Apoptosis via p53 and Bim. Neoplasia 2018, 20, 411–424. [Google Scholar] [CrossRef]
- Yu, W.; Li, J.; Wang, Q.; Wang, B.; Zhang, L.; Liu, Y.; Tang, M.; Xu, G.; Yang, Z.; Wang, X.; et al. Targeting POH1 inhibits prostate cancer cell growth and enhances the suppressive efficacy of androgen deprivation and docetaxel. Prostate 2019, 79, 1304–1315. [Google Scholar] [CrossRef]
- Hayashi, M.; Jono, H.; Shinriki, S.; Nakamura, T.; Guo, J.; Sueta, A.; Tomiguchi, M.; Fujiwara, S.; Yamamoto-Ibusuki, M.; Murakami, K.-I.; et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res. Treat. 2014, 143, 447–457. [Google Scholar] [CrossRef]
- Yuan, L.; Lv, Y.; Li, H.; Gao, H.; Song, S.; Zhang, Y.; Xing, G.; Kong, X.; Wang, L.; Li, Y.; et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat. Cell Biol. 2015, 17, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, P.; Wei, Y.; Piao, H.-L.; Wang, W.; Maddika, S.; Wang, M.; Chen, D.; Sun, Y.; Hung, M.-C.; et al. Deubiquitylation and stabilization of PTEN by USP13. Nat. Cell Biol. 2013, 15, 1486–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Men’s Health 2019, 37, 288–295. [Google Scholar] [CrossRef] [PubMed]
- McClurg, U.L.; Harle, V.J.; Nabbi, A.; Batalha-Pereira, A.; Walker, S.; Coffey, K.; Gaughan, L.; McCracken, S.R.; Robson, C.N. Ubiquitin-specific protease 12 interacting partners Uaf-1 and WDR20 are potential therapeutic targets in prostate cancer. Oncotarget 2015, 6, 37724–37736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Liu, N.; Hua, X.; Cai, J.; Xia, X.; Wang, X.; Huang, H.; Liu, J. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis. 2017, 8, e2585. [Google Scholar] [CrossRef]
- Dirac, A.M.; Bernards, R. The Deubiquitinating Enzyme USP26 Is a Regulator of Androgen Receptor Signaling. Mol. Cancer Res. 2010, 8, 844–854. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-T.; Okada, M.; Nakato, R.; Izumi, K.; Bando, M.; Shirahige, K. The Deubiquitinating Enzyme USP7 Regulates Androgen Receptor Activity by Modulating Its Binding to Chromatin. J. Biol. Chem. 2015, 290, 21713–21723. [Google Scholar] [CrossRef] [Green Version]
- Schrecengost, R.S.; Dean, J.L.; Goodwin, J.F.; Schiewer, M.J.; Urban, M.W.; Stanek, T.J.; Sussman, R.T.; Hicks, J.L.; Birbe, R.C.; Draganova-Tacheva, R.A.; et al. USP22 Regulates Oncogenic Signaling Pathways to Drive Lethal Cancer Progression. Cancer Res. 2014, 74, 272–286. [Google Scholar] [CrossRef] [Green Version]
- Faus, H.; Meyer, H.-A.; Huber, M.; Bahr, I.; Haendler, B. The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol. Cell. Endocrinol. 2005, 245, 138–146. [Google Scholar] [CrossRef]
- Ge, J.; Yu, W.; Li, J.; Ma, H.; Wang, P.; Zhou, Y.; Wang, Y.; Zhang, J.; Shi, G. USP16 regulates castration-resistant prostate cancer cell proliferation by deubiquitinating and stablizing c-Myc. J. Exp. Clin. Cancer Res. 2021, 40, 59. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Bedard, N.; Chevalier, S.; Wing, S.S. Identification of Distinctive Patterns of USP19-Mediated Growth Regulation in Normal and Malignant Cells. PLoS ONE 2011, 6, e15936. [Google Scholar] [CrossRef] [PubMed]
- Ummanni, R.; Jost, E.; Braig, M.; Lohmann, F.; Mundt, F.; Barett, C.; Schlomm, T.; Sauter, G.; Senff, T.; Bokemeyer, C.; et al. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol. Cancer 2011, 10, 129. [Google Scholar] [CrossRef] [Green Version]
- Iglesias-Gato, D.; Chuan, Y.-C.; Jiang, N.; Svensson, C.; Bao, J.; Paul, I.; Egevad, L.; Kessler, B.M.; Wikström, P.; Niu, Y.; et al. OTUB1 de-ubiquitinating enzyme promotes prostate cancer cell invasion in vitro and tumorigenesis in vivo. Mol. Cancer 2015, 14, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Kim, Y.M.; Lim, S.; Nam, Y.K.; Jeong, J.; Lee, K.-J. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene 2009, 28, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Jang, M.J.; Baek, S.H.; Kim, J.H. UCH-L1 promotes cancer metastasis in prostate cancer cells through EMT induction. Cancer Lett. 2011, 302, 128–135. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Luan, T.; Zuo, Y.; Chen, J.; Zhang, H.; Ye, Z.; Wang, H.; Hai, B. Deubiquitinase USP9X regulates the invasion of prostate cancer cells by regulating the ERK pathway and mitochondrial dynamics. Oncol. Rep. 2019, 41, 3292–3304. [Google Scholar] [CrossRef]
- Song, H.M.; Lee, J.E.; Kim, J.H. Ubiquitin C-terminal hydrolase-L3 regulates EMT process and cancer metastasis in prostate cell lines. Biochem. Biophys. Res. Commun. 2014, 452, 722–727. [Google Scholar] [CrossRef]
- Rossi, S.; Graner, E.; Febbo, P.; Weinstein, L.; Bhattacharya, N.; Onody, T.; Bubley, G.; Balk, S.; Loda, M. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol. Cancer Res. 2003, 1, 707–715. [Google Scholar]
- Pflug, B.R.; Pecher, S.M.; Brink, A.W.; Nelson, J.B.; Foster, B.A. Increased fatty acid synthase expression and activity during progression of prostate cancer in the TRAMP model. Prostate 2003, 57, 245–254. [Google Scholar] [CrossRef]
- Priolo, C.; Tang, D.; Brahamandan, M.; Benassi, B.; Sicinska, E.; Ogino, S.; Farsetti, A.; Porrello, A.; Finn, S.; Zimmermann, J.; et al. The Isopeptidase USP2a Protects Human Prostate Cancer from Apoptosis. Cancer Res. 2006, 66, 8625–8632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCann, J.J.; Vasilevskaya, I.A.; Neupane, N.P.; Shafi, A.A.; McNair, C.; Dylgjeri, E.; Mandigo, A.C.; Schiewer, M.J.; Schrecengost, R.S.; Gallagher, P.; et al. USP22 Functions as an Oncogenic Driver in Prostate Cancer by Regulating Cell Proliferation and DNA Repair. Cancer Res. 2020, 80, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κb, and up-regulating expression of microrna-21. Gastroenterology 2017, 152, 851–866.e824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abed, J.; Emgård, J.E.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galnac. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Sun, X.; Shi, X.; Wang, H.; Wu, G.; Jiang, C.; Yu, D.; Zhang, W.; Xue, B.; Ding, Y. USP39 promotes colorectal cancer growth and metastasis through the Wnt/β-catenin pathway. Oncol. Rep. 2017, 37, 2398–2404. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Kong, L.; Yang, Q.; Duan, A.; Ju, X.; Cai, B.; Chen, L.; An, T.; Li, Y. Parthenolide inhibits ubiquitin-specific peptidase 7 (USP7), Wnt signaling, and colorectal cancer cell growth. J. Biol. Chem. 2020, 295, 3576–3589. [Google Scholar] [CrossRef]
- Sun, K.; He, S.B.; Yao, Y.Z.; Qu, J.G.; Xie, R.; Ma, Y.Q.; Zong, M.H.; Chen, J.X. Tre2 (usp6nl) promotes colorectal cancer cell proliferation via wnt/beta-catenin pathway. Cancer Cell Int. 2019, 19, 102. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, Q.; Ren, W.; Yan, B.; Yi, L.; Tang, T.; Lin, H.; Zhang, Y. USP44 suppresses proliferation and enhances apoptosis in colorectal cancer cells by inactivating the Wnt/β-catenin pathway via Axin1 deubiquitination. Cell Biol. Int. 2020, 44, 1651–1659. [Google Scholar] [CrossRef] [Green Version]
- Sloane, M.A.; Wong, J.W.; Perera, D.; Nunez, A.C.; Pimanda, J.E.; Hawkins, N.J.; Sieber, O.M.; Bourke, M.J.; Hesson, L.B.; Ward, R.L. Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia. Epigenetics 2014, 9, 1092–1100. [Google Scholar] [CrossRef] [Green Version]
- Giebel, N.; de Jaime-Soguero, A.; García Del Arco, A.; Landry, J.J.M.; Tietje, M.; Villacorta, L.; Benes, V.; Fernández-Sáiz, V.; Acebrón, S.P. Usp42 protects znrf3/rnf43 from r-spondin-dependent clearance and inhibits wnt signalling. EMBO Rep. 2021, 22, e51415. [Google Scholar] [CrossRef]
- Miao, D.; Wang, Y.; Jia, Y.; Tong, J.; Jiang, S.; Liu, L. ZRANB1 enhances stem-cell-like features and accelerates tumor progression by regulating Sox9-mediated USP22/Wnt/β-catenin pathway in colorectal cancer. Cell. Signal. 2021, 90, 110200. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liu, J.; Peng, Y.; Zhang, J.; Dai, X.; Zhang, S.; Wang, Y.; Liu, J.; Long, J. Deubiquitinase OTUD6A promotes proliferation of cancer cells via regulating Drp1 stability and mitochondrial fission. Mol. Oncol. 2020, 14, 3169–3183. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.-X.; Wang, S.-S.; Huang, Y.; Wang, X.-J.; Chi, P. USP43 directly regulates ZEB1 protein, mediating proliferation and metastasis of colorectal cancer. J. Cancer 2021, 12, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Huang, A.; Cui, X.; Han, K.; Hou, X.; Wang, Q.; Cui, L.; Yang, Y. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics 2019, 9, 4208–4220. [Google Scholar] [CrossRef]
- Zhao, J.; Tuo, Y.; Luo, W.; He, S.; Chen, Y. Prognostic and Clinicopathological Significance of SATB1 in Colorectal Cancer: A Meta-Analysis. Front. Physiol. 2018, 9, 535. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Dong, L.; Wang, Y.; Liu, L.; Long, H.; Li, H.; Li, J.; Yang, X.; Liu, Z.; Duan, G.; et al. Reversible regulation of SATB1 ubiquitination by USP47 and SMURF2 mediates colon cancer cell proliferation and tumor progression. Cancer Lett. 2019, 448, 40–51. [Google Scholar] [CrossRef]
- Gennaro, V.J.; Stanek, T.J.; Peck, A.R.; Sun, Y.; Wang, F.; Qie, S.; Knudsen, K.E.; Rui, H.; Butt, T.; Diehl, J.A.; et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, E9298–E9307. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Li, S.; Cui, X.; Han, K.; Wang, J.; Hou, X.; Cui, L.; He, S.; Xiao, J.; Yang, Y. Inhibition of Ubiquitin Specific Protease 1 Sensitizes Colorectal Cancer Cells to DNA-Damaging Chemotherapeutics. Front. Oncol. 2019, 9, 1406. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Gu, L.; Lin, X.; Zhou, X.; Lu, B.; Liu, C.; Lei, C.; Zhou, F.; Zhao, Q.; Prochownik, E.V.; et al. USP19 exacerbates lipogenesis and colorectal carcinogenesis by stabilizing ME1. Cell Rep. 2021, 37, 110174. [Google Scholar] [CrossRef]
- Sheng, Y.; Saridakis, V.; Sarkari, F.; Duan, S.; Wu, T.; Arrowsmith, C.H.; Frappier, L. Molecular recognition of p53 and mdm2 by usp7/hausp. Nat. Struct. Mol. Biol. 2006, 13, 285–291. [Google Scholar] [CrossRef]
- Dykstra, M.A.; Gimon, T.I.; Ronksley, P.E.; Buie, W.D.; MacLean, A.R. Classic and Novel Histopathologic Risk Factors for Lymph Node Metastasis in T1 Colorectal Cancer: A Systematic Review and Meta-analysis. Dis. Colon Rectum 2021, 64, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, J.; Fu, X.; Du, W.; Zhou, L.; Meng, X.; Yu, H.; Lin, J.; Ye, W.; Liu, J.; et al. OTUB1 promotes metastasis and serves as a marker of poor prognosis in colorectal cancer. Mol. Cancer 2014, 13, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zheng, Y.; Li, X.; Dong, X.; Chen, W.; Guan, Z.; Zhang, C. UCHL3 promotes proliferation of colorectal cancer cells by regulating SOX12 via AKT/mTOR signaling pathway. Am. J. Transl. Res. 2020, 12, 6445–6454. [Google Scholar] [PubMed]
- Li, B.; Qi, Z.-P.; He, D.-L.; Chen, Z.-H.; Liu, J.-Y.; Wong, M.-W.; Zhang, J.-W.; Xu, E.-P.; Shi, Q.; Cai, S.-L.; et al. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J. Exp. Clin. Cancer Res. 2021, 40, 126. [Google Scholar] [CrossRef]
- Sun, H.; Ou, B.; Zhao, S.; Liu, X.; Song, L.; Liu, X.; Wang, R.; Peng, Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. eBioMedicine 2019, 48, 236–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.-I.; Hong, H.K.; Yeo, S.-Y.; Kim, S.-H.; Cho, Y.B.; Kim, K.K. Ubiquitin-Specific Protease 21 Promotes Colorectal Cancer Metastasis by Acting as a Fra-1 Deubiquitinase. Cancers 2020, 12, 207. [Google Scholar] [CrossRef] [Green Version]
- Seo, D.; Jung, S.M.; Park, J.S.; Lee, J.; Ha, J.; Kim, M.; Park, S.H. The deubiquitinating enzyme PSMD14 facilitates tumor growth and chemoresistance through stabilizing the ALK2 receptor in the initiation of BMP6 signaling pathway. eBioMedicine 2019, 49, 55–71. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Liu, T.; Yin, Y.; Zhao, W.; Lin, Z.; Yin, Y.; Lu, D.; You, F. The deubiquitinase OTUD1 enhances iron transport and potentiates host antitumor immunity. EMBO Rep. 2021, 22, e51162. [Google Scholar] [CrossRef]
- Kosinsky, R.L.; Zerche, M.; Saul, D.; Wang, X.; Wohn, L.; Wegwitz, F.; Begus-Nahrmann, Y.; Johnsen, S.A. USP22 exerts tumor-suppressive functions in colorectal cancer by decreasing mTOR activity. Cell Death Differ. 2020, 27, 1328–1340. [Google Scholar] [CrossRef]
- Hou, X.; Xia, J.; Feng, Y.; Cui, L.; Yang, Y.; Yang, P.; Xu, X. USP47-Mediated Deubiquitination and Stabilization of TCEA3 Attenuates Pyroptosis and Apoptosis of Colorectal Cancer Cells Induced by Chemotherapeutic Doxorubicin. Front. Pharmacol. 2021, 12, 713322. [Google Scholar] [CrossRef]
- Hezel, A.F.; Kimmelman, A.C.; Stanger, B.Z.; Bardeesy, N.; DePinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006, 20, 1218–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietri, E.; Balsano, R.; Coriano, M.; Gelsomino, F.; Leonardi, F.; Bui, S.; Gnetti, L.; Valle, R.D.; Garajová, I. The implication of liquid biopsies to predict chemoresistance in pancreatic cancer. Cancer Drug Resist. 2021, 4, 559–572. [Google Scholar] [CrossRef]
- Pan, Y.; Tang, H.; Li, Q.; Chen, G.; Li, D. Exosomes and their roles in the chemoresistance of pancreatic cancer. Cancer Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Li, Y.; Yin, Y.; Li, L.; Wu, C.; Chen, Y.; Nowsheen, S.; Hu, Q.; Zhang, L.; Lou, Z.; et al. USP49 negatively regulates tumorigenesis and chemoresistance through FKBP51-AKT signaling. EMBO J. 2017, 36, 1434–1446. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Yu, K.; Chen, K.; Zhu, X.; Yang, Z.; Wang, Q.; Gao, J.; Wang, Y.; Cao, T.; Xu, H.; et al. Fbxo45 facilitates pancreatic carcinoma progression by targeting USP49 for ubiquitination and degradation. Cell Death Dis. 2022, 13, 231. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Huo, Y.; Yang, J.; Fu, X.; Yang, M.; Tao, L.; Liu, D.; Zhang, J.; Hua, R.; Sun, Y. Decreased expression of USP9X is associated with poor prognosis in Chinese pancreatic ductal adenocarcinoma patients. Oncol. Lett. 2018, 15, 9287–9292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Mancera, P.A.; Initiative, A.P.C.G.; Rust, A.; Van Der Weyden, L.; Kristiansen, G.; Li, A.; Sarver, A.L.; Silverstein, K.A.T.; Grützmann, R.; Aust, D.; et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 2012, 486, 266–270. [Google Scholar] [CrossRef]
- Liu, L.; Yao, D.; Zhang, P.; Ding, W.; Zhang, X.; Zhang, C.; Gong, S.; Zhang, Y.; Wang, J.; Sun, T.; et al. Deubiquitinase USP9X promotes cell migration, invasion and inhibits apoptosis of human pancreatic cancer. Oncol. Rep. 2017, 38, 3531–3537. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.L.; Wilder, P.J.; Wuebben, E.L.; Ouellette, M.M.; Hollingsworth, M.A.; Rizzino, A. Context-dependent function of the deubiquitinating enzyme USP9X in pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2014, 15, 1042–1052. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.; Dziubinski, M.; Di Magliano, M.P.; Simeone, D.M.; Owens, S.; Thomas, D.; Peterson, L.; Potu, H.; Talpaz, M.; Donato, N.J. Usp9x Promotes Survival in Human Pancreatic Cancer and Its Inhibition Suppresses Pancreatic Ductal Adenocarcinoma In Vivo Tumor Growth. Neoplasia 2018, 20, 152–164. [Google Scholar] [CrossRef]
- Hou, P.; Ma, X.; Zhang, Q.; Wu, C.-J.; Liao, W.; Li, J.; Wang, H.; Zhao, J.; Zhou, X.; Guan, C.; et al. USP21 deubiquitinase promotes pancreas cancer cell stemness via Wnt pathway activation. Genes Dev. 2019, 33, 1361–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, H.; Zhu, W.; Zhou, B.; Ying, J.; Wu, J.; Zhang, H.; Sun, H.; Gao, S. Deubiquitinase inhibitor degrasyn suppresses metastasis by targeting USP5-WT1-E-cadherin signalling pathway in pancreatic ductal adenocarcinoma. J. Cell. Mol. Med. 2020, 24, 1370–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Xu, Z.; Li, Q.; Feng, Q.; Zheng, C.; Du, Y.; Yuan, R.; Peng, X. USP28 facilitates pancreatic cancer progression through activation of Wnt/β-catenin pathway via stabilising FOXM1. Cell Death Dis. 2021, 12, 887. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-Y.; Zhai, L.; Yang, C.; Nie, S.; Erdjument-Bromage, H.; Tempst, P.; Chang, C.; Wang, H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 2007, 449, 1068–1072. [Google Scholar] [CrossRef]
- Zhuo, X.; Guo, X.; Zhang, X.; Jing, G.; Wang, Y.; Chen, Q.; Jiang, Q.; Liu, J.; Zhang, C. Usp16 regulates kinetochore localization of Plk1 to promote proper chromosome alignment in mitosis. J. Cell Biol. 2015, 210, 727–735. [Google Scholar] [CrossRef]
- Ren, D.; Sun, Y.; Li, D.; Wu, H.; Jin, X. USP22-mediated deubiquitination of PTEN inhibits pancreatic cancer progression by inducing p21 expression. Mol. Oncol. 2022, 16, 1200–1217. [Google Scholar] [CrossRef]
- Feng, L.; Wang, K.; Tang, P.; Chen, S.; Liu, T.; Lei, J.; Yuan, R.; Hu, Z.; Li, W.; Yu, X. Deubiquitinase USP18 promotes the progression of pancreatic cancer via enhancing the Notch1-c-Myc axis. Aging 2020, 12, 19273–19292. [Google Scholar] [CrossRef]
- Ma, Y.; Xia, P.; Wang, Z.; Xu, J.; Zhang, L.; Jiang, Y. PDIA6 promotes pancreatic cancer progression and immune escape through CSN5-mediated deubiquitination of β-catenin and PD-L1. Neoplasia 2021, 23, 912–928. [Google Scholar] [CrossRef]
- Fan, Y.; Hu, D.; Li, D.; Ma, C.; Tang, Y.; Tao, Q.; Deng, L.; Tang, D. Uchl3 promotes aerobic glycolysis of pancreatic cancer through upregulating ldha expression. Clin. Transl. Oncol. 2021, 23, 1637–1645. [Google Scholar] [CrossRef]
- Grattarola, M.; Cucci, M.A.; Roetto, A.; Dianzani, C.; Barrera, G.; Pizzimenti, S. Post-translational down-regulation of Nrf2 and YAP proteins, by targeting deubiquitinases, reduces growth and chemoresistance in pancreatic cancer cells. Free Radic. Biol. Med. 2021, 174, 202–210. [Google Scholar] [CrossRef]
- Chen, H.; Zhu, X.; Sun, R.; Ma, P.; Zhang, E.; Wang, Z.; Fan, Y.; Zhou, G.; Mao, R. Ubiquitin-specific protease 7 is a druggable target that is essential for pancreatic cancer growth and chemoresistance. Investig. New Drugs 2020, 38, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, T.; Li, H.; Fan, Y.; Yuan, L.; Guo, X.; Zhu, Q.; Yao, Y.; Li, X.; Liu, C.; Yu, X.; et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat. Commun. 2019, 10, 2914. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Deng, T.; Ma, H.; Liu, Y.; Feng, P.; Wei, D.; Ling, N.; Li, L.; Qiu, S.; Zhang, L.; et al. Deubiquitinase DUB3 Regulates Cell Cycle Progression via Stabilizing Cyclin A for Proliferation of Non-Small Cell Lung Cancer Cells. Cells 2019, 8, 297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.H.; Yeh, D.W.; Lai, C.Y.; Liu, Y.L.; Huang, L.R.; Lee, A.Y.; Jin, S.C.; Chuang, T.H. Usp17 mediates macrophage-promoted inflammation and stemness in lung cancer cells by regulating traf2/traf3 complex formation. Oncogene 2018, 37, 6327–6340. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Zhang, L.; Yang, Z.; Chen, X.; Luo, J.; Zhou, Z.; Mei, X.; Yu, X.; Shao, Z.; et al. Targeting deubiquitinase USP28 for cancer therapy. Cell Death Dis. 2018, 9, 186. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Li, C.; Li, H.; Yuan, L.; Dai, H.; Peng, Z.; Deng, Z.; Chang, Z.; Cui, C.-P.; Zhang, L. Ubiquitin ligase CHIP regulates OTUD3 stability and suppresses tumour metastasis in lung cancer. Cell Death Differ. 2020, 27, 3177–3195. [Google Scholar] [CrossRef]
- Mustachio, L.M.; Lu, Y.; Tafe, L.J.; Memoli, V.; Rodriguez-Canales, J.; Mino, B.; Villalobos, P.A.; Wistuba, I.; Katayama, H.; Hanash, S.M.; et al. Deubiquitinase USP18 Loss Mislocalizes and Destabilizes KRAS in Lung Cancer. Mol. Cancer Res. 2017, 15, 905–914. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Xiao, H.; Yang, Q.; Hu, R.; Jiang, L.; Bi, R.; Jiang, X.; Wang, L.; Mei, J.; Ding, F.; et al. The usp21/yy1/snhg16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp. Mol. Med. 2020, 52, 41–55. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Jiang, S.; Mao, C.; Zheng, H.; Cao, B.; Zhang, Z.; Zhao, J.; Zeng, Y.; Mao, X. The deubiquitinase USP10 restores PTEN activity and inhibits non-small cell lung cancer cell proliferation. J. Biol. Chem. 2021, 297, 101088. [Google Scholar] [CrossRef]
- Sun, J.; Li, T.; Zhao, Y.; Huang, L.; Sun, H.; Wu, H.; Jiang, X. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol. Cell. Biochem. 2018, 441, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Wu, Y.S.; Hung, C.Y.; Wang, S.A.; Young, M.J.; Hsu, T.I.; Hung, J.J. Usp24 induces il-6 in tumor-associated microenvironment by stabilizing p300 and β-trcp and promotes cancer malignancy. Nat. Commun. 2018, 9, 3996. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-A.; Young, M.-J.; Jeng, W.-Y.; Liu, C.-Y.; Hung, J.-J. USP24 stabilizes bromodomain containing proteins to promote lung cancer malignancy. Sci. Rep. 2020, 10, 20870. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xia, S.; Li, H.; Wang, X.; Li, C.; Chao, Y.; Zhang, L.; Han, C. The deubiquitinase USP10 regulates KLF4 stability and suppresses lung tumorigenesis. Cell Death Differ. 2020, 27, 1747–1764. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, D.; Zhu, M.; Yu, H.; Pan, Z.; Liu, L.; Geng, Q.; Pan, H.; Yan, M.; Yao, M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics 2019, 9, 179–195. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Yeh, D.-W.; Lu, C.-H.; Liu, Y.-L.; Chuang, Y.-C.; Ruan, J.-W.; Kao, C.-Y.; Huang, L.-R.; Chuang, T.-H. Epigenetic Silencing of Ubiquitin Specific Protease 4 by Snail1 Contributes to Macrophage-Dependent Inflammation and Therapeutic Resistance in Lung Cancer. Cancers 2020, 12, 148. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Hu, Q.; He, T.; Xu, J.; Yi, Y.; Xie, S.; Ding, L.; Fu, M.; Guo, R.; Xiao, Z.-X.J.; et al. The Deubiquitinase USP4 Stabilizes Twist1 Protein to Promote Lung Cancer Cell Stemness. Cancers 2020, 12, 1582. [Google Scholar] [CrossRef]
- Pan, J.; Deng, Q.; Jiang, C.; Wang, X.; Niu, T.; Li, H.; Chen, T.; Jin, J.; Pan, W.; Cai, X.; et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene 2015, 34, 3957–3967. [Google Scholar] [CrossRef]
- García-Santisteban, I.; Peters, G.J.; Giovannetti, E.; Rodríguez, J.A. USP1 deubiquitinase: Cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol. Cancer 2013, 12, 91. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wang, M.; Xiao, Y.; Wang, Y.; Ma, L.; Guo, L.; Wu, X.; Lin, X.; Zhang, P. USP35 mitigates endoplasmic reticulum stress-induced apoptosis by stabilizing RRBP1 in non-small cell lung cancer. Mol. Oncol. 2022, 16, 1572–1590. [Google Scholar] [CrossRef]
- Tang, Z.; Jiang, W.; Mao, M.; Zhao, J.; Chen, J.; Cheng, N. Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. Clin. Transl. Med. 2021, 11, e390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, B.; Qiang, Y.; Huang, H.; Wang, C.; Li, D.; Qian, J. Overexpression of deubiquitinating enzyme USP 28 promoted non-small cell lung cancer growth. J. Cell. Mol. Med. 2015, 19, 799–805. [Google Scholar] [CrossRef]
- Dai, X.; Lu, L.; Deng, S.; Meng, J.; Wan, C.; Huang, J.; Sun, Y.; Hu, Y.; Wu, B.; Wu, G.; et al. USP7 targeting modulates anti-tumor immune response by reprogramming Tumor-associated Macrophages in Lung Cancer. Theranostics 2020, 10, 9332–9347. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhang, M.; Moses, N.; Hu, C.-L.; Polin, L.; Chen, W.; Jang, H.; Heyza, J.; Malysa, A.; Caruso, J.A.; et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020, 11, 328. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.; Han, S.Y.; Choi, C.H.; Cho, H.; Lee, M.-S.; Kim, S.-Y.; Song, J.S.; Hong, K.-M.; Lee, H.-W.; Hewitt, S.M.; et al. Oncogene-induced senescence mediated by c-Myc requires USP10 dependent deubiquitination and stabilization of p14ARF. Cell Death Differ. 2018, 25, 1050–1062. [Google Scholar] [CrossRef] [Green Version]
| Name | Targets | Functions |
|---|---|---|
| BAP1 | KFL5 | P |
| COPS5 | β-catenin | M |
| CYLD | NEMO; TAK1; p53 | P;M;A |
| OTUB1 | SNAIL; p53 | M |
| OTUB2 | U2AF2 | M |
| OTUD1 | IREB2; p53 | M;A |
| OTUD3 | GRP78; PTEN | P;M |
| OTUD6A | DRP1 | P |
| PSMD14 | GRB2; TGFR; CAV1; CCND1 | P;M;A |
| TRABID | Twist | M |
| UCHL1 | EGFR; SMAD2; TGFR; p53 | P;M;A |
| UCHL3 | FOXM1 | M |
| UCHL5 | PRP19 | M |
| USP1 | ERα | P;A |
| USP10 | KLF4; HDAC6; MSI2; NLPR7; PTEN; p53 | P;M;A |
| USP11 | Erα; CCND1; PPP1CA | P;M |
| USP12 | AR | P |
| USP13 | PTEN | M |
| USP14 | AR | P;A |
| USP16 | c-Myc; H2A | P |
| USP17 | cyclin A; cMyc; p21 | P |
| USP18 | KRAS | P;M |
| USP19 | p27; RPF1 | P |
| USP20 | SNAI2 | M |
| USP21 | MEK2; FOXM1; Fra-1; TCSF7; YY1 | P;M |
| USP22 | AR; CCND1; ERα; c-Myc | P;A |
| USP24 | p300; BRDs | M |
| USP26 | AR | P |
| USP28 | FOXM1; p53; p21 | P;A |
| USP29 | p53 | P |
| USP2a | MDM2; p53 | A |
| USP35 | RRBP1 | A |
| USP37 | SNAIL; c-Myc | M |
| USP4 | PI3K; Twist1; p53; β-catenin | M |
| USP43 | ZEB1 | P |
| USP44 | Axin1 | A |
| USP47 | SATB1; TCEA3 | P;A |
| USP49 | FKBP51 | P |
| USP5 | EF-Tu; WT1; p53 | P;M |
| USP7 | AR; ERα; MDM2; p53; β-catenin | P;A |
| USP9X | ASK1; PTGES; p53; β-catenin; survivin | P;M;A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
LIU, J.; LEUNG, C.T.; LIANG, L.; WANG, Y.; CHEN, J.; LAI, K.P.; TSE, W.K.F. Deubiquitinases in Cancers: Aspects of Proliferation, Metastasis, and Apoptosis. Cancers 2022, 14, 3547. https://doi.org/10.3390/cancers14143547
LIU J, LEUNG CT, LIANG L, WANG Y, CHEN J, LAI KP, TSE WKF. Deubiquitinases in Cancers: Aspects of Proliferation, Metastasis, and Apoptosis. Cancers. 2022; 14(14):3547. https://doi.org/10.3390/cancers14143547
Chicago/Turabian StyleLIU, Jiaqi, Chi Tim LEUNG, Luyun LIANG, Yuqin WANG, Jian CHEN, Keng Po LAI, and William Ka Fai TSE. 2022. "Deubiquitinases in Cancers: Aspects of Proliferation, Metastasis, and Apoptosis" Cancers 14, no. 14: 3547. https://doi.org/10.3390/cancers14143547






