Genomic Landscape of Vinflunine Response in Metastatic Urothelial Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Next Generation Sequencing and Variant Calling
2.3. Gene Expression Profiles
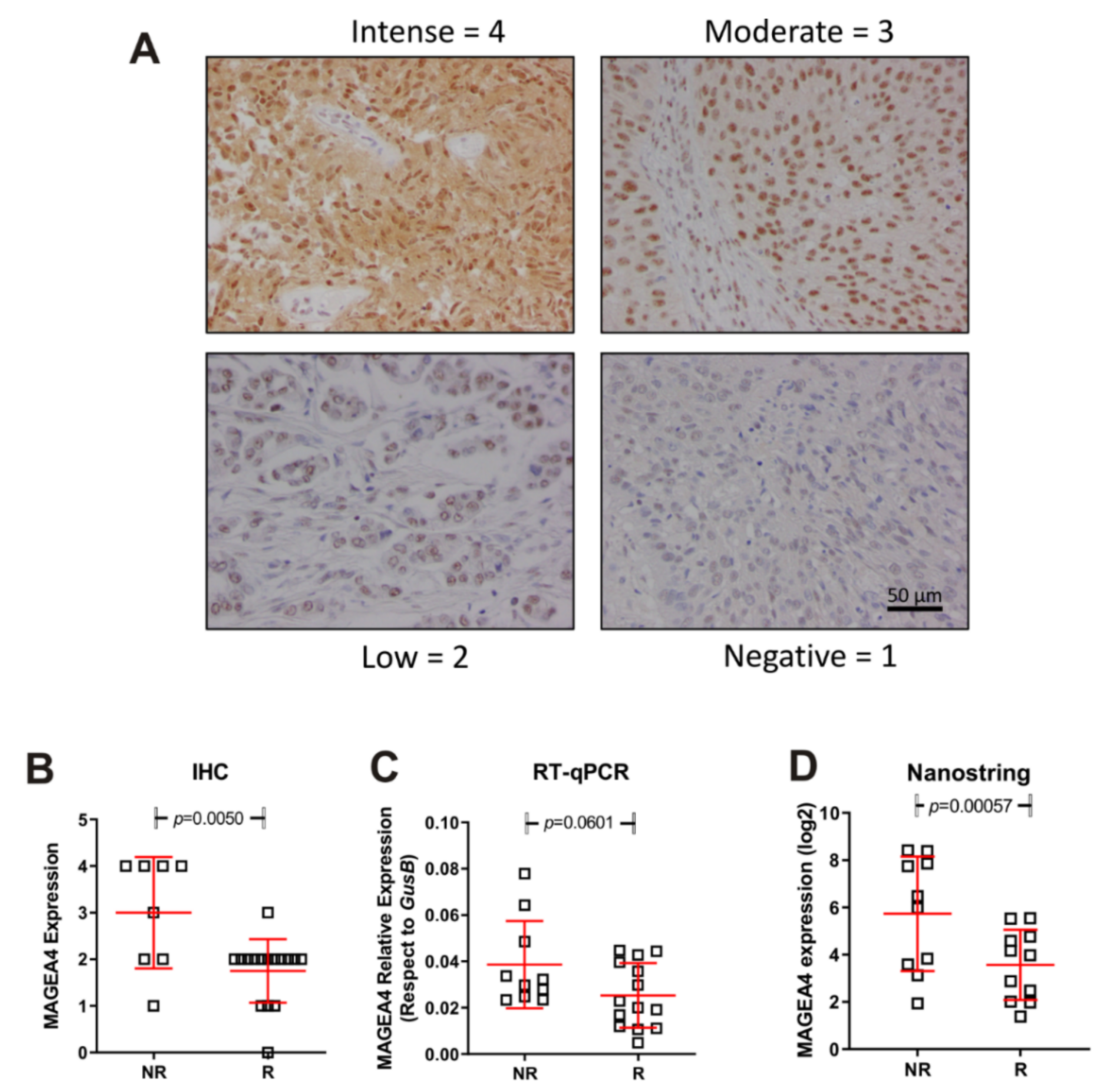
2.4. Immunohistochemistry and Real Time Quantitative PCR for MAGEA4
2.5. Statistical Analysis
2.6. Data Availability
3. Results
3.1. Clinical Characteristics
3.2. Somatic Genetic Alterations
3.3. Tumour Immune Expression Is Associated with Differences in Vinflunine Efficacy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellmunt, J.; Orsola, A.; Leow, J.J.; Wiegel, T.; De Santis, M.; Horwich, A.; On behalf of the ESMO Guidelines Working Group. Bladder cancer: ESMO Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. 3), iii40–iii48. [Google Scholar] [CrossRef]
- Bellmunt, J.; von der Maase, H.; Mead, G.M.; Skoneczna, I.; De Santis, M.; Daugaard, G.; Boehle, A.; Chevreau, C.; Paz-Ares, L.; Laufman, L.R.; et al. Randomized phase III study comparing paclitaxel/cisplatin/gemcitabine and gemcitabine/cisplatin in patients with locally advanced or metastatic urothelial cancer without prior systemic therapy: EORTC Intergroup Study 30987. J. Clin. Oncol. 2012, 30, 1107–1113. [Google Scholar] [CrossRef]
- Bellmunt, J.; De Wit, R.; Vaughn, D.J.; Fradet, Y.; Lee, J.L.; Fong, L.; Vogelzang, N.J.; Climent, M.A.; Petrylak, D.P.; Choueiri, T.K.; et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N. Engl. J. Med. 2017, 376, 1015–1026. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; Van Der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef] [Green Version]
- Powles, T.; Park, S.H.; Voog, E.; Caserta, C.; Valderrama, B.P.; Gurney, H.; Kalofonos, H.; Radulović, S.; Demey, W.; Ullén, A.; et al. Avelumab Maintenance Therapy for Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2020, 383, 1218–1230. [Google Scholar] [CrossRef]
- Spiess, P.E.; Agarwal, N.; Bangs, R.; Boorjian, S.A.; Buyyounouski, M.K.; Clark, P.E.; Downs, T.M.; Efstathiou, J.A.; Flaig, T.W.; Friedlander, T.; et al. Bladder Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2017, 15, 1240–1267. [Google Scholar] [CrossRef] [Green Version]
- Powles, T.; Rosenberg, J.E.; Sonpavde, G.P.; Loriot, Y.; Durán, I.; Lee, J.L.; Matsubara, N.; Vulsteke, C.; Castellano, D.; Wu, C.; et al. Enfortumab Vedotin in Previously Treated Advanced Urothelial Carcinoma. N. Engl. J. Med. 2021, 384, 1125–1135. [Google Scholar] [CrossRef]
- Bellmunt, J.; Théodore, C.; Demkov, T.; Komyakov, B.; Sengelov, L.; Daugaard, G.; Caty, A.; Carles, J.; Jagiello-Gruszfeld, A.; Karyakin, O.; et al. Phase III trial of vinflunine plus best supportive care compared with best supportive care alone after a platinum-containing regimen in patients with advanced transitional cell carcinoma of the urothelial tract. J. Clin. Oncol. 2009, 27, 4454–4461. [Google Scholar] [CrossRef]
- Ng, J.S. Vinflunine: Review of a new vinca alkaloid and its potential role in oncology. J. Oncol. Pharm. Pract. 2011, 17, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Bellmunt, J.; Fougeray, R.; Rosenberg, J.E.; Von der Maase, H.; Schutz, F.A.; Salhi, Y.; Culine, S.; Choueiri, T.K. Long-term survival results of a randomized phase III trial of vinflunine plus best supportive care versus best supportive care alone in advanced urothelial carcinoma patients after failure of platinum-based chemotherapy. Ann. Oncol. 2013, 24, 1466–1472. [Google Scholar] [CrossRef]
- Brousell, S.C.; Fantony, J.J.; Van Noord, M.G.; Harrison, M.R.; Inman, B.A. Vinflunine for the treatment of advanced or metastatic transitional cell carcinoma of the urothelial tract: An evidence-based review of safety, efficacy, and place in therapy. Core Evid. 2018, 13, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Ranjbar, M.R.N.; Wu, Z.; DiCarlo, J.; Wang, Y. Detecting very low allele fraction variants using targeted DNA sequencing and a novel molecular barcode-aware variant caller. BMC Genom. 2017, 18, 5. [Google Scholar] [CrossRef] [Green Version]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Mutational Signatures in Cancer (MuSiCa): A Web Application to Implement Mutational Signatures Analysis in Cancer Samples n.d. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6001047/ (accessed on 18 January 2021).
- Cesano, A. nCounter® PanCancer Immune Profiling Panel (NanoString Technologies, Inc., Seattle, WA). J. Immunother. Cancer 2015, 3, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellmunt, J.; Paz-Ares, L.; Cuello, M.; Cecere, F.L.; Albiol, S.; Guillem, V.; Gallardo, E.; Carles, J.; Mendez, P.; De la Cruz, J.J.; et al. Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Ann. Oncol. 2007, 18, 522–528. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Mouw, K.W.; Kim, P.; Iyer, G.; Wagle, N.; Al-Ahmadie, H.; Zhu, C.; Ostrovnaya, I.; Kryukov, G.V.; O’Connor, K.W.; et al. Somatic ERCC2 Mutations Correlate with Cisplatin Sensitivity in Muscle-Invasive Urothelial Carcinoma. Cancer Discov. 2014, 4, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Shen, Y.; Wen, S.; Yamada, S.; Jungbluth, A.A.; Gnjatic, S.; Bajorin, D.F.; Reuter, V.E.; Herr, H.; Old, L.J.; et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc. Natl. Acad. Sci. USA 2007, 104, 3967–3972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.S.; Su, H.Y.L.; Li, P.H.; Chiang, P.H.; Huang, C.H.; Chen, C.H.; Hsieh, M.C. Prognostic impact of tumor infiltrating lymphocytes on patients with metastatic urothelial carcinoma receiving platinum based chemotherapy. Sci. Rep. 2018, 8, 7485. [Google Scholar] [CrossRef] [PubMed]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogunovic, D.; Byun, M.; Durfee, L.A.; Abhyankar, A.; Sanal, O.; Mansouri, D.; Salem, S.; Radovanovic, I.; Grant, A.V.; Adimi, P.; et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 2012, 337, 1684–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Bao, L.; Dunham, K.; Lucas, K. MAGE-A1, MAGE-A3, and NY-ESO-1 can be upregulated on neuroblastoma cells to facilitate cytotoxic T lymphocyte-mediated tumor cell killing. Cancer Immunol. Immunother. 2011, 60, 1299. [Google Scholar] [CrossRef] [PubMed]
- de Jong, J.J.; Boormans, J.L.; van Rhijn, B.W.; Seiler, R.; Boorjian, S.A.; Konety, B.; Bivalacqua, T.J.; Wheeler, T.; Svatek, R.S.; Douglas, J.; et al. Distribution of Molecular Subtypes in Muscle-invasive Bladder Cancer Is Driven by Sex-specific Differences. Eur. Urol. Oncol. 2020, 3, 420–423. [Google Scholar] [CrossRef] [PubMed]



| Responders | Non-Responders (n = 11) | Global | p Value | |
|---|---|---|---|---|
| (n = 12) | (n = 23) | |||
| Male (%)/Female (%) | 11 (19%)/1 (9%) | 8 (72%)/3 (28%) | 19 (82%)/4 (18%) | 0.23 |
| Age media (SD) | 67 (9.2) | 66 (11.4) | 66 | 0.57 |
| Smokers | 8 (66%) | 7 (63%) | 15 (65%) | 0.54 |
| Urothelial carcinoma | 12 (100%) | 11 (100) | 100 (100%) | |
| 1 Line | ||||
| 0 | 6 (50%) | 5 (45%) | 11 (48%) | |
| 1 | 6 (50%) | 5 (45%) | 11 (48%) | |
| 2 | 0 | 1 (10%) | 1 (4%) | 0.56 |
| Prior platinum based CT | ||||
| Cisplatin | 10 (83%) | 5 (45%) | 15 (65%) | |
| Carboplatin | 2 (17%) | 6 (55%) | 8 (35%) | 0.06 |
| Response to platinum based CT | ||||
| Partial response | 1 (8%) | 5 (45%) | 6 (26%) | |
| Stable disease | 6 (50%) | 5 (45%) | 11 (48%) | |
| Progressive disease | 5 (42%) | 1 (9%) | 6 (26%) | 0.06 |
| Number of line for vinflunine | ||||
| 2 | 9 (81%) | 10 (90%) | 19 (83%) | |
| 3 | 1 (8%) | 1 (9%) | 2 (8.6%) | |
| 4 | 2 (16%) | 0 | 2 (8.6%) | 0.36 |
| Location of metastases | ||||
| Lung | 5 (42%) | 4 (36%) | 9 (39%) | 0.79 |
| Lymph nodes | 9 (81%) | 7 (27%) | 16 (69%) | 0.55 |
| Liver | 1 (8%) | 3 (45%) | 4 (17%) | 0.23 |
| Bone | 4 (36%) | 5 (27/) | 9 (39%) | 0.52 |
| Median PFS (months) of vinflunine treatment (CI 95%) | 10 (8.7–11.5) | 2.2 (1.8–2.5) | 6 (0–15.5) | 0.02 |
| Median OS (months) of vinflunine treatment | 19.9 (10.7–29.0) | 2.5 (2.1–2.8) | 11.5 (3.4–19.5) | 0.06 |
| Gene | Log2 Fold Change | Std Error (log2) | Lower Confidence Limit (log2) | Upper Confidence Limit (log2) | p-Value | BY.p-Value | Gene.Sets |
|---|---|---|---|---|---|---|---|
| MAGEA12- | 3.58 | 0.726 | 2.16 | 5 | 0.0000927 | 0.485 | CT Antigen |
| IFIT1- | 1.99 | 0.458 | 1.09 | 2.89 | 0.000311 | 0.497 | Chemokines |
| ISG15- | 1.58 | 0.375 | 0.84 | 2.31 | 0.000442 | 0.497 | |
| IFITM1 | 1.89 | 0.452 | 1 | 2.77 | 0.000464 | 0.497 | Regulation |
| IFI27- | 2.27 | 0.555 | 1.19 | 3.36 | 0.000557 | 0.497 | Chemokines |
| MAGEA4- | 2.69 | 0.652 | 1.41 | 3.97 | 0.00057 | 0.497 | CT Antigen |
| Immune Pathway | Signature Scores (NR/R) |
|---|---|
| Regulation | 3.52 |
| Chemokines | 3.22 |
| Cytokines | 2.06 |
| Cell Functions | 1.94 |
| T-Cell Functions | 1.83 |
| TNF Superfamily | 1.59 |
| Antigen Processing | 1.43 |
| Pathogen Defense | 1.40 |
| Interleukins | 1.30 |
| Adhesion | 1.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardini, A.; Dueñas, M.; Martín-Soberon, M.C.; Rubio, C.; Suarez-Cabrera, C.; Ruiz-Palomares, R.; Munera-Maravilla, E.; Lázaro, S.; Lodewijk, I.; Rueda, D.; et al. Genomic Landscape of Vinflunine Response in Metastatic Urothelial Cancer. Cancers 2022, 14, 378. https://doi.org/10.3390/cancers14020378
Bernardini A, Dueñas M, Martín-Soberon MC, Rubio C, Suarez-Cabrera C, Ruiz-Palomares R, Munera-Maravilla E, Lázaro S, Lodewijk I, Rueda D, et al. Genomic Landscape of Vinflunine Response in Metastatic Urothelial Cancer. Cancers. 2022; 14(2):378. https://doi.org/10.3390/cancers14020378
Chicago/Turabian StyleBernardini, Alejandra, Marta Dueñas, María Cruz Martín-Soberon, Carolina Rubio, Cristian Suarez-Cabrera, Raquel Ruiz-Palomares, Ester Munera-Maravilla, Sara Lázaro, Iris Lodewijk, Daniel Rueda, and et al. 2022. "Genomic Landscape of Vinflunine Response in Metastatic Urothelial Cancer" Cancers 14, no. 2: 378. https://doi.org/10.3390/cancers14020378







