Prognostic Capability of TNBC 3-Gene Score among Triple-Negative Breast Cancer Subtypes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Subtype Identification
2.3. Elaboration of the Metabase
2.4. Prognostic Capability of the TNBC 3-Gene Score According to TNBC Subtypes
2.5. Evaluation of the Relation between TNBC 3-Gene Score and TILs
3. Results
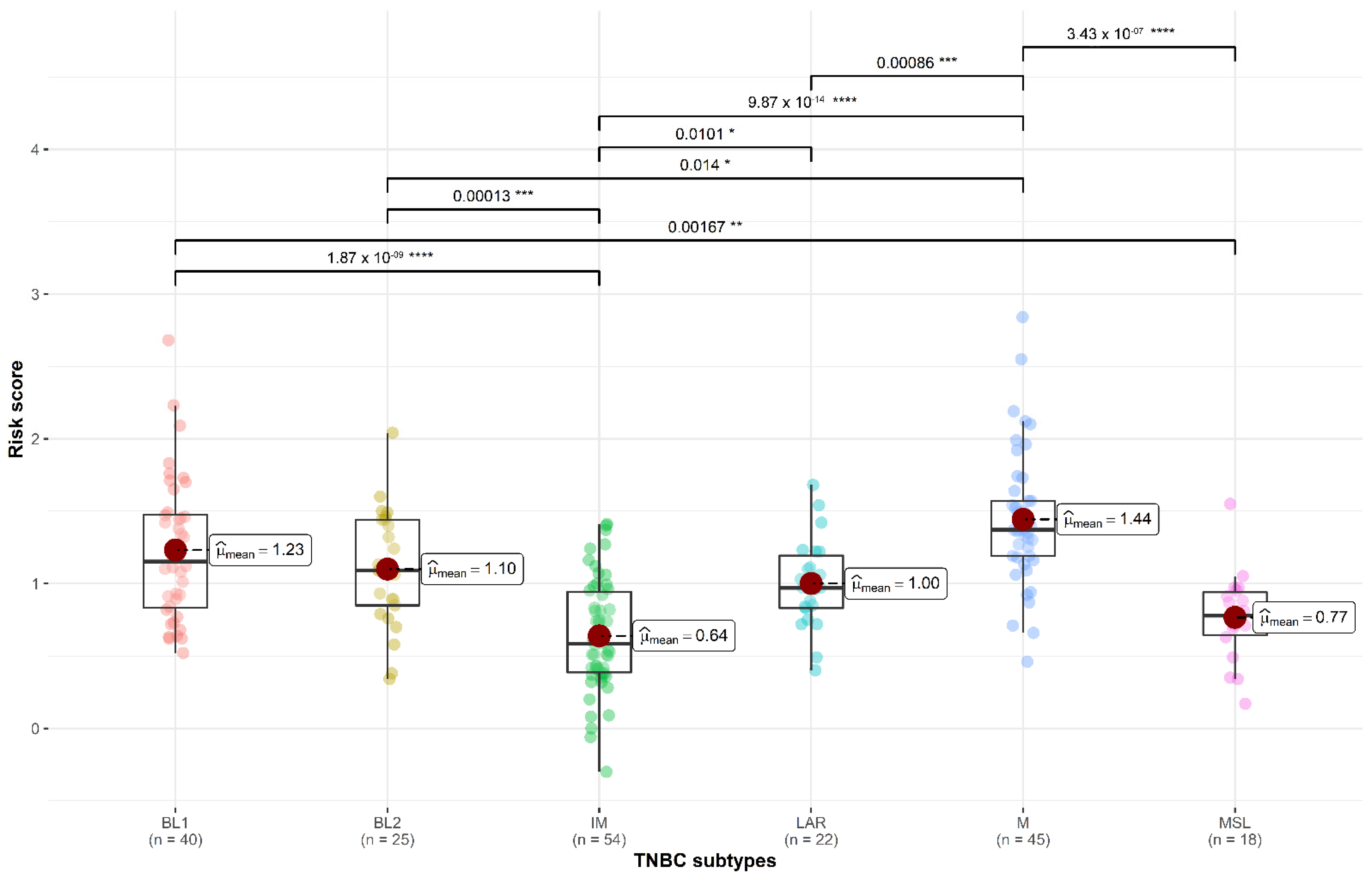
3.1. TNBC Subtypes
3.2. Gene Expression of CCL5, DDIT4, and POLR1C in the Metabase
3.3. Predictive Value of the TNBC 3-Gene Score in the Metabase
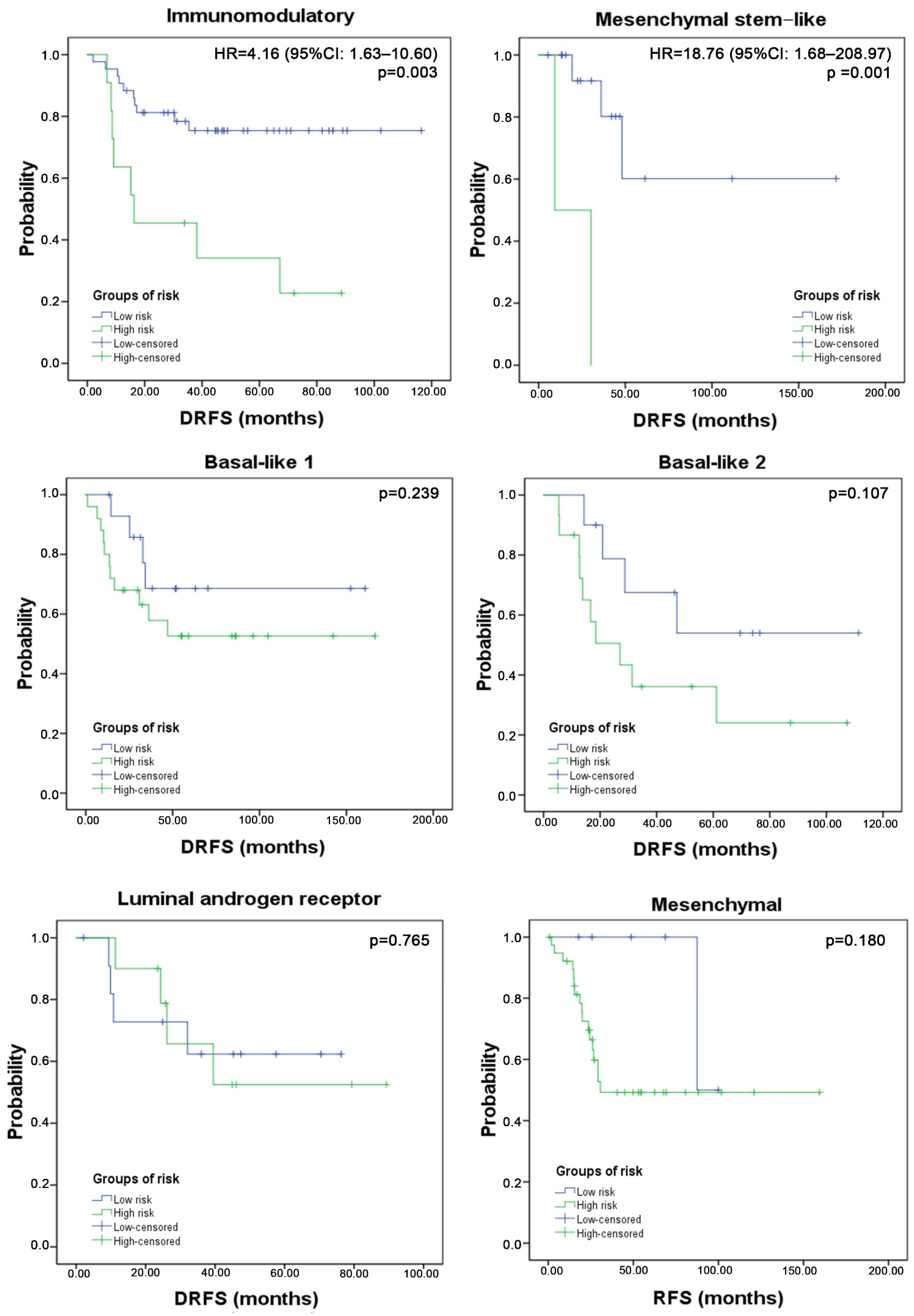
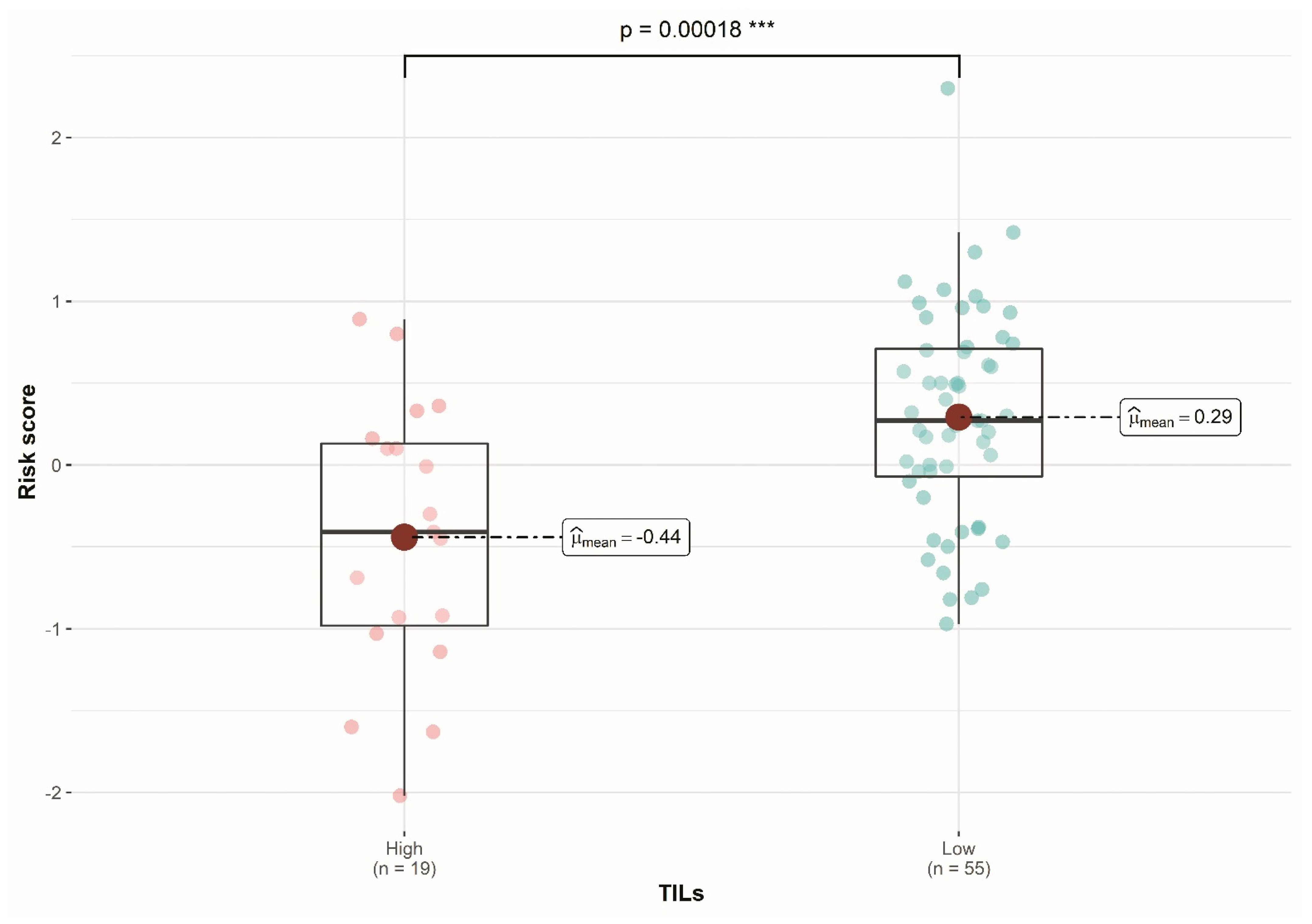
3.4. Three-Gene Score Predictive Value in TNBC Subtypes and Relation with TILs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Bauer, K.R.; Brown, M.; Cress, R.D.; Parise, C.A.; Caggiano, V. Descriptive Analysis of Estrogen Receptor (ER)-Negative, Progesterone Receptor (PR)-Negative, and HER2-Negative Invasive Breast Cancer, the so-Called Triple-Negative Phenotype. Cancer 2007, 109, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Haffty, B.G.; Yang, Q.; Reiss, M.; Kearney, T.; Higgins, S.A.; Weidhaas, J.; Harris, L.; Hait, W.; Toppmeyer, D. Locoregional Relapse and Distant Metastasis in Conservatively Managed Triple Negative Early-Stage Breast Cancer. J. Clin. Oncol. 2006, 24, 5652–5657. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, C.S.; Gómez, H.L.; Cruz, W.R.; Pinto, J.A.; Dyer, R.R.; Velarde, R.; Suazo, J.F.; Neciosup, S.P.; León, M.; de la Cruz, M.A.; et al. Breast Cancer Classification According to Immunohistochemistry Markers: Subtypes and Association With Clinicopathologic Variables in a Peruvian Hospital Database. Clin. Breast Cancer 2010, 10, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Chiu, K.T.; Edmiston, S.; et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef]
- Morris, G.J.; Naidu, S.; Topham, A.K.; Guiles, F.; Xu, Y.; McCue, P.; Schwartz, G.F.; Park, P.K.; Rosenberg, A.L.; Brill, K.; et al. Differences in Breast Carcinoma Characteristics in Newly Diagnosed African–American and Caucasian Patients. Cancer 2007, 110, 876–884. [Google Scholar] [CrossRef]
- Yao, L.; Pang, Z.; Wang, M.; Wang, M.; Sun, X.; Cui, M.; Zheng, Y.; Li, X.; Dong, H.; Zhang, Q.; et al. The Choice of a Neoadjuvant Chemotherapy Cycle for Breast Cancer Has Significance in Clinical Practice: Results from a Population-Based, Real World Study. Cancer Biol. Med. 2022, 19, 755–767. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-Negative Breast Cancer: Clinical Features and Patterns of Recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to Neoadjuvant Therapy and Long-Term Survival in Patients With Triple-Negative Breast Cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of Human Triple-Negative Breast Cancer Subtypes and Preclinical Models for Selection of Targeted Therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.W.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive Genomic Analysis Identifies Novel Subtypes and Targets of Triple-Negative Breast Cancer. Clin. Cancer Res. 2015, 21, 1688. [Google Scholar] [CrossRef] [PubMed]
- Hubalek, M.; Czech, T.; Müller, H. Biological Subtypes of Triple-Negative Breast Cancer. Breast Care 2017, 12, 8. [Google Scholar] [CrossRef]
- Ensenyat-Mendez, M.; Llinàs-Arias, P.; Orozco, J.I.J.; Íñiguez-Muñoz, S.; Salomon, M.P.; Sesé, B.; DiNome, M.L.; Marzese, D.M. Current Triple-Negative Breast Cancer Subtypes: Dissecting the Most Aggressive Form of Breast Cancer. Front. Oncol. 2021, 11, 681476. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-Negative Breast Cancer Molecular Subtyping and Treatment Progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Santonja, A.; Sánchez-Muñoz, A.; Lluch, A.; Chica-Parrado, M.R.; Albanell, J.; Chacón, J.I.; Antolín, S.; Jerez, J.M.; de la Haba, J.; de Luque, V.; et al. Triple Negative Breast Cancer Subtypes and Pathologic Complete Response Rate to Neoadjuvant Chemotherapy. Oncotarget 2018, 9, 26406–26416. [Google Scholar] [CrossRef]
- Pinto, J.A.; Araujo, J.; Cardenas, N.K.; Morante, Z.; Doimi, F.; Vidaurre, T.; Balko, J.M.; Gomez, H.L. A Prognostic Signature Based on Three-Genes Expression in Triple-Negative Breast Tumours with Residual Disease. NPJ Genomic Med. 2016, 1, 15015. [Google Scholar] [CrossRef]
- Pinto, J.A.; Rolfo, C.; Raez, L.E.; Prado, A.; Araujo, J.M.; Bravo, L.; Fajardo, W.; Morante, Z.D.; Aguilar, A.; Neciosup, S.P.; et al. In Silico Evaluation of DNA Damage Inducible Transcript 4 Gene (DDIT4) as Prognostic Biomarker in Several Malignancies. Sci. Rep. 2017, 7, 1526. [Google Scholar] [CrossRef]
- Araujo, J.M.; Gomez, A.C.; Aguilar, A.; Salgado, R.; Balko, J.M.; Bravo, L.; Doimi, F.; Bretel, D.; Morante, Z.; Flores, C.; et al. Effect of CCL5 Expression in the Recruitment of Immune Cells in Triple Negative Breast Cancer. Sci. Rep. 2018, 8, 4899. [Google Scholar] [CrossRef] [PubMed]
- El Bairi, K.; Haynes, H.R.; Blackley, E.; Fineberg, S.; Shear, J.; Turner, S.; de Freitas, J.R.; Sur, D.; Amendola, L.C.; Gharib, M.; et al. The Tale of TILs in Breast Cancer: A Report from The International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer 2021, 7, 150. [Google Scholar] [CrossRef]
- Gao, G.; Wang, Z.; Qu, X.; Zhang, Z. Prognostic Value of Tumor-Infiltrating Lymphocytes in Patients with Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. BMC Cancer 2020, 20, 179. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Hatzis, C.; Pusztai, L.; Valero, V.; Booser, D.J.; Esserman, L.; Lluch, A.; Vidaurre, T.; Holmes, F.; Souchon, E.; Wang, H.; et al. A Genomic Predictor of Response and Survival Following Taxane-Anthracycline Chemotherapy for Invasive Breast Cancer. JAMA 2011, 305, 1873. [Google Scholar] [CrossRef] [PubMed]
- Jézéquel, P.; Loussouarn, D.; Guérin-Charbonnel, C.; Campion, L.; Vanier, A.; Gouraud, W.; Lasla, H.; Guette, C.; Valo, I.; Verrièle, V.; et al. Gene-Expression Molecular Subtyping of Triple-Negative Breast Cancer Tumours: Importance of Immune Response. Breast Cancer Res. 2015, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Desmedt, C.; Di Leo, A.; de Azambuja, E.; Larsimont, D.; Haibe-Kains, B.; Selleslags, J.; Delaloge, S.; Duhem, C.; Kains, J.-P.; Carly, B.; et al. Multifactorial Approach to Predicting Resistance to Anthracyclines. J. Clin. Oncol. 2011, 29, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Gray, W.H.; Lehmann, B.D.; Bauer, J.A.; Shyr, Y.; Pietenpol, J.A. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012, 11, CIN.S9983. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Sun, P.; He, J.; Chao, X.; Chen, K.; Xu, Y.; Huang, Q.; Yun, J.; Li, M.; Luo, R.; Kuang, J.; et al. A Computational Tumor-Infiltrating Lymphocyte Assessment Method Comparable with Visual Reporting Guidelines for Triple-Negative Breast Cancer. EBioMedicine 2021, 70, 103492. [Google Scholar] [CrossRef]
- Pruneri, G.; Vingiani, A.; Bagnardi, V.; Rotmensz, N.; De Rose, A.; Palazzo, A.; Colleoni, A.M.; Goldhirsch, A.; Viale, G. Clinical Validity of Tumor-Infiltrating Lymphocytes Analysis in Patients with Triple-Negative Breast Cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Espinosa Fernandez, J.R.; Eckhardt, B.L.; Lee, J.; Lim, B.; Pearson, T.; Seitz, R.S.; Hout, D.R.; Schweitzer, B.L.; Nielsen, T.J.; Rayne Lawrence, O.; et al. Identification of Triple-Negative Breast Cancer Cell Lines Classified under the Same Molecular Subtype Using Different Molecular Characterization Techniques: Implications for Translational Research. PLoS ONE 2020, 15, 231953. [Google Scholar] [CrossRef]
- Zheng, H.; Siddharth, S.; Parida, S.; Wu, X.; Sharma, D. Tumor Microenvironment: Key Players in Triple Negative Breast Cancer Immunomodulation. Cancers 2021, 13, 3357. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, S.M. Genome Instability-Derived Genes Are Novel Prognostic Biomarkers for Triple-Negative Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 701073. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Deng, J.; Zhang, L.; Yuan, J.; Yang, H.; Li, Q. Tumor Microenvironment Characterization in Triple-Negative Breast Cancer Identifies Prognostic Gene Signature. Aging (Albany NY) 2021, 13, 5485–5505. [Google Scholar] [CrossRef]
- Wang, X.; Su, W.; Tang, D.; Jing, J.; Xiong, J.; Deng, Y.; Liu, H.; Ma, W.; Liu, Z.; Zhang, Q. An Immune-Related Gene Prognostic Index for Triple-Negative Breast Cancer Integrates Multiple Aspects of Tumor-Immune Microenvironment. Cancers 2021, 13, 5342. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Drubay, D.; Adams, S.; Pruneri, G.; Francis, P.A.; Lacroix-Triki, M.; Joensuu, H.; Dieci, M.V.; Badve, S.; Demaria, S.; et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J. Clin. Oncol. 2019, 37, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; He, S. The Characteristics of Tumor Microenvironment in Triple Negative Breast Cancer. Cancer Manag. Res. 2022, 14, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Inoue, N.; Morimoto, K.; Watanabe, T.; Hirota, S.; Imamura, M.; Matsushita, Y.; Katagiri, T.; Okamura, H.; Miyoshi, Y. Significant Association between High Serum CCL5 Levels and Better Disease-Free Survival of Patients with Early Breast Cancer. Cancer Sci. 2020, 111, 209–218. [Google Scholar] [CrossRef]
- Gerratana, L.; Basile, D.; Buono, G.; De Placido, S.; Giuliano, M.; Minichillo, S.; Coinu, A.; Martorana, F.; De Santo, I.; Del Mastro, L.; et al. Androgen Receptor in Triple Negative Breast Cancer: A Potential Target for the Targetless Subtype. Cancer Treat. Rev. 2018, 68, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.T.; Kim, Y.A.; Kim, J.; Park, J.H.; Choi, I.S.; Hwang, K.R.; Chai, Y.J.; Park, J.H. Influence of Androgen Receptor on the Prognosis of Breast Cancer. J. Clin. Med. 2020, 9, 83. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Miremadi, A.; Pinder, S.E.; Ellis, I.O.; Caldas, C. An Immune Response Gene Expression Module Identifies a Good Prognosis Subtype in Estrogen Receptor Negative Breast Cancer. Genome Biol. 2007, 8, R157. [Google Scholar] [CrossRef] [Green Version]
- Rody, A.; Karn, T.; Liedtke, C.; Pusztai, L.; Ruckhaeberle, E.; Hanker, L.; Gaetje, R.; Solbach, C.; Ahr, A.; Metzler, D.; et al. A Clinically Relevant Gene Signature in Triple Negative and Basal-like Breast Cancer. Breast Cancer Res. 2011, 13, R97. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, C.; Bayar, M.A.; Curigliano, G.; Symmans, F.W.; Desmedt, C.; Bonnefoi, H.; Sinn, B.; Pruneri, G.; Vicier, C.; Pierga, J.Y.; et al. A Gene Signature to Predict High Tumor-Infiltrating Lymphocytes after Neoadjuvant Chemotherapy and Outcome in Patients with Triple-Negative Breast Cancer. Ann. Oncol. 2018, 29, 162–169. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araujo, J.M.; De la Cruz-Ku, G.; Cornejo, M.; Doimi, F.; Dyer, R.; Gomez, H.L.; Pinto, J.A. Prognostic Capability of TNBC 3-Gene Score among Triple-Negative Breast Cancer Subtypes. Cancers 2022, 14, 4286. https://doi.org/10.3390/cancers14174286
Araujo JM, De la Cruz-Ku G, Cornejo M, Doimi F, Dyer R, Gomez HL, Pinto JA. Prognostic Capability of TNBC 3-Gene Score among Triple-Negative Breast Cancer Subtypes. Cancers. 2022; 14(17):4286. https://doi.org/10.3390/cancers14174286
Chicago/Turabian StyleAraujo, Jhajaira M., Gabriel De la Cruz-Ku, Melanie Cornejo, Franco Doimi, Richard Dyer, Henry L. Gomez, and Joseph A. Pinto. 2022. "Prognostic Capability of TNBC 3-Gene Score among Triple-Negative Breast Cancer Subtypes" Cancers 14, no. 17: 4286. https://doi.org/10.3390/cancers14174286
APA StyleAraujo, J. M., De la Cruz-Ku, G., Cornejo, M., Doimi, F., Dyer, R., Gomez, H. L., & Pinto, J. A. (2022). Prognostic Capability of TNBC 3-Gene Score among Triple-Negative Breast Cancer Subtypes. Cancers, 14(17), 4286. https://doi.org/10.3390/cancers14174286





