Impact of Denoising on Deep-Learning-Based Automatic Segmentation Framework for Breast Cancer Radiotherapy Planning
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Delineation
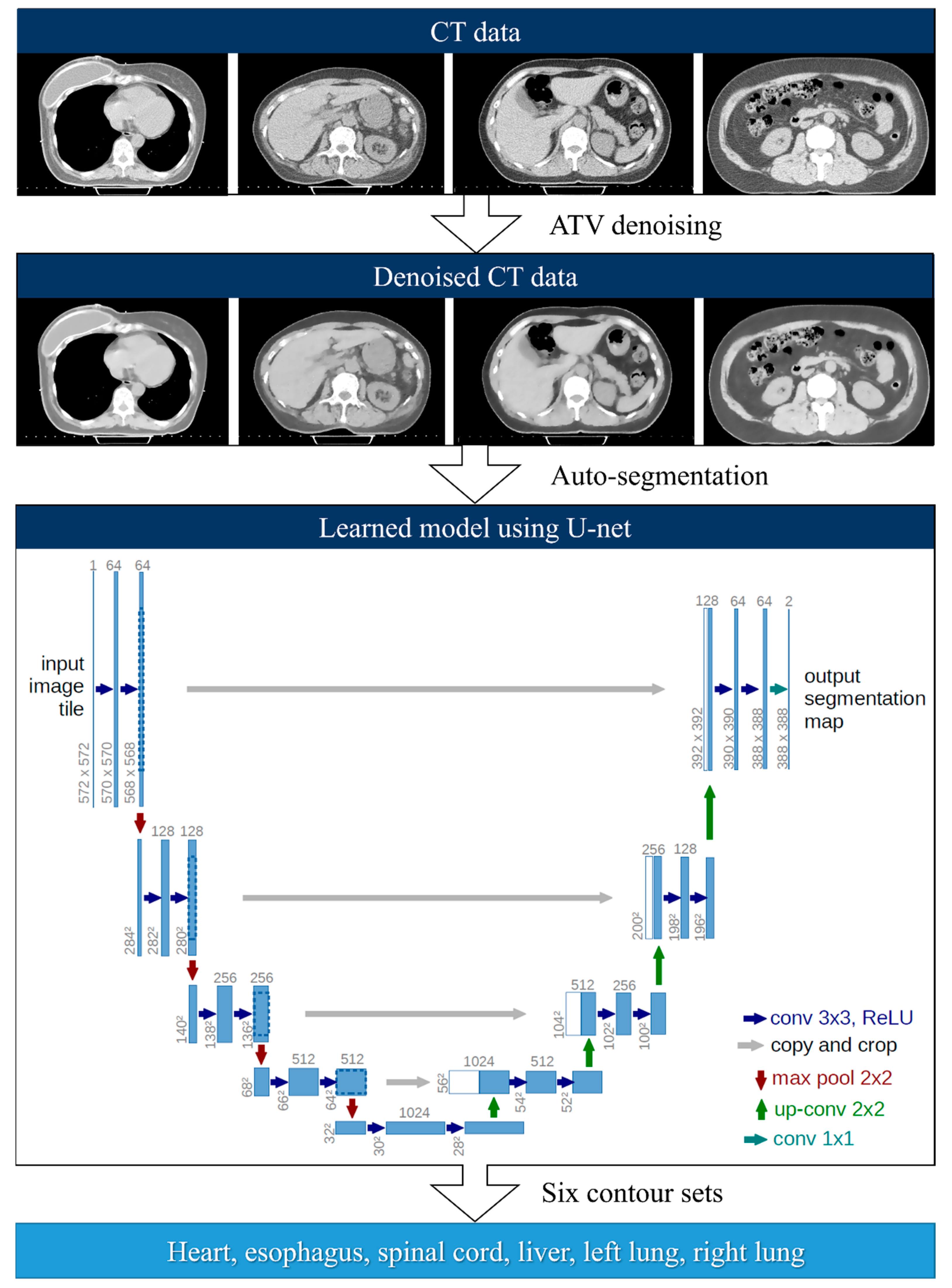
2.2. Deep-Learning-Based Auto-Segmentation
2.3. Anisotropic Total Variation Denoiser-Based Auto-Segmentation
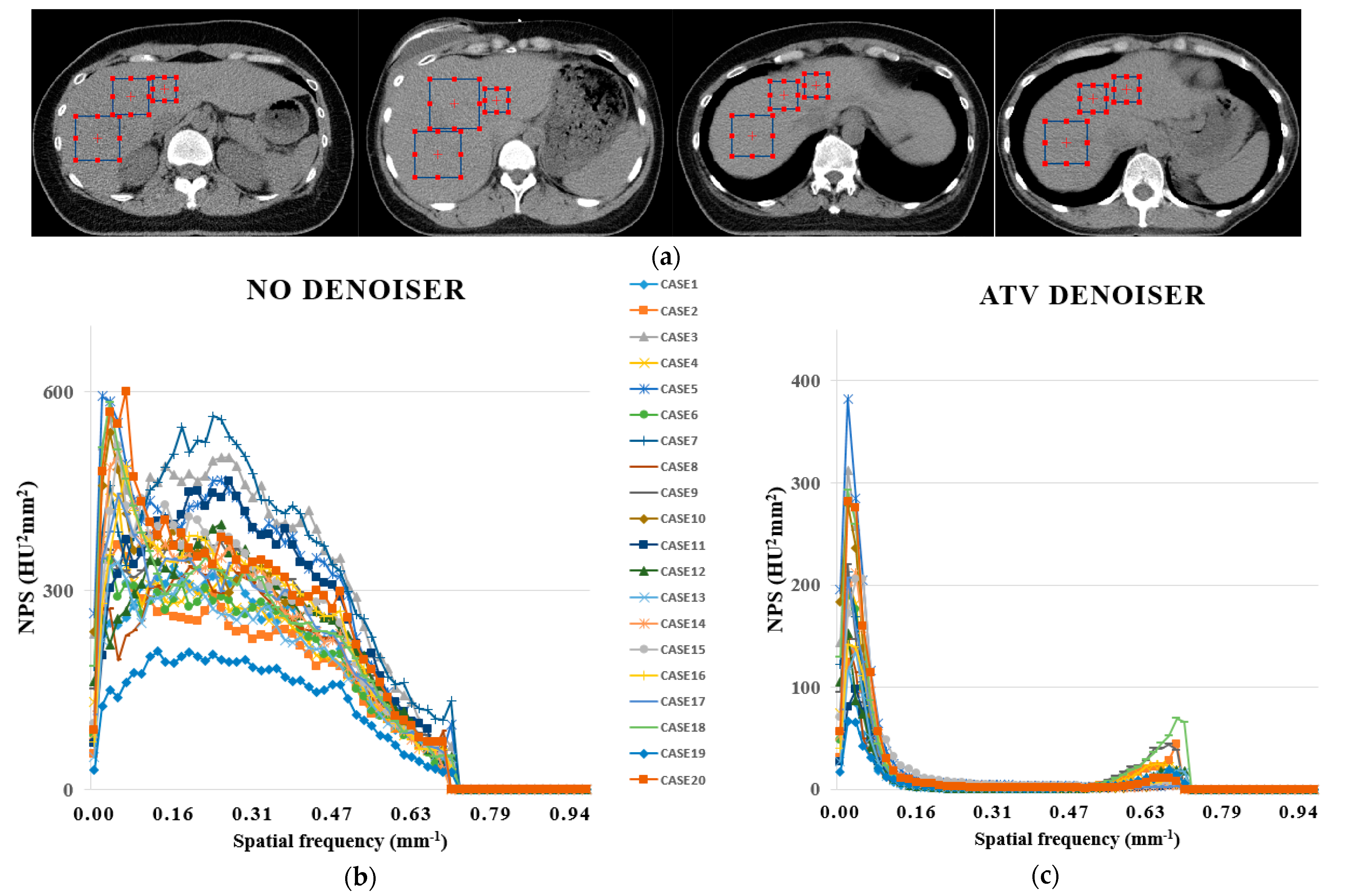
2.4. Quantitative Analysis
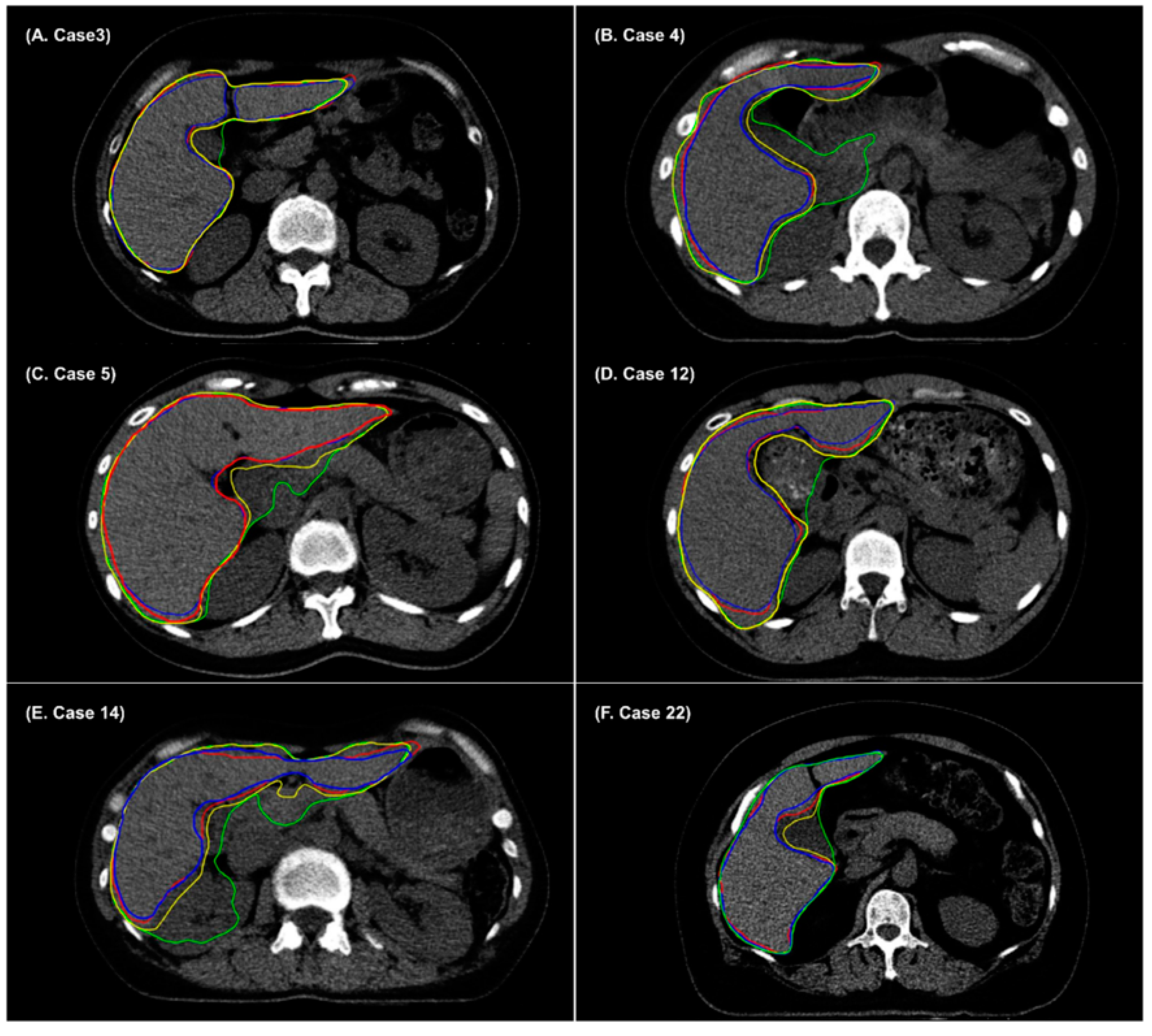
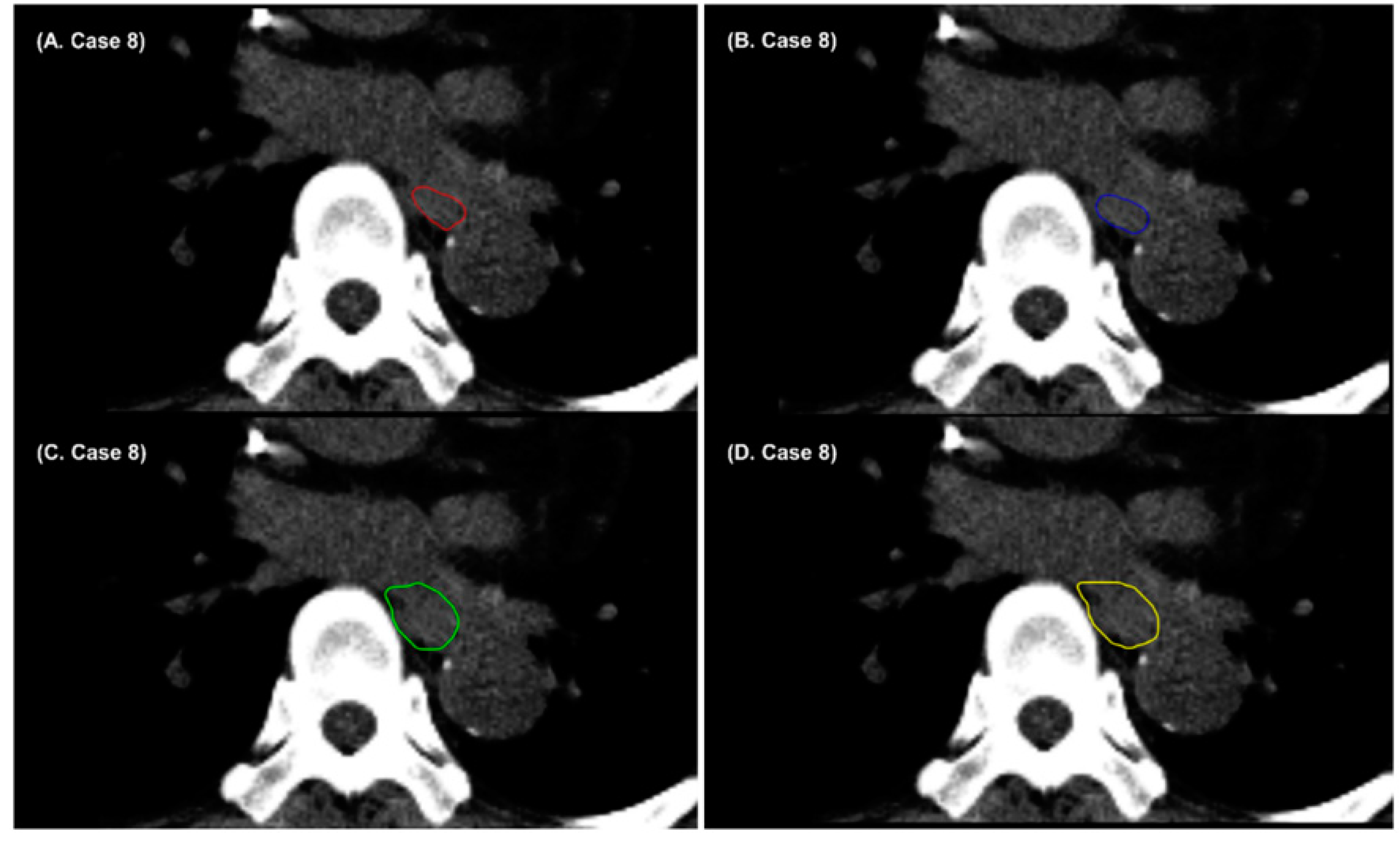
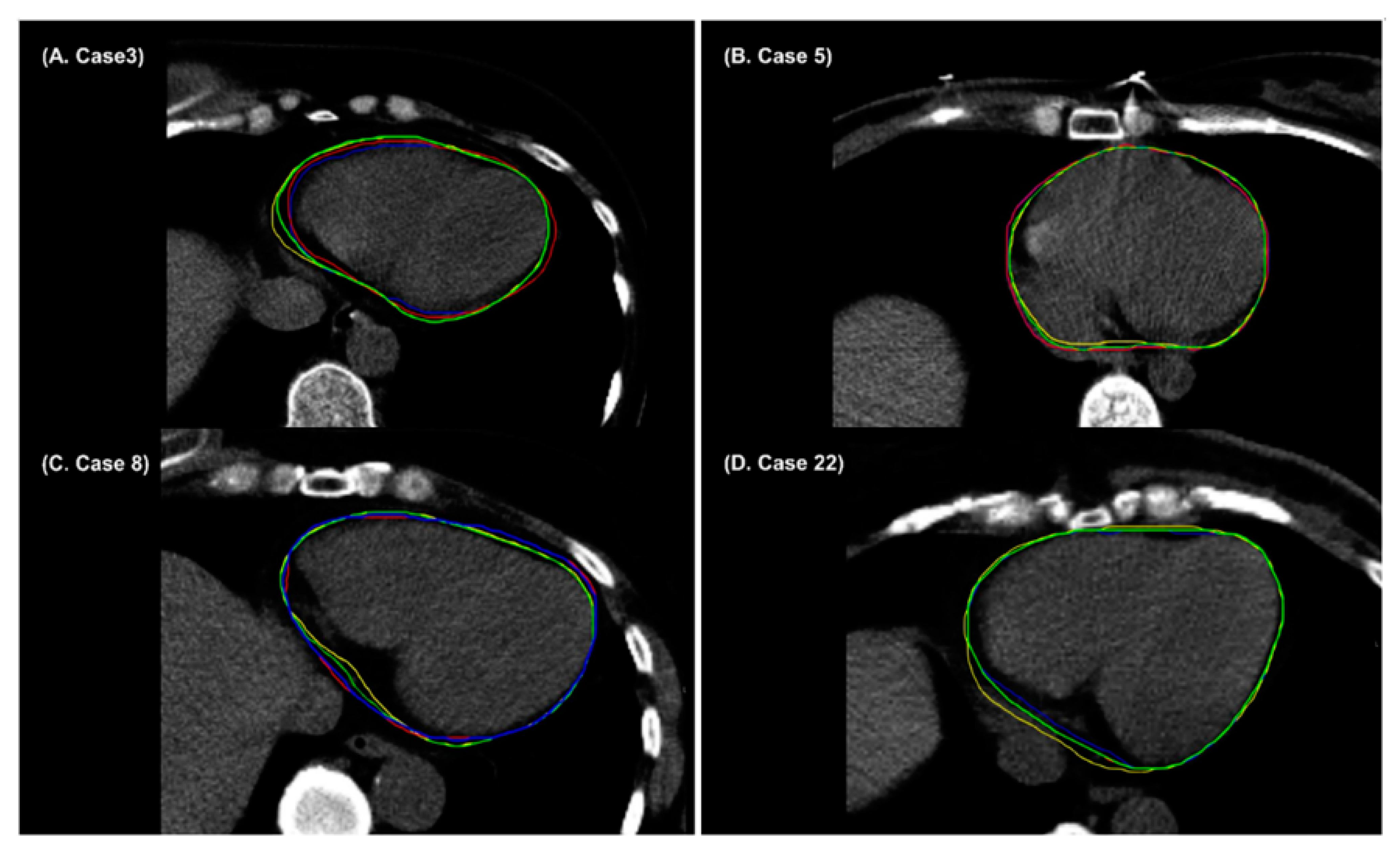
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Create gradient CDF histogram using End For End For Calculateusing Equation (2) End For End For Calculateusing Equation (2) End For Calculateusing While R( Calculateusing Equation (2) End For Calculate End While Update End For End For |
References
- Cardenas, C.E.; Yang, J.; Anderson, B.M.; Court, L.E.; Brock, K.B. Advances in auto-segmentation. In Seminars in Radiation Oncology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 185–197. [Google Scholar]
- Kosmin, M.; Ledsam, J.; Romera-Paredes, B.; Mendes, R.; Moinuddin, S.; de Souza, D.; Gunn, L.; Kelly, C.; Hughes, C.; Karthikesalingam, A. Rapid advances in auto-segmentation of organs at risk and target volumes in head and neck cancer. Radiother. Oncol. 2019, 135, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Ayyalusamy, A.; Vellaiyan, S.; Subramanian, S.; Ilamurugu, A.; Satpathy, S.; Nauman, M.; Katta, G.; Madineni, A. Auto-segmentation of head and neck organs at risk in radiotherapy and its dependence on anatomic similarity. Radiat. Oncol. J. 2019, 37, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwee, J.; Louie, A.V.; Gaede, S.; Bauman, G.; D’Souza, D.; Sexton, T.; Lock, M.; Ahmad, B.; Rodrigues, G. Technology assessment of automated atlas based segmentation in prostate bed contouring. Radiat. Oncol. 2011, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Xu, C.; Kuang, X.; Wang, H.; Chang, E.I.C.; Huang, W.; Fan, Y. 3d-sift-flow for atlas-based ct liver image segmentation. Med. Phys. 2016, 43, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Daisne, J.-F.; Blumhofer, A. Atlas-based automatic segmentation of head and neck organs at risk and nodal target volumes: A clinical validation. Radiat. Oncol. 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, N.; Gou, S.; Yang, S.; Ruan, D.; Sheng, K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med. Phys. 2018, 45, 4558–4567. [Google Scholar] [CrossRef] [Green Version]
- Kiljunen, T.; Akram, S.; Niemelä, J.; Löyttyniemi, E.; Seppälä, J.; Heikkilä, J.; Vuolukka, K.; Kääriäinen, O.-S.; Heikkilä, V.-P.; Lehtiö, K. A deep learning-based automated ct segmentation of prostate cancer anatomy for radiation therapy planning-a retrospective multicenter study. Diagnostics 2020, 10, 959. [Google Scholar] [CrossRef]
- Windisch, P.; Koechli, C.; Rogers, S.; Schröder, C.; Förster, R.; Zwahlen, D.R.; Bodis, S. Machine learning for the detection and segmentation of benign tumors of the central nervous system: A systematic review. Cancers 2022, 14, 2676. [Google Scholar] [CrossRef]
- Kim, N.; Chun, J.; Chang, J.S.; Lee, C.G.; Keum, K.C.; Kim, J.S. Feasibility of continual deep learning-based segmentation for personalized adaptive radiation therapy in head and neck area. Cancers 2021, 13, 702. [Google Scholar] [CrossRef]
- Yoo, S.K.; Kim, T.H.; Chun, J.; Choi, B.S.; Kim, H.; Yang, S.; Yoon, H.I.; Kim, J.S. Deep-learning-based automatic detection and segmentation of brain metastases with small volume for stereotactic ablative radiotherapy. Cancers 2022, 14, 2555. [Google Scholar] [CrossRef]
- Comelli, A.; Dahiya, N.; Stefano, A.; Vernuccio, F.; Portoghese, M.; Cutaia, G.; Bruno, A.; Salvaggio, G.; Yezzi, A. Deep learning-based methods for prostate segmentation in magnetic resonance imaging. Appl. Sci. 2021, 11, 782. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, K.-W.; Yang, R.; Geng, L.-S. Review of deep learning based automatic segmentation for lung cancer radiotherapy. Front. Oncol. 2021, 11, 2599. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Thomas, M.A.; Dise, J.; Kavanaugh, J.; Hilliard, J.; Zoberi, I.; Robinson, C.G.; Hugo, G.D. Robustness of deep learning segmentation of cardiac substructures in noncontrast computed tomography for breast cancer radiotherapy. Med. Phys. 2021, 48, 7172–7188. [Google Scholar] [CrossRef] [PubMed]
- van Rooij, W.; Dahele, M.; Brandao, H.R.; Delaney, A.R.; Slotman, B.J.; Verbakel, W.F. Deep learning-based delineation of head and neck organs at risk: Geometric and dosimetric evaluation. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 677–684. [Google Scholar] [CrossRef]
- Van der Veen, J.; Willems, S.; Deschuymer, S.; Robben, D.; Crijns, W.; Maes, F.; Nuyts, S. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother. Oncol. 2019, 138, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Elguindi, S.; Zelefsky, M.J.; Jiang, J.; Veeraraghavan, H.; Deasy, J.O.; Hunt, M.A.; Tyagi, N. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiotherapy. Phys. Imaging Radiat. Oncol. 2019, 12, 80–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savenije, M.H.; Maspero, M.; Sikkes, G.G.; van der Voort van Zyp, J.; Kotte, T.; Alexis, N.; Bol, G.H.; van den Berg, T.; Cornelis, A. Clinical implementation of mri-based organs-at-risk auto-segmentation with convolutional networks for prostate radiotherapy. Radiat. Oncol. 2020, 15, 1–12. [Google Scholar] [CrossRef]
- Zabel, W.J.; Conway, J.L.; Gladwish, A.; Skliarenko, J.; Didiodato, G.; Goorts-Matthews, L.; Michalak, A.; Reistetter, S.; King, J.; Nakonechny, K. Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Pract. Radiat. Oncol. 2021, 11, e80–e89. [Google Scholar] [CrossRef]
- Diniz, J.O.B.; Ferreira, J.L.; Diniz, P.H.B.; Silva, A.C.; de Paiva, A.C. Esophagus segmentation from planning ct images using an atlas-based deep learning approach. Comput. Methods Programs Biomed. 2020, 197, 105685. [Google Scholar] [CrossRef]
- Liu, Y.; Lei, Y.; Fu, Y.; Wang, T.; Tang, X.; Jiang, X.; Curran, W.J.; Liu, T.; Patel, P.; Yang, X. Ct-based multi-organ segmentation using a 3d self-attention u-net network for pancreatic radiotherapy. Med. Phys. 2020, 47, 4316–4324. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, X.; Yang, B.; Bi, N.; Zhang, T.; Men, K.; Dai, J. Evaluation of automatic segmentation model with dosimetric metrics for radiotherapy of esophageal cancer. Front. Oncol. 2020, 10, 1843. [Google Scholar] [CrossRef] [PubMed]
- van der Heyden, B.; Wohlfahrt, P.; Eekers, D.B.; Richter, C.; Terhaag, K.; Troost, E.G.; Verhaegen, F. Dual-energy ct for automatic organs-at-risk segmentation in brain-tumor patients using a multi-atlas and deep-learning approach. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Hu, J.; Wu, Q.; Xu, F.; Nie, S.; Zhao, Y.; Bai, S.; Yi, Z. Automatic delineation of the clinical target volume and organs at risk by deep learning for rectal cancer postoperative radiotherapy. Radiother. Oncol. 2020, 145, 186–192. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Guan, H.; Zhen, H.; Sun, Y.; Chen, Q.; Chen, Y.; Wang, S.; Qiu, J. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother. Oncol. 2020, 153, 172–179. [Google Scholar] [CrossRef]
- Kim, N.; Chang, J.S.; Kim, Y.B.; Kim, J.S. Atlas-based auto-segmentation for postoperative radiotherapy planning in endometrial and cervical cancers. Radiat. Oncol. 2020, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A survey of brain tumor segmentation and classification algorithms. J. Imaging 2021, 7, 179. [Google Scholar] [CrossRef]
- Minogue, S.; Gillham, C.; Kearney, M.; Mullaney, L. Intravenous contrast media in radiation therapy planning computed tomography scans–current practice in ireland. Tech. Innov. Patient Support Radiat. Oncol. 2019, 12, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Spampinato, M.V.; Abid, A.; Matheus, M.G. Current radiographic iodinated contrast agents. Magn. Reson. Imaging Clin. 2017, 25, 697–704. [Google Scholar] [CrossRef]
- Huynh, K.; Baghdanian, A.H.; Baghdanian, A.A.; Sun, D.S.; Kolli, K.P.; Zagoria, R.J. Updated guidelines for intravenous contrast use for ct and mri. Emerg. Radiol. 2020, 27, 115–126. [Google Scholar] [CrossRef]
- Lee, H.; Park, J.; Choi, Y.; Park, K.R.; Min, B.J.; Lee, I.J. Low-dose cbct reconstruction via joint non-local total variation denoising and cubic b-spline interpolation. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Lee, H.; Sung, J.; Choi, Y.; Kim, J.W.; Lee, I.J. Mutual information-based non-local total variation denoiser for low-dose cone-beam computed tomography. Front. Oncol. 2021, 11, 751057. [Google Scholar] [CrossRef] [PubMed]
- Kollem, S.; Reddy, K.R.L.; Rao, D.S. A review of image denoising and segmentation methods based on medical images. Int. J. Mach. Learn. Comput. 2019, 9, 288–295. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Fahimian, B.P.; Xing, L. Binary moving-blocker-based scatter correction in cone-beam computed tomography with width-truncated projections: Proof of concept. Phys. Med. Biol. 2017, 62, 2176. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Xing, L.; Lee, R.; Fahimian, B.P. Scatter correction in cone-beam ct via a half beam blocker technique allowing simultaneous acquisition of scatter and image information. Med. Phys. 2012, 39, 2386–2395. [Google Scholar] [CrossRef]
- Lee, H.; Yoon, J.; Lee, E. Anisotropic total variation denoising technique for low-dose cone-beam computed tomography imaging. Prog. Med. Phys. 2018, 29, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Chang, J.S.; Shin, K.H.; Kim, J.H.; Park, W.; Kim, H.; Kim, K.; Lee, I.J.; Yoon, W.S.; Cha, J. Post-mastectomy radiation therapy in breast reconstruction: A patterns of care study of the korean radiation oncology group. Radiat. Oncol. J. 2020, 38, 236. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Dong, X.; Lei, Y.; Wang, T.; Thomas, M.; Tang, L.; Curran, W.J.; Liu, T.; Yang, X. Automatic multiorgan segmentation in thorax ct images using u-net-gan. Med. Phys. 2019, 46, 2157–2168. [Google Scholar] [CrossRef] [Green Version]
- Perona, P.; Malik, J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 629–639. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, T.; Xing, L. Iterative image reconstruction for cbct using edge-preserving prior. Med. Phys. 2009, 36, 252–260. [Google Scholar] [CrossRef] [Green Version]
- Greffier, J.; Hamard, A.; Pereira, F.; Barrau, C.; Pasquier, H.; Beregi, J.P.; Frandon, J. Image quality and dose reduction opportunity of deep learning image reconstruction algorithm for ct: A phantom study. Eur. Radiol. 2020, 30, 3951–3959. [Google Scholar] [CrossRef] [PubMed]
- Samei, E.; Bakalyar, D.; Boedeker, K.L.; Brady, S.; Fan, J.; Leng, S.; Myers, K.J.; Popescu, L.M.; Ramirez Giraldo, J.C.; Ranallo, F. Performance evaluation of computed tomography systems: Summary of aapm task group 233. Med. Phys. 2019, 46, e735–e756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, S.Y.; Chang, J.S.; Choi, M.S.; Chang, Y.; Choi, B.S.; Chun, J.; Keum, K.C.; Kim, J.S.; Kim, Y.B. Clinical feasibility of deep learning-based auto-segmentation of target volumes and organs-at-risk in breast cancer patients after breast-conserving surgery. Radiat. Oncol. 2021, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic multi-organ segmentation on abdominal ct with dense v-networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef] [Green Version]
- Vu, C.C.; Siddiqui, Z.A.; Zamdborg, L.; Thompson, A.B.; Quinn, T.J.; Castillo, E.; Guerrero, T.M. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology–use of non-domain transfer learning. J. Appl. Clin. Med. Phys. 2020, 21, 108–113. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, J.; Qiu, B.; Liu, Y.; Liu, X.; Chen, L. Comparison of the automatic segmentation of multiple organs at risk in ct images of lung cancer between deep convolutional neural network-based and atlas-based techniques. Acta Oncol. 2019, 58, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Men, K.; Geng, H.; Biswas, T.; Liao, Z.; Xiao, Y. Automated quality assurance of oar contouring for lung cancer based on segmentation with deep active learning. Front. Oncol. 2020, 10, 986. [Google Scholar] [CrossRef]
- Choi, M.S.; Choi, B.S.; Chung, S.Y.; Kim, N.; Chun, J.; Kim, Y.B.; Chang, J.S.; Kim, J.S. Clinical evaluation of atlas-and deep learning-based automatic segmentation of multiple organs and clinical target volumes for breast cancer. Radiother. Oncol. 2020, 153, 139–145. [Google Scholar] [CrossRef]
- Feng, X.; Qing, K.; Tustison, N.J.; Meyer, C.H.; Chen, Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3d images. Med. Phys. 2019, 46, 2169–2180. [Google Scholar] [CrossRef] [Green Version]


| Case | Heart | Right Lung | Left Lung | Esophagus | Spinal Cord | Liver | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| * Manteia | † Denoiser | Manteia | Denoiser | Manteia | Denoiser | Manteia | Denoiser | Manteia | Denoiser | Manteia | Denoiser | |
| Case 1 | 0.964 | 0.960 | 0.982 | 0.983 | 0.981 | 0.981 | 0.685 | 0.742 | 0.777 | 0.824 | 0.953 | 0.954 |
| Case 2 | 0.951 | 0.947 | 0.980 | 0.980 | 0.979 | 0.979 | 0.670 | 0.723 | 0.891 | 0.873 | 0.936 | 0.939 |
| Case 3 | 0.958 | 0.952 | 0.982 | 0.982 | 0.979 | 0.979 | 0.660 | 0.602 | 0.881 | 0.884 | 0.957 | 0.963 |
| Case 4 | 0.931 | 0.928 | 0.971 | 0.970 | 0.975 | 0.975 | 0.744 | 0.719 | 0.866 | 0.858 | 0.937 | 0.938 |
| Case 5 | 0.908 | 0.894 | 0.971 | 0.971 | 0.975 | 0.975 | 0.745 | 0.672 | 0.877 | 0.877 | 0.948 | 0.952 |
| Case 6 | 0.930 | 0.929 | 0.982 | 0.982 | 0.973 | 0.974 | 0.573 | 0.691 | 0.877 | 0.877 | 0.890 | 0.926 |
| Case 7 | 0.978 | 0.977 | 0.951 | 0.951 | 0.954 | 0.955 | 0.735 | 0.730 | 0.876 | 0.870 | 0.874 | 0.864 |
| Case 8 | 0.929 | 0.926 | 0.971 | 0.972 | 0.970 | 0.970 | 0.669 | 0.662 | 0.859 | 0.856 | 0.964 | 0.963 |
| Case 9 | 0.938 | 0.936 | 0.978 | 0.978 | 0.972 | 0.972 | 0.664 | 0.732 | 0.814 | 0.819 | 0.959 | 0.960 |
| Case 10 | 0.952 | 0.947 | 0.982 | 0.982 | 0.981 | 0.981 | 0.765 | 0.786 | 0.883 | 0.877 | 0.959 | 0.963 |
| Case 11 | 0.945 | 0.944 | 0.979 | 0.979 | 0.977 | 0.978 | 0.746 | 0.737 | 0.838 | 0.831 | 0.920 | 0.924 |
| Case 12 | 0.946 | 0.941 | 0.962 | 0.963 | 0.964 | 0.966 | 0.680 | 0.634 | 0.868 | 0.848 | 0.933 | 0.933 |
| Case 13 | 0.952 | 0.950 | 0.984 | 0.984 | 0.981 | 0.982 | 0.681 | 0.695 | 0.865 | 0.854 | 0.926 | 0.932 |
| Case 14 | 0.934 | 0.930 | 0.981 | 0.981 | 0.981 | 0.981 | 0.570 | 0.633 | 0.858 | 0.854 | 0.909 | 0.933 |
| Case 15 | 0.930 | 0.926 | 0.975 | 0.976 | 0.973 | 0.973 | 0.702 | 0.744 | 0.859 | 0.865 | 0.893 | 0.931 |
| Case 16 | 0.906 | 0.898 | 0.934 | 0.934 | 0.940 | 0.940 | 0.691 | 0.637 | 0.870 | 0.870 | 0.956 | 0.959 |
| Case 17 | 0.939 | 0.932 | 0.977 | 0.977 | 0.972 | 0.972 | 0.697 | 0.697 | 0.857 | 0.863 | 0.954 | 0.955 |
| Case 18 | 0.950 | 0.948 | 0.976 | 0.976 | 0.979 | 0.979 | 0.725 | 0.697 | 0.878 | 0.882 | 0.939 | 0.942 |
| Case 19 | 0.910 | 0.887 | 0.981 | 0.981 | 0.981 | 0.979 | 0.653 | 0.617 | 0.853 | 0.859 | 0.873 | 0.885 |
| Case 20 | 0.877 | 0.875 | 0.983 | 0.984 | 0.975 | 0.975 | 0.730 | 0.775 | 0.853 | 0.846 | 0.954 | 0.956 |
| Case 21 | 0.919 | 0.921 | 0.981 | 0.981 | 0.978 | 0.978 | 0.727 | 0.719 | 0.819 | 0.801 | 0.940 | 0.941 |
| Case 22 | 0.999 | 0.968 | 0.998 | 0.992 | 0.998 | 0.992 | 0.990 | 0.797 | 0.805 | 0.807 | 0.935 | 0.951 |
| Case 23 | 0.956 | 0.954 | 0.977 | 0.977 | 0.980 | 0.979 | 0.625 | 0.636 | 0.878 | 0.872 | 0.954 | 0.957 |
| Case 24 | 0.963 | 0.962 | 0.986 | 0.986 | 0.979 | 0.980 | 0.728 | 0.725 | 0.872 | 0.857 | 0.955 | 0.956 |
| Case 25 | 0.919 | 0.920 | 0.987 | 0.987 | 0.979 | 0.978 | 0.767 | 0.763 | 0.872 | 0.874 | 0.964 | 0.967 |
| Case 26 | 0.966 | 0.966 | 0.990 | 0.989 | 0.986 | 0.987 | 0.813 | 0.821 | 0.892 | 0.876 | 0.964 | 0.966 |
| Case 27 | 0.963 | 0.961 | 0.992 | 0.992 | 0.989 | 0.989 | 0.650 | 0.638 | 0.797 | 0.815 | 0.959 | 0.960 |
| Case 28 | 0.963 | 0.963 | 0.991 | 0.991 | 0.984 | 0.985 | 0.785 | 0.789 | 0.853 | 0.861 | 0.959 | 0.960 |
| Case 29 | 0.967 | 0.967 | 0.985 | 0.985 | 0.976 | 0.978 | 0.765 | 0.768 | 0.823 | 0.836 | 0.934 | 0.937 |
| Case 30 | 0.951 | 0.951 | 0.985 | 0.985 | 0.980 | 0.980 | 0.661 | 0.671 | 0.818 | 0.835 | 0.927 | 0.930 |
| Case 31 | 0.950 | 0.950 | 0.986 | 0.986 | 0.984 | 0.984 | 0.668 | 0.650 | 0.841 | 0.844 | 0.896 | 0.899 |
| Case 32 | 0.947 | 0.946 | 0.986 | 0.986 | 0.980 | 0.980 | 0.791 | 0.778 | 0.865 | 0.863 | 0.911 | 0.910 |
| Case 33 | 0.956 | 0.956 | 0.962 | 0.962 | 0.975 | 0.975 | 0.771 | 0.780 | 0.831 | 0.846 | 0.948 | 0.950 |
| Case 34 | 0.950 | 0.950 | 0.978 | 0.978 | 0.979 | 0.980 | 0.740 | 0.725 | 0.861 | 0.857 | 0.943 | 0.946 |
| Case 35 | 0.947 | 0.949 | 0.977 | 0.977 | 0.981 | 0.981 | 0.749 | 0.739 | 0.887 | 0.880 | 0.956 | 0.959 |
| Case 36 | 0.937 | 0.938 | 0.985 | 0.985 | 0.977 | 0.978 | 0.788 | 0.782 | 0.881 | 0.883 | 0.950 | 0.952 |
| Case 37 | 0.945 | 0.945 | 0.984 | 0.984 | 0.980 | 0.981 | 0.730 | 0.727 | 0.879 | 0.882 | 0.939 | 0.938 |
| Case 38 | 0.933 | 0.934 | 0.987 | 0.987 | 0.983 | 0.982 | 0.809 | 0.819 | 0.872 | 0.878 | 0.947 | 0.949 |
| Case 39 | 0.945 | 0.945 | 0.968 | 0.962 | 0.974 | 0.975 | 0.759 | 0.743 | 0.887 | 0.894 | 0.952 | 0.953 |
| Case 40 | 0.911 | 0.911 | 0.982 | 0.982 | 0.977 | 0.978 | 0.664 | 0.672 | 0.876 | 0.868 | 0.947 | 0.947 |
| Average | 0.943 | 0.940 | 0.979 | 0.978 | 0.977 | 0.977 | 0.719 | 0.717 | 0.858 | 0.868 | 0.938 | 0.943 |
| p-value | 0.000 | 0.091 | 0.095 | 0.705 | 0.762 | 0.000 | ||||||
| Case | Heart | Right Lung | Left Lung | Esophagus | Spinal Cord | Liver | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| * Manteia | † Denoiser | Manteia | Denoiser | Manteia | Denoiser | Manteia | Denoiser | Manteia | Denoiser | Manteia | Denoiser | |
| Case 1 | 0.964 | 0.960 | 0.981 | 0.981 | 0.980 | 0.980 | 0.685 | 0.742 | 0.749 | 0.785 | 0.953 | 0.955 |
| Case 2 | 0.937 | 0.934 | 0.985 | 0.985 | 0.982 | 0.982 | 0.696 | 0.730 | 0.872 | 0.844 | 0.932 | 0.934 |
| Case 3 | 0.964 | 0.962 | 0.984 | 0.984 | 0.980 | 0.981 | 0.638 | 0.597 | 0.865 | 0.867 | 0.949 | 0.956 |
| Case 4 | 0.912 | 0.909 | 0.973 | 0.972 | 0.979 | 0.979 | 0.741 | 0.697 | 0.857 | 0.864 | 0.927 | 0.929 |
| Case 5 | 0.907 | 0.895 | 0.977 | 0.977 | 0.977 | 0.976 | 0.721 | 0.625 | 0.848 | 0.848 | 0.941 | 0.947 |
| Case 6 | 0.927 | 0.927 | 0.987 | 0.987 | 0.980 | 0.980 | 0.577 | 0.693 | 0.887 | 0.892 | 0.884 | 0.920 |
| Case 7 | 0.986 | 0.989 | 0.996 | 0.997 | 0.995 | 0.996 | 0.808 | 0.839 | 0.870 | 0.873 | 0.849 | 0.859 |
| Case 8 | 0.925 | 0.922 | 0.964 | 0.963 | 0.973 | 0.973 | 0.658 | 0.689 | 0.872 | 0.851 | 0.954 | 0.953 |
| Case 9 | 0.934 | 0.933 | 0.979 | 0.978 | 0.974 | 0.974 | 0.702 | 0.747 | 0.851 | 0.852 | 0.952 | 0.952 |
| Case 10 | 0.916 | 0.909 | 0.984 | 0.984 | 0.984 | 0.983 | 0.732 | 0.756 | 0.869 | 0.852 | 0.957 | 0.959 |
| Case 11 | 0.938 | 0.939 | 0.986 | 0.986 | 0.974 | 0.974 | 0.683 | 0.683 | 0.786 | 0.749 | 0.912 | 0.915 |
| Case 12 | 0.949 | 0.943 | 0.977 | 0.977 | 0.967 | 0.969 | 0.707 | 0.655 | 0.778 | 0.747 | 0.925 | 0.925 |
| Case 13 | 0.935 | 0.935 | 0.985 | 0.985 | 0.982 | 0.982 | 0.665 | 0.692 | 0.844 | 0.823 | 0.921 | 0.925 |
| Case 14 | 0.935 | 0.933 | 0.984 | 0.984 | 0.979 | 0.979 | 0.633 | 0.695 | 0.823 | 0.790 | 0.902 | 0.927 |
| Case 15 | 0.907 | 0.909 | 0.983 | 0.983 | 0.976 | 0.976 | 0.660 | 0.648 | 0.835 | 0.841 | 0.921 | 0.880 |
| Case 16 | 0.926 | 0.936 | 0.985 | 0.985 | 0.979 | 0.979 | 0.681 | 0.637 | 0.781 | 0.787 | 0.953 | 0.956 |
| Case 17 | 0.939 | 0.932 | 0.977 | 0.977 | 0.972 | 0.972 | 0.697 | 0.697 | 0.804 | 0.822 | 0.949 | 0.949 |
| Case 18 | 0.950 | 0.951 | 0.982 | 0.982 | 0.982 | 0.982 | 0.734 | 0.755 | 0.870 | 0.880 | 0.932 | 0.936 |
| Case 19 | 0.890 | 0.867 | 0.985 | 0.985 | 0.983 | 0.981 | 0.630 | 0.604 | 0.860 | 0.865 | 0.857 | 0.869 |
| Case 20 | 0.850 | 0.848 | 0.986 | 0.986 | 0.979 | 0.978 | 0.738 | 0.743 | 0.885 | 0.860 | 0.949 | 0.950 |
| Case 21 | 0.954 | 0.955 | 0.991 | 0.991 | 0.986 | 0.986 | 0.730 | 0.731 | 0.853 | 0.835 | 0.946 | 0.947 |
| Case 22 | 0.999 | 0.968 | 0.995 | 0.990 | 0.995 | 0.989 | 0.902 | 0.752 | 0.790 | 0.827 | 0.941 | 0.952 |
| Case 23 | 0.956 | 0.954 | 0.975 | 0.975 | 0.978 | 0.978 | 0.622 | 0.632 | 0.838 | 0.816 | 0.953 | 0.955 |
| Case 24 | 0.963 | 0.962 | 0.984 | 0.984 | 0.977 | 0.978 | 0.726 | 0.724 | 0.838 | 0.820 | 0.956 | 0.958 |
| Case 25 | 0.924 | 0.925 | 0.986 | 0.985 | 0.978 | 0.980 | 0.767 | 0.763 | 0.860 | 0.864 | 0.962 | 0.965 |
| Case 26 | 0.966 | 0.966 | 0.986 | 0.986 | 0.983 | 0.984 | 0.813 | 0.821 | 0.868 | 0.861 | 0.960 | 0.961 |
| Case 27 | 0.963 | 0.961 | 0.990 | 0.990 | 0.987 | 0.988 | 0.636 | 0.627 | 0.840 | 0.837 | 0.959 | 0.959 |
| Case 28 | 0.963 | 0.963 | 0.991 | 0.991 | 0.984 | 0.985 | 0.785 | 0.790 | 0.853 | 0.862 | 0.951 | 0.951 |
| Case 29 | 0.967 | 0.967 | 0.984 | 0.983 | 0.974 | 0.976 | 0.765 | 0.768 | 0.847 | 0.827 | 0.934 | 0.936 |
| Case 30 | 0.951 | 0.951 | 0.984 | 0.984 | 0.979 | 0.980 | 0.661 | 0.671 | 0.857 | 0.840 | 0.938 | 0.939 |
| Case 31 | 0.932 | 0.932 | 0.986 | 0.986 | 0.983 | 0.983 | 0.668 | 0.650 | 0.852 | 0.843 | 0.895 | 0.897 |
| Case 32 | 0.939 | 0.938 | 0.985 | 0.985 | 0.978 | 0.978 | 0.791 | 0.778 | 0.857 | 0.855 | 0.912 | 0.911 |
| Case 33 | 0.956 | 0.956 | 0.960 | 0.960 | 0.975 | 0.975 | 0.710 | 0.709 | 0.857 | 0.839 | 0.948 | 0.950 |
| Case 34 | 0.949 | 0.948 | 0.976 | 0.976 | 0.976 | 0.977 | 0.739 | 0.723 | 0.876 | 0.861 | 0.947 | 0.949 |
| Case 35 | 0.935 | 0.934 | 0.982 | 0.982 | 0.980 | 0.980 | 0.749 | 0.739 | 0.880 | 0.874 | 0.953 | 0.955 |
| Case 36 | 0.936 | 0.936 | 0.984 | 0.984 | 0.976 | 0.976 | 0.793 | 0.786 | 0.860 | 0.840 | 0.951 | 0.952 |
| Case 37 | 0.945 | 0.945 | 0.982 | 0.982 | 0.978 | 0.979 | 0.669 | 0.662 | 0.812 | 0.821 | 0.938 | 0.936 |
| Case 38 | 0.944 | 0.945 | 0.986 | 0.986 | 0.981 | 0.981 | 0.809 | 0.819 | 0.870 | 0.874 | 0.950 | 0.951 |
| Case 39 | 0.955 | 0.955 | 0.965 | 0.960 | 0.974 | 0.975 | 0.682 | 0.663 | 0.884 | 0.892 | 0.953 | 0.953 |
| Case 40 | 0.911 | 0.911 | 0.980 | 0.980 | 0.977 | 0.977 | 0.624 | 0.630 | 0.874 | 0.875 | 0.948 | 0.948 |
| Average | 0.940 | 0.938 | 0.982 | 0.982 | 0.979 | 0.980 | 0.711 | 0.709 | 0.847 | 0.841 | 0.935 | 0.938 |
| p-value | 0.008 | 0.091 | 0.097 | 0.984 | 0.082 | 0.000 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Im, J.H.; Lee, I.J.; Choi, Y.; Sung, J.; Ha, J.S.; Lee, H. Impact of Denoising on Deep-Learning-Based Automatic Segmentation Framework for Breast Cancer Radiotherapy Planning. Cancers 2022, 14, 3581. https://doi.org/10.3390/cancers14153581
Im JH, Lee IJ, Choi Y, Sung J, Ha JS, Lee H. Impact of Denoising on Deep-Learning-Based Automatic Segmentation Framework for Breast Cancer Radiotherapy Planning. Cancers. 2022; 14(15):3581. https://doi.org/10.3390/cancers14153581
Chicago/Turabian StyleIm, Jung Ho, Ik Jae Lee, Yeonho Choi, Jiwon Sung, Jin Sook Ha, and Ho Lee. 2022. "Impact of Denoising on Deep-Learning-Based Automatic Segmentation Framework for Breast Cancer Radiotherapy Planning" Cancers 14, no. 15: 3581. https://doi.org/10.3390/cancers14153581
APA StyleIm, J. H., Lee, I. J., Choi, Y., Sung, J., Ha, J. S., & Lee, H. (2022). Impact of Denoising on Deep-Learning-Based Automatic Segmentation Framework for Breast Cancer Radiotherapy Planning. Cancers, 14(15), 3581. https://doi.org/10.3390/cancers14153581






