The Biological and Clinical Consequences of RNA Splicing Factor U2AF1 Mutation in Myeloid Malignancies
Abstract
Simple Summary
Abstract
1. Introduction
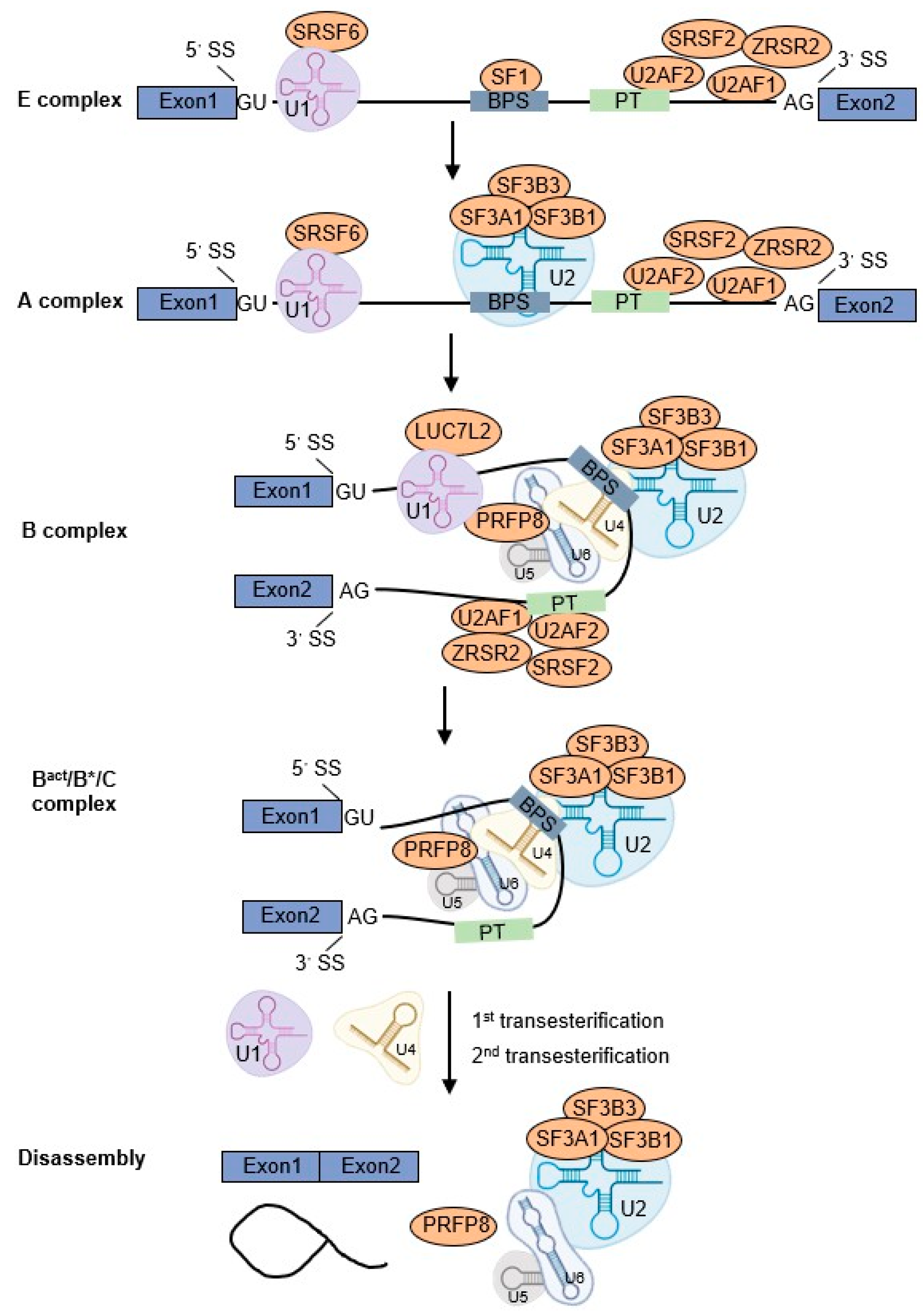
2. U2AF1 Participates in Pre-mRNA Splicing Process
2.1. RNA Splicing Cycle
2.2. Alternative Splicing Patterns
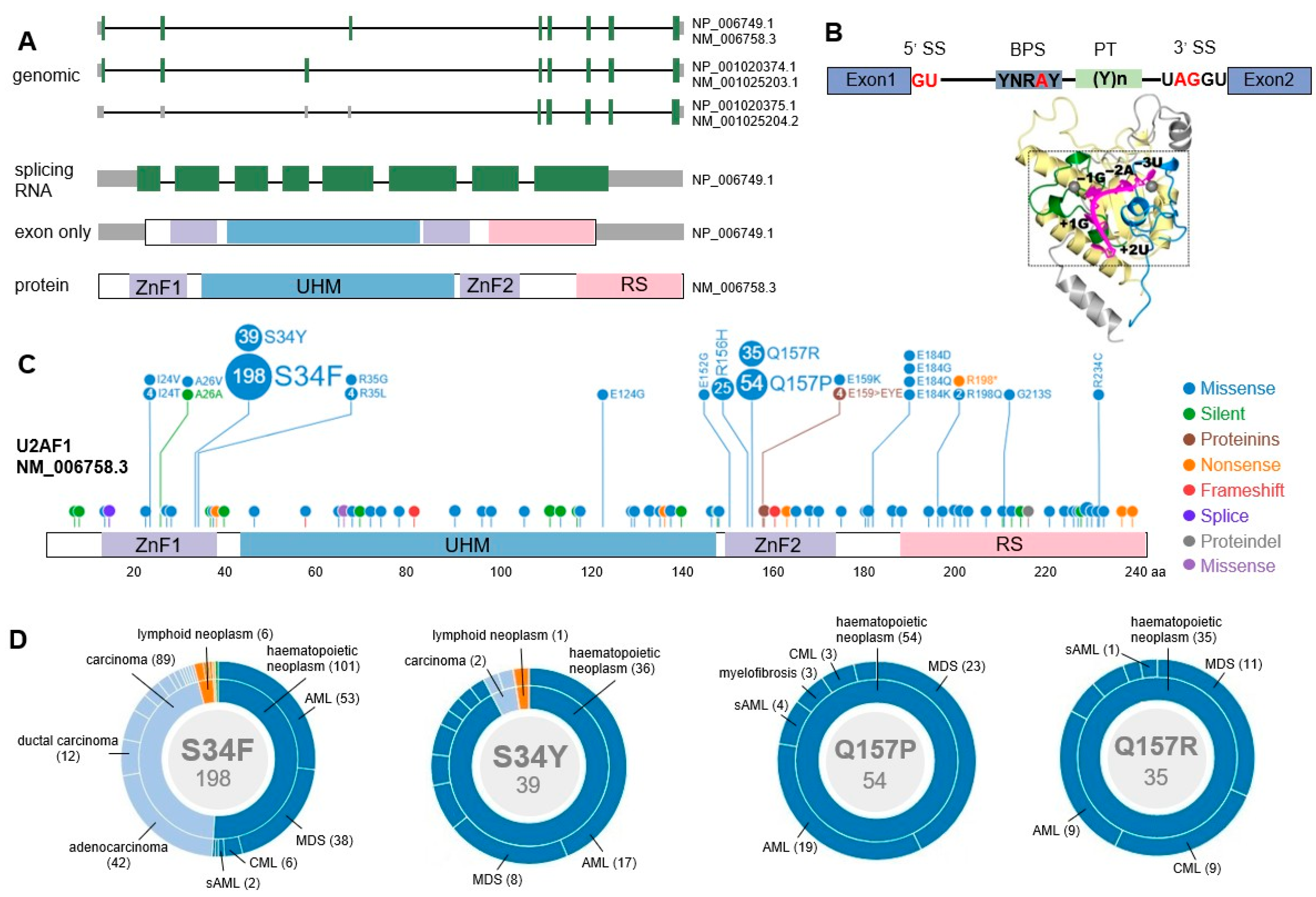
2.3. U2AF1 Gene Structure
3. U2AF1 Mutations in Myeloid Malignancies
3.1. Mutational Patterns of U2AF1
3.2. Correlations between U2AF1 Mutations and Clinical Features
3.3. Impacts of U2AF1 Mutation on Prognosis and Leukemic Transformation
4. U2AF1 Mutation Affects Hematopoietic Function and Target Genes
4.1. U2AF1 Mutation Impairs Hematopoietic Function
4.2. U2AF1 Mutation Affects Alternative Splicing
4.3. U2AF1 Mutation Alters Downstream Genes
4.4. Noncanonical Functions of U2AF1 Mutation
5. U2AF1 Mutation as a New Therapeutic Target
6. Discussion and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cazzola, M.; Della, P.M.; Malcovati, L. The genetic basis of myelodysplasia and its clinical relevance. Blood 2013, 122, 4021–4034. [Google Scholar] [CrossRef] [PubMed]
- Cazzola, M. Myelodysplastic syndromes. N. Engl. J. Med. 2020, 383, 1358–1374. [Google Scholar] [CrossRef] [PubMed]
- Menssen, A.J.; Walter, M.J. Genetics of progression from MDS to secondary leukemia. Blood 2020, 136, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango Ossa, J.E.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022, 1. [Google Scholar] [CrossRef]
- Jenkins, J.L.; Kielkopf, C.L. Splicing factor mutations in myelodysplasias: Insights from spliceosome structures. Trends Genet. 2017, 33, 336–348. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Zhou, Z.; Gong, Q.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark. Res. 2020, 8, 38. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005, 386, 713–724. [Google Scholar] [CrossRef]
- Chen, S.; Benbarche, S.; Abdel-Wahab, O. Splicing factor mutations in hematologic malignancies. Blood 2021, 138, 599–612. [Google Scholar] [CrossRef]
- Brooks, A.N.; Choi, P.S.; de Waal, L.; Sharifnia, T.; Imielinski, M.; Saksena, G.; Pedamallu, C.S.; Sivachenko, A.; Rosenberg, M.; Chmielecki, J.; et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE 2014, 9, e87361. [Google Scholar] [CrossRef] [PubMed]
- Je, E.M.; Yoo, N.J.; Kim, Y.J.; Kim, M.S.; Lee, S.H. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int. J. Cancer 2013, 133, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Przychodzen, B.; Jerez, A.; Guinta, K.; Sekeres, M.A.; Padgett, R.; Maciejewski, J.P.; Makishima, H. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood 2013, 122, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.C.; Lührmann, R. SnapShot: Spliceosome dynamics I. Cell 2015, 161, 1474. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wan, R.; Shi, Y. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Csh. Perspect Biol. 2019, 11, a32409. [Google Scholar] [CrossRef] [PubMed]
- Visconte, V.; Nakashima, M.O.; Rogers, H.J. Mutations in splicing factor genes in myeloid malignancies: Significance and impact on clinical features. Cancers 2019, 11, 1844. [Google Scholar] [CrossRef]
- Wahl, M.C.; Lührmann, R. SnapShot: Spliceosome dynamics II. Cell 2015, 162, 456. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.; Manley, J.L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013, 3, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.N.; Venugopal, P.; Scott, H.S.; Hiwase, D.K. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol. Rev. 2015, 263, 257–278. [Google Scholar] [CrossRef]
- Yoshida, H.; Park, S.; Sakashita, G.; Nariai, Y.; Kuwasako, K.; Muto, Y.; Urano, T.; Obayashi, E. Elucidation of the aberrant 3′ splice site selection by cancer-associated mutations on the U2AF1. Nat. Commun. 2020, 11, 4744. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Okeyo-Owuor, T.; White, B.S.; Chatrikhi, R.; Mohan, D.R.; Kim, S.; Griffith, M.; Ding, L.; Ketkar-Kulkarni, S.; Hundal, J.; Laird, K.M.; et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 2015, 29, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Adema, V.; Hirsch, C.M.; Przychodzen, B.P.; Nazha, A.; Kuzmanovic, T.; Negoro, E.; You, D.; Makishima, H.; Clemente, M.J.; Carraway, H.E.; et al. U2AF1 mutations in S34 and Q157 create distinct molecular and clinical contexts. Blood 2016, 128, 3155. [Google Scholar] [CrossRef]
- Damm, F.; Kosmider, O.; Gelsi-Boyer, V.; Renneville, A.; Carbuccia, N.; Hidalgo-Curtis, C.; Della, V.V.; Couronne, L.; Scourzic, L.; Chesnais, V.; et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 2012, 119, 3211–3218. [Google Scholar] [CrossRef]
- Pellagatti, A.; Armstrong, R.N.; Steeples, V.; Sharma, E.; Repapi, E.; Singh, S.; Sanchi, A.; Radujkovic, A.; Horn, P.; Dolatshad, H.; et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: Dysregulated genes/pathways and clinical associations. Blood 2018, 132, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Kade, S.; Schlarmann, C.; Löffeld, P.; Morgan, M.; Krauter, J.; Wlodarski, M.W.; Kölking, B.; Wichmann, M.; Görlich, K.; et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 2012, 119, 3578–3584. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tang, J.; Lin, C.; Kuo, Y.; Li, L.; Tseng, M.; Huang, C.; Lai, Y.; Lee, F.; Liu, M.; et al. Clinical implications of U2AF1 mutation in patients with myelodysplastic syndrome and its stability during disease progression. Am. J. Hematol. 2013, 88, E277–E282. [Google Scholar] [CrossRef] [PubMed]
- Shingai, N.; Harada, Y.; Iizuka, H.; Ogata, Y.; Doki, N.; Ohashi, K.; Hagihara, M.; Komatsu, N.; Harada, H. Impact of splicing factor mutations on clinical features in patients with myelodysplastic syndromes. Int. J. Hematol. 2018, 108, 598–606. [Google Scholar] [CrossRef]
- Kang, M.; Kim, H.; Seo, B.; Lee, J.H.; Choi, S.; Kim, S.; Shin, J.; Suh, S.; Ahn, J.; Shin, M. The prognostic impact of mutations in spliceosomal genes for myelodysplastic syndrome patients without ring sideroblasts. BMC Cancer 2015, 15. [Google Scholar] [CrossRef]
- Wu, L.; Song, L.; Xu, L.; Chang, C.; Xu, F.; Wu, D.; He, Q.; Su, J.; Zhou, L.; Xiao, C.; et al. Genetic landscape of recurrent ASXL1, U2AF1, SF3B1, SRSF2, and EZH2 mutations in 304 Chinese patients with myelodysplastic syndromes. Tumor Biol. 2016, 37, 4633–4640. [Google Scholar] [CrossRef]
- Graubert, T.A.; Shen, D.; Ding, L.; Okeyo-Owuor, T.; Lunn, C.L.; Shao, J.; Krysiak, K.; Harris, C.C.; Koboldt, D.C.; Larson, D.E.; et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2012, 44, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, N.; Wu, X.; Zheng, X.; Ling, Y.; Gong, Y. Prognostic value of U2AF1 mutant in patients with de novo myelodysplastic syndromes: A meta-analysis. Ann. Hematol. 2019, 98, 2629–2639. [Google Scholar] [CrossRef]
- Li, B.; Zou, D.; Yang, S.; Ouyang, G.; Mu, Q. Prognostic significance of U2AF1 mutations in myelodysplastic syndromes: A meta-analysis. J. Int. Med. Res. 2020, 48, 1410459245. [Google Scholar] [CrossRef] [PubMed]
- Gangat, N.; Mudireddy, M.; Lasho, T.L.; Finke, C.M.; Nicolosi, M.; Szuber, N.; Patnaik, M.M.; Pardanani, A.; Hanson, C.A.; Ketterling, R.P.; et al. Mutations and prognosis in myelodysplastic syndromes: Karyotype-adjusted analysis of targeted sequencing in 300 consecutive cases and development of a genetic risk model. Am. J. Hematol. 2018, 93, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Mudireddy, M.; Finke, C.M.; Nicolosi, M.; Lasho, T.L.; Hanson, C.A.; Patnaik, M.M.; Pardanani, A.; Gangat, N. U2AF1 mutation variants in myelodysplastic syndromes and their clinical correlates. Am. J. Hematol. 2018, 93, E146–E148. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Patnaik, M.M.; Saeed, L.; Mudireddy, M.; Idossa, D.; Finke, C.; Ketterling, R.P.; Pardanani, A.; Gangat, N. Targeted next-generation sequencing in myelodysplastic syndromes and prognostic interaction between mutations and IPSS-R. Am. J. Hematol. 2017, 92, 1311–1317. [Google Scholar] [CrossRef]
- Bejar, R.; Stevenson, K.E.; Caughey, B.A.; Abdel-Wahab, O.; Steensma, D.P.; Galili, N.; Raza, A.; Kantarjian, H.; Levine, R.L.; Neuberg, D.; et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 2012, 30, 3376–3382. [Google Scholar] [CrossRef]
- Li, B.; Liu, J.; Jia, Y.; Wang, J.; Xu, Z.; Qin, T.; Shi, Z.; Song, Z.; Peng, S.; Huang, H.; et al. Clinical features and biological implications of different U2AF1 mutation types in myelodysplastic syndromes. Genes Chromosomes Cancer 2018, 57, 80–88. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Dong, Z.; Li, T.; Xie, X.; Wan, D.; Jiang, Z.; Yu, J.; Guo, R. Differential U2AF1 mutation sites, burden and co-mutation genes can predict prognosis in patients with myelodysplastic syndrome. Sci. Rep. UK 2020, 10, 18622. [Google Scholar] [CrossRef]
- Bamopoulos, S.A.; Batcha, A.M.N.; Jurinovic, V.; Rothenberg-Thurley, M.; Janke, H.; Ksienzyk, B.; Philippou-Massier, J.; Graf, A.; Krebs, S.; Blum, H.; et al. Clinical presentation and differential splicing of SRSF2, U2AF1 and SF3B1 mutations in patients with acute myeloid leukemia. Leukemia 2020, 34, 2621–2634. [Google Scholar] [CrossRef]
- Ohgami, R.S.; Ma, L.; Merker, J.D.; Gotlib, J.R.; Schrijver, I.; Zehnder, J.L.; Arber, D.A. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod. Pathol. 2015, 28, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Saygin, C.; Hirsch, C.; Przychodzen, B.; Sekeres, M.A.; Hamilton, B.K.; Kalaycio, M.; Carraway, H.E.; Gerds, A.T.; Mukherjee, S.; Nazha, A.; et al. Mutations in DNMT3A, U2AF1, and EZH2 identify intermediate-risk acute myeloid leukemia patients with poor outcome after CR1. Blood Cancer J. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Yao, D.M.; Lin, J.; Qian, W.; Wang, C.Z.; Chai, H.Y.; Yang, J.; Li, Y.; Deng, Z.Q.; Ma, J.C.; et al. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE 2012, 7, e45760. [Google Scholar] [CrossRef] [PubMed]
- Meggendorfer, M.; Eder, C.; Jeromin, S.; Haferlach, C.; Kern, W.; Haferlach, T.; Schnittger, S. Comprehensive analysis of U2AF1 mutations in 843 patients with myeloid neoplasms with respect to other genetic alterations. Blood 2013, 122, 4973. [Google Scholar] [CrossRef]
- Meggendorfer, M.; Haferlach, C.; Kern, W.; Haferlach, T.; Schnittger, S. The two mutation hot spots Ser34 and Gln157 in U2AF1 show different occurrence, correlation and clinical features in myeloid malignancies: An analysis of 785 cases. Blood 2012, 120, 3789. [Google Scholar] [CrossRef]
- Xie, Z.; Nanaa, A.; Saliba, A.N.; He, R.; Viswanatha, D.; Nguyen, P.; Jevremovic, D.; Greipp, P.; Salama, M.E.; Gangat, N.; et al. Treatment outcome of clonal cytopenias of undetermined significance: A single-institution retrospective study. Blood Cancer J. 2021, 11. [Google Scholar] [CrossRef]
- Malcovati, L.; Gallì, A.; Travaglino, E.; Ambaglio, I.; Rizzo, E.; Molteni, E.; Elena, C.; Ferretti, V.V.; Catricalà, S.; Bono, E.; et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood 2017, 129, 3371–3378. [Google Scholar] [CrossRef]
- Komrokji, R.S.; Al Ali, N.; Hussaini, M.O.; Sallman, D.A.; Rollison, D.E.; Padron, E. U2AF1 and EZH2 mutations are associated with non-immune hemolytic anemia in myelodysplastic syndromes. Blood 2020, 136, 35–36. [Google Scholar] [CrossRef]
- Tefferi, A.; Finke, C.M.; Lasho, T.L.; Wassie, E.A.; Knudson, R.; Ketterling, R.P.; Hanson, C.A.; Pardanani, A. U2AF1 mutations in primary myelofibrosis are strongly associated with anemia and thrombocytopenia despite clustering with JAK2V617F and normal karyotype. Leukemia 2014, 28, 431–433. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, K.; Hwang, B.; Im, K.; Park, S.N.; Kim, J.; Hwang, S.M.; Bang, D.; Lee, D.S. The high frequency of the U2AF1 S34Y mutation and its association with isolated trisomy 8 in myelodysplastic syndrome in Asians, but not in Caucasians. Leuk. Res. 2017, 61, 96–103. [Google Scholar] [CrossRef]
- Hwang, S.M.; Kim, S.Y.; Kim, J.A.; Park, H.; Park, S.N.; Im, K.; Kim, K.; Kim, S.; Lee, D.S. Short telomere length and its correlation with gene mutations in myelodysplastic syndrome. J. Hematol. Oncol. 2016, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Song, D.; Guo, J.; Jin, J.; Tao, Y.; Zhang, Z.; Xu, F.; He, Q.; Li, X.; Chang, C.; et al. U2AF1 mutation promotes tumorigenicity through facilitating autophagy flux mediated by FOXO3a activation in myelodysplastic syndromes. Cell Death Dis. 2021, 12, 655. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.Y.; Seo, J.Y.; Kim, S.H.; Jung, H.A.; Park, S.; Kim, K.; Jung, C.W.; Kim, J.S.; Park, J.S.; Kim, H.J.; et al. Mutations in the spliceosomal machinery genes SRSF2, U2AF1, and ZRSR2 and response to decitabine in myelodysplastic syndrome. Anticancer. Res. 2015, 35, 3081–3089. [Google Scholar] [PubMed]
- Yip, B.H.; Steeples, V.; Repapi, E.; Armstrong, R.N.; Llorian, M.; Roy, S.; Shaw, J.; Dolatshad, H.; Taylor, S.; Verma, A.; et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J. Clin. Investig. 2017, 127, 2206–2221. [Google Scholar] [CrossRef]
- Fei, D.L.; Zhen, T.; Durham, B.; Ferrarone, J.; Zhang, T.; Garrett, L.; Yoshimi, A.; Abdel-Wahab, O.; Bradley, R.K.; Liu, P.; et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor geneU2af1. Proc. Natl. Acad. Sci. USA 2018, 115, E10437–E10446. [Google Scholar] [CrossRef]
- Shirai, C.L.; Ley, J.N.; White, B.S.; Kim, S.; Tibbitts, J.; Shao, J.; Ndonwi, M.; Wadugu, B.; Duncavage, E.J.; Okeyo-Owuor, T.; et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015, 27, 631–643. [Google Scholar] [CrossRef]
- Alberti, M.O.; Zhu, M.; Nonavinkere Srivatsan, S.; Ahmed, T.; Shao, J.J.; Wadugu, B.; Shirai, C.L.; Walter, M.J. Mutant U2AF1S34F and U2AF1Q157P induce distinct RNA splicing and hematopoietic phenotypes in vivo. Blood 2019, 134, 770. [Google Scholar]
- Dutta, A.; Yang, Y.; Le, B.; Mohi, G. The RNA splicing factor U2AF1 controls hematopoietic stem cell survival and function. Blood 2020, 136, 9. [Google Scholar] [CrossRef]
- Dutta, A.; Yang, Y.; Le, B.T.; Zhang, Y.; Abdel-Wahab, O.; Zang, C.; Mohi, G. U2af1 is required for survival and function of hematopoietic stem/progenitor cells. Leukemia 2021, 35, 2382–2398. [Google Scholar] [CrossRef]
- Martínez-Valiente, C.; Garcia-Ruiz, C.; Rosón, B.; Liquori, A.; González-Romero, E.; Fernández-González, R.; Gómez-Redondo, I.; Cervera, J.; Gutiérrez-Adán, A.; Sanjuan-Pla, A. Aberrant alternative splicing in U2af1/Tet2 double mutant mice contributes to major hematological phenotypes. Int. J. Mol. Sci. 2021, 22, 6963. [Google Scholar] [CrossRef]
- Biancon, G.; Joshi, P.; Zimmer, J.T.; Hunck, T.; Gao, Y.; Lessard, M.D.; Courchaine, E.; Barentine, A.; Machyna, M.; Botti, V.; et al. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol. Cell 2022, 82, 1107–1122. [Google Scholar] [CrossRef] [PubMed]
- Ilagan, J.O.; Ramakrishnan, A.; Hayes, B.; Murphy, M.E.; Zebari, A.S.; Bradley, P.; Bradley, R.K. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015, 25, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, X.; Cao, Z.; Qiu, S.; Li, Y.; Zhong, M.; Xue, Z.; Xu, Y.; Xing, H.; Tang, K.; et al. Mutant U2AF1-induced differential alternative splicing causes an oxidative stress in bone marrow stromal cells. Exp. Biol. Med. 2021, 246, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Cheruiyot, A.; Li, S.; Nonavinkere Srivatsan, S.; Ahmed, T.; Chen, Y.; Lemacon, D.S.; Li, Y.; Yang, Z.; Wadugu, B.A.; Warner, W.A.; et al. Nonsense-mediated RNA decay is a unique vulnerability of cancer cells harboring SF3B1 or U2AF1 mutations. Cancer Res. 2021, 81, 4499–4513. [Google Scholar] [CrossRef]
- Kim, S.P.; Srivatsan, S.N.; Chavez, M.; Shirai, C.L.; White, B.S.; Ahmed, T.; Alberti, M.O.; Shao, J.; Nunley, R.; White, L.S.; et al. Mutant U2AF1-induced alternative splicing of H2afy (macroH2A1) regulates B-lymphopoiesis in mice. Cell Rep. 2021, 36, 109626. [Google Scholar] [CrossRef]
- Park, S.M.; Ou, J.; Chamberlain, L.; Simone, T.M.; Yang, H.; Virbasius, C.; Ali, A.M.; Zhu, L.J.; Mukherjee, S.; Raza, A.; et al. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3′ end formation. Mol. Cell 2016, 62, 479–490. [Google Scholar] [CrossRef]
- Basiorka, A.A.; Mcgraw, K.L.; Eksioglu, E.A.; Chen, X.; Johnson, J.; Zhang, L.; Zhang, Q.; Irvine, B.A.; Cluzeau, T.; Sallman, D.A.; et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 2016, 128, 2960–2975. [Google Scholar] [CrossRef]
- Smith, M.A.; Choudhary, G.S.; Pellagatti, A.; Choi, K.; Bolanos, L.C.; Bhagat, T.D.; Gordon-Mitchell, S.; Von Ahrens, D.; Pradhan, K.; Steeples, V.; et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 2019, 21, 640–650. [Google Scholar] [CrossRef]
- Akef, A.; Mcgraw, K.; Cappell, S.D.; Larson, D.R. Ribosome biogenesis is a downstream effector of the oncogenic U2AF1-S34F mutation. PLoS Biol. 2020, 18, e3000920. [Google Scholar] [CrossRef]
- Palangat, M.; Anastasakis, D.G.; Fei, D.L.; Lindblad, K.E.; Bradley, R.; Hourigan, C.S.; Hafner, M.; Larson, D.R. The splicing factor U2AF1 contributes to cancer progression through a noncanonical role in translation regulation. Genes Dev. 2019, 33, 482–497. [Google Scholar]
- Wadugu, B.A.; Nonavinkere Srivatsan, S.; Heard, A.; Alberti, M.O.; Ndonwi, M.; Liu, J.; Grieb, S.; Bradley, J.; Shao, J.; Ahmed, T.; et al. U2af1 is a haplo-essential gene required for hematopoietic cancer cell survival in mice. J. Clin. Investig. 2021, 131, e141401. [Google Scholar] [CrossRef] [PubMed]
- Shirai, C.L.; White, B.S.; Tripathi, M.; Tapia, R.; Ley, J.N.; Ndonwi, M.; Kim, S.; Shao, J.; Carver, A.; Saez, B.; et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun. 2017, 8, 14060. [Google Scholar] [CrossRef] [PubMed]
| Disease (Frequency) | Mutation Types | Sample | Clinical Consequences | Ref. |
|---|---|---|---|---|
| MDS (5%) | S34, Q157, Q84, E124, E152, R156 | Targeted deep sequencing of 1700 patients with myeloid neoplasms | Ancestral U2AF1 mutations predict shorter OS compared to secondary mutations; S34 co-occurs with ETV6, BCOR, and CUX1; Q157 co-occurs with ASXL1 and DNMT3A | [23] |
| MDS (5.4%) | S34F/Y, Q157R/P/fs | Mutation analysis of 221 MDS patients | Associated with chromosome 20 deletions and ASXL1 mutation; Not associated with the risk of progression to sAML | [24] |
| MDS (7.1%) | S34, Q157, R156 | Targeted next-generation sequencing of 84 MDS patients | Associated with a high proportion of exon skipping and retained introns events | [25] |
| MDS (7.3%) | S34F, Q157P, E159fs | Sequencing analysis of 193 MDS patients | Positively correlated with ASXL1 and DNMT3A mutation; Not associated with the presence of ring sideroblasts; Have no impact on patient survival; Have a trend toward a more rapid progression to AML; (Patients received treatments including all-trans retinoic acid, antithymocyte globulin, deferasirox, lenalidomide, or thalidomide) | [26] |
| MDS (7.5%) | S34F/Y, Q157R/P | Direct sequencing in 478 patients with de novo MDS | Positively correlated with isolated −20/20q-, ASXL1, RUNX1, and DNMT3A mutations, negatively correlated with SRSF2 mutation; Independent poor-risk factor for OS in MDS patients; Predict shorter time-to-leukemia transformation | [27] |
| MDS (7.5%) | S34F/Y, Q157P | Sanger sequencing of 106 MDS patients | Associated with low mean corpuscular volume and myeloid to erythroid ratio; Have no impact on OS | [28] |
| MDS (7.8%) | S34F/Y, Q157P | Mutation analyses of 129 de novo MDS patients without ring sideroblasts | Associated with low hemoglobin levels and high-risk MDS; Predict poor PFS; Associated with inferior OS in low-risk MDS patients | [29] |
| MDS (8.6%) | S34F/Y, Q157P | Next-generation sequencing of 304 Chinese MDS patients | More common in patients with trisomy 8 or 20q deletions; Predict poor OS in MDS patients | [30] |
| MDS (8.7%) | S34F/Y | Sanger sequencing of 150 patients with de novo MDS | Enhance splicing and exon skipping; Increased risk of progression to sAML | [31] |
| MDS (5–17%, 11.7%) | S34, Q157, R156 | Meta-analysis of 14 studies with 3322 MDS patients | Independent, detrimental prognostic factors for OS and AML transformation | [32] |
| MDS (11.7%) | S34, Q157 | Meta-analysis of 13 studies with 3038 MDS patients | Associated with poor OS, but not DFS; Q157 mutation predicts worse OS than S34; Have no impact on hypomethylating therapy | [33] |
| MDS (14%) | - | Targeted capture assays of 300 primary MDS patients | More prevalent in the intermediate-risk cytogenetic category; Unfavorable survival impact | [34] |
| MDS (15%) | S34F/Y, Q157R/P, R156H | Next-generation sequencing of 357 primary MDS patients | Not associated with anemia; Q157 positively correlated with ASXL1 mutation; Q157 mutation has adverse survival impact | [35] |
| MDS (16%) | - | Next-generation sequencing of 179 primary MDS patients | Associated with lower-risk karyotype and platelet count; Adverse survival impact | [36] |
| MDS with lower risk (16%) | - | DNA sequencing of 288 patients with MDS | Associated with low platelet count and shorter overall survival | [37] |
| MDS (17%) | S34F/Y, Q157R/P, R156H | Targeted gene sequencing of 511 MDS patients | Associated with anemia, thrombocytopenia, ASXL1 mutation, isolated +8, and poor survival; Inversely associated with TP53, SF3B1 mutations, and complex karyotypes | [38] |
| MDS (21.7%) | S34F/Y, Q157P | Retrospective analysis of the next-generation sequencing data of 234 MDS patients | Positively correlated with ASXL1, RUNX1, and SETBP1 mutation; negatively correlated with SF3B1 and NPM1 mutation; VAF > 40% of U2AF1 is an independent indicator for poor OS of MDS patients | [39] |
| AML (3.4%) | S34F/Y, R35Q | Nanopore sequencing of 1119 AML patients | Predict poor OS in AML patients | [40] |
| AML (4%) | S34F/Y | Somatic mutation data from TCGA AML patients | Preferentially exhibit alterations in cassette exon and alternative 3′SS; Preferentially splices to CAG rather than UAG | [11] |
| AML (6.5%) | - | Targeted next-generation sequencing of 93 AML patients | Associated with AML with myelodysplasia-related changes and trilineage morphologic dysplasia; Associated with the absence of clinical remission, poor OS and DFS | [41] |
| AML (11%) | - | Targeted sequencing in 100 intermediate-risk AML patients | Predict poor OS and RFS | [42] |
| MDS (19.7%) sAML (4.6%) | S34F/Y | DNA sequencing of 2345 tumor tissues | - | [12] |
| MDS (6.3%) AML (2.5%) CML (0%) | S34F/Y, Q157R/P | Mutation scanning of 275 primary AML, 96 primary MDS, and 81 CML Chinese patients | Predict poor OS, but not DFS, in AML patients; Have no impact on OS in MDS patients | [43] |
| MDS without RS (11.6%) CML (8%) sAML (9.7%) AML (1.3%) MPN (1.9%) | S34F/Y, Q157R/P, A26V | Whole-exome sequencing of paired tumor/control DNA from 29 patients with myelodysplasia | Suppress cell proliferation and induce apoptosis; Induce abnormal RNA splicing and compromised hematopoiesis | [21] |
| MDS (10%) sAML and AML (7%) MPN (8%) MDS/MPN (14.5%) MDS with high risk (14%) | S34F/Y, Q157R/P, A26V, R35L, R156Q, G213A | Sanger sequencing and exome sequencing of 524 patients with hematologic malignancies | Associated with exon skipping; Induce abnormal splicing of genes in important pathways | [13] |
| MDS (10.9%) AML (9.5%) MDS/MPN (7.1%) MPN (1.2%) | S34, Q157 | Melting curve analyses or next-generation sequencing of 843 patients | Associated with lower hemoglobin levels and platelet counts; Associated with del(20q) in MDS, AML, and MDS/MPN | [44] |
| MDS (S34 14.6%, Q157 1.1%) AML (S34 12.3%, Q157 0%) MDS/MPN (S34 2.2%, Q157 3.5%) MPN (S34 0.6%, Q157 0.6%) | S34, Q157 | Melting curve analysis of 785 patients | S34 mutation associated with low hemoglobin level and platelet count; Associated with del(20q) in MDS | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Cai, W.; Hua, Y.; Yang, X.; Zhou, J. The Biological and Clinical Consequences of RNA Splicing Factor U2AF1 Mutation in Myeloid Malignancies. Cancers 2022, 14, 4406. https://doi.org/10.3390/cancers14184406
Zhao Y, Cai W, Hua Y, Yang X, Zhou J. The Biological and Clinical Consequences of RNA Splicing Factor U2AF1 Mutation in Myeloid Malignancies. Cancers. 2022; 14(18):4406. https://doi.org/10.3390/cancers14184406
Chicago/Turabian StyleZhao, Yangjing, Weili Cai, Ye Hua, Xiaochen Yang, and Jingdong Zhou. 2022. "The Biological and Clinical Consequences of RNA Splicing Factor U2AF1 Mutation in Myeloid Malignancies" Cancers 14, no. 18: 4406. https://doi.org/10.3390/cancers14184406
APA StyleZhao, Y., Cai, W., Hua, Y., Yang, X., & Zhou, J. (2022). The Biological and Clinical Consequences of RNA Splicing Factor U2AF1 Mutation in Myeloid Malignancies. Cancers, 14(18), 4406. https://doi.org/10.3390/cancers14184406






