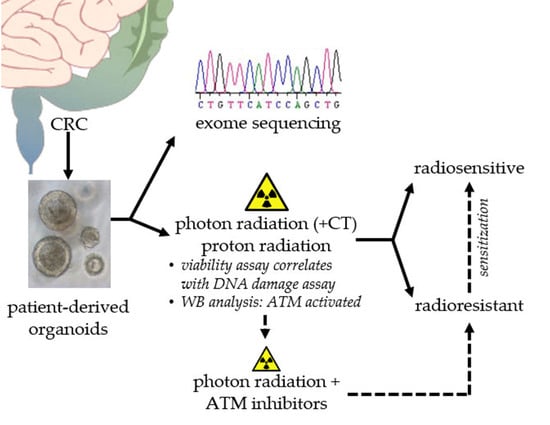
Sensitization of Patient-Derived Colorectal Cancer Organoids to Photon and Proton Radiation by Targeting DNA Damage Response Mechanisms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Tissues
2.2. Establishment and Culture of Human CRC Organoids
2.3. Next-Generation Sequencing
2.4. Treatment with (Chemo-)Radiotherapy and Inhibitors
2.5. Viability Assay
2.6. DNA Damage Repair Assay
2.7. Western Blot Analysis
- Phospho-ATM (Ser1981) (D6H9) rabbit monoclonal antibody #5883;
- ATM (D2E2) rabbit monoclonal antibody #2873;
- GAPDH (14C10) rabbit monoclonal antibody #2118;
- anti-mouse IgG HRP linked secondary antibody #7076;
- anti-rabbit IgG HRP linked secondary antibody #7074.
2.8. Statistical Analysis
- Interaction effects < 5% = additivity;
- Effects between 5% and 10% = marginal additivity;
- Higher than 10% = antagonism or synergy.
3. Results
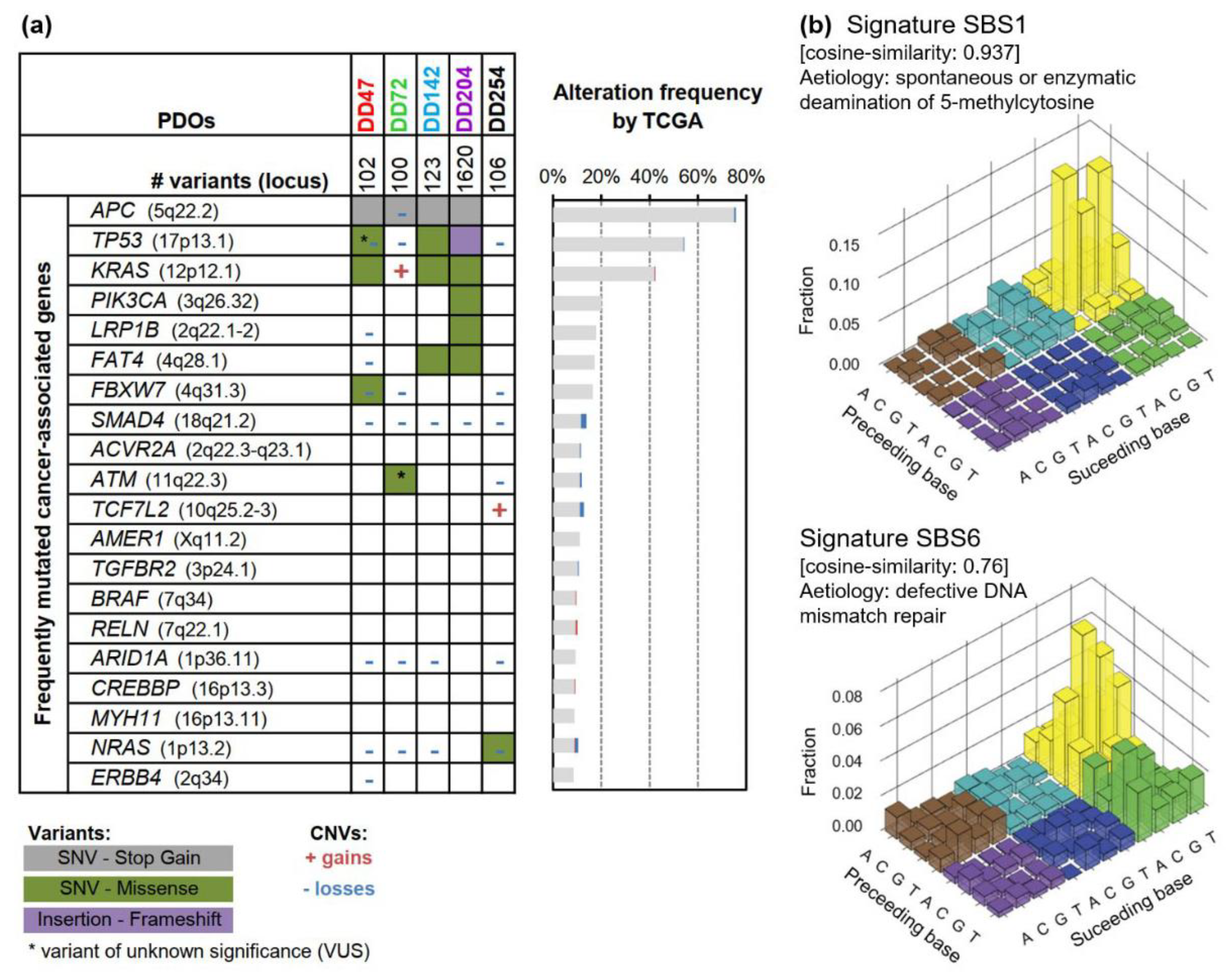
3.1. Genetic Characterization of Patient-Derived CRC Organoids
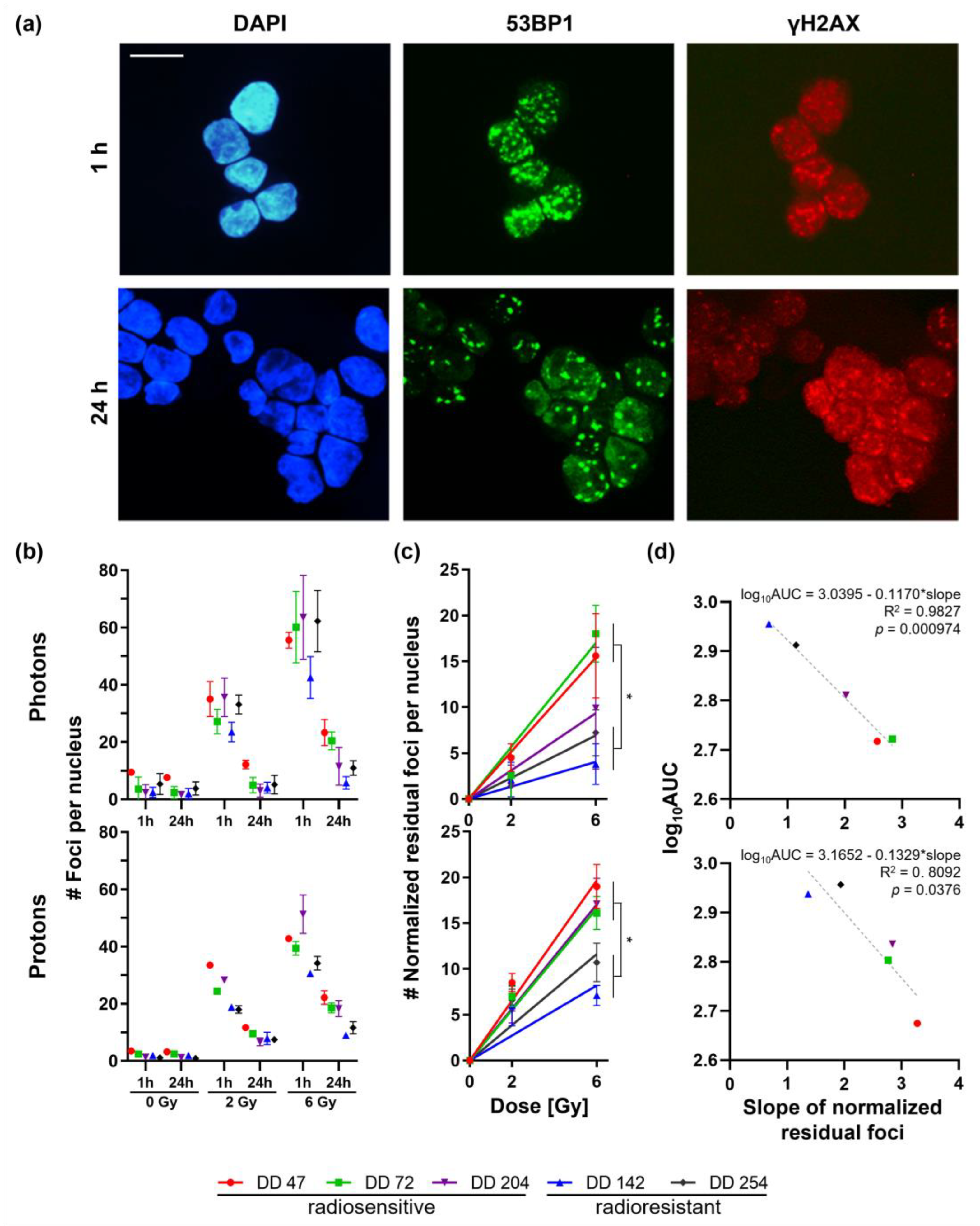
3.2. Heterogeneous Response of CRC PDOs upon Irradiation
3.3. CRC PDOs Repair DNA Damage to Different Degrees
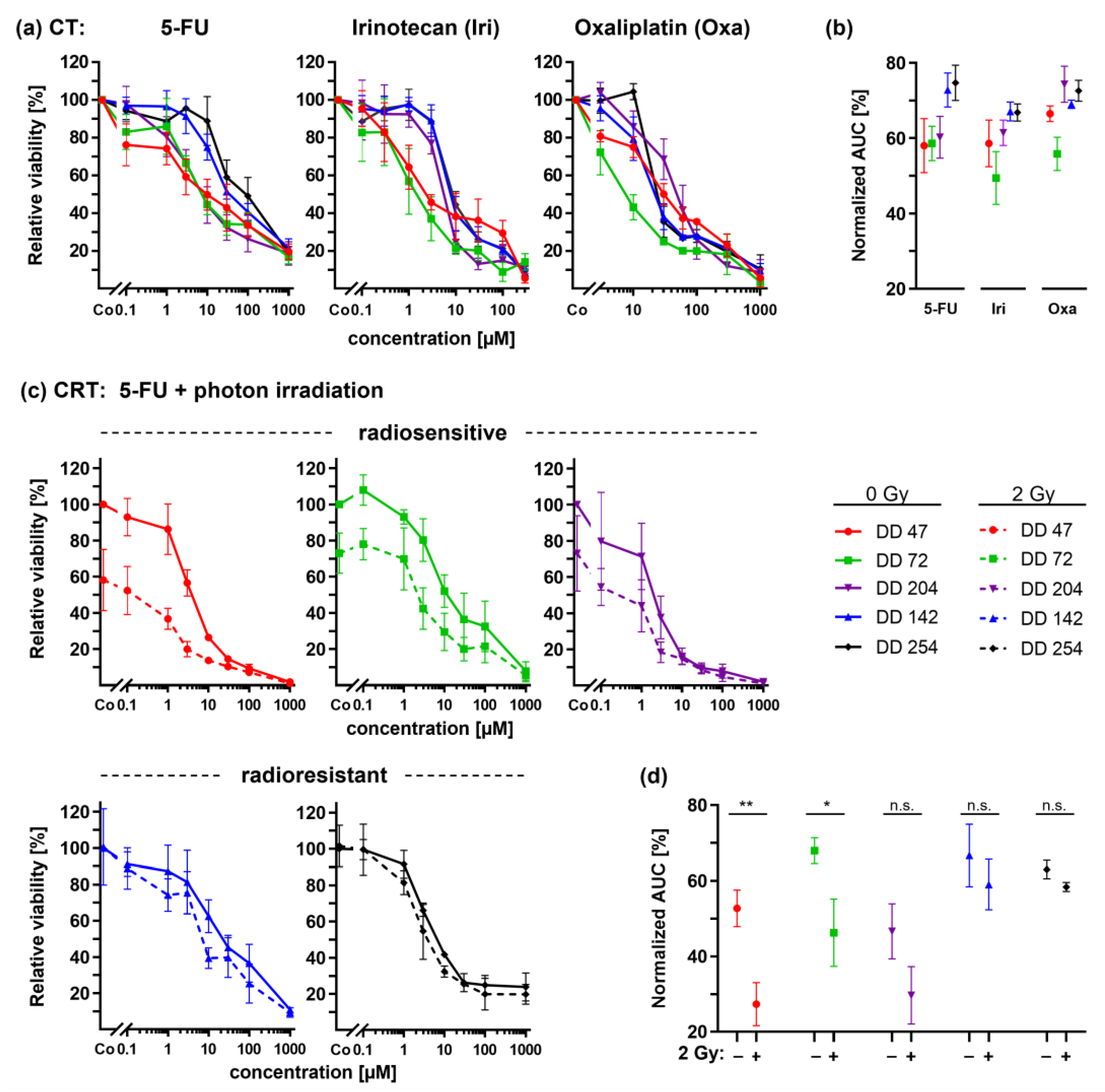
3.4. CRC PDOs Cannot Be Sensitized to Irradiation by 5-FU
3.5. ATM Inhibitors Can Sensitize Radioresistant CRC PDOs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sauer, R.; Liersch, T.; Merkel, S.; Fietkau, R.; Hohenberger, W.; Hess, C.; Becker, H.; Raab, H.R.; Villanueva, M.T.; Witzigmann, H.; et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: Results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J. Clin. Oncol. 2012, 30, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Bosset, J.F.; Calais, G.; Mineur, L.; Maingon, P.; Stojanovic-Rundic, S.; Bensadoun, R.J.; Bardet, E.; Beny, A.; Ollier, J.C.; Bolla, M.; et al. Fluorouracil-based adjuvant chemotherapy after preoperative chemoradiotherapy in rectal cancer: Long-term results of the EORTC 22921 randomised study. Lancet Oncol. 2014, 15, 184–190. [Google Scholar] [CrossRef]
- Gerard, J.P.; Conroy, T.; Bonnetain, F.; Bouche, O.; Chapet, O.; Closon-Dejardin, M.T.; Untereiner, M.; Leduc, B.; Francois, E.; Maurel, J.; et al. Preoperative radiotherapy with or without concurrent fluorouracil and leucovorin in T3-4 rectal cancers: Results of FFCD 9203. J. Clin. Oncol. 2006, 24, 4620–4625. [Google Scholar] [CrossRef]
- Folkesson, J.; Birgisson, H.; Pahlman, L.; Cedermark, B.; Glimelius, B.; Gunnarsson, U. Swedish Rectal Cancer Trial: Long lasting benefits from radiotherapy on survival and local recurrence rate. J. Clin. Oncol. 2005, 23, 5644–5650. [Google Scholar] [CrossRef] [PubMed]
- Van Gijn, W.; Marijnen, C.A.; Nagtegaal, I.D.; Kranenbarg, E.M.; Putter, H.; Wiggers, T.; Rutten, H.J.; Pahlman, L.; Glimelius, B.; van de Velde, C.J.; et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011, 12, 575–582. [Google Scholar] [CrossRef]
- Tomasello, G.; Petrelli, F.; Ghidini, M.; Pezzica, E.; Passalacqua, R.; Steccanella, F.; Turati, L.; Sgroi, G.; Barni, S. Tumor regression grade and survival after neoadjuvant treatment in gastro-esophageal cancer: A meta-analysis of 17 published studies. Eur. J. Surg. Oncol. 2017, 43, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Coinu, A.; Cabiddu, M.; Ghilardi, M.; Vavassori, I.; Barni, S. Correlation of pathologic complete response with survival after neoadjuvant chemotherapy in bladder cancer treated with cystectomy: A meta-analysis. Eur. Urol. 2014, 65, 350–357. [Google Scholar] [CrossRef] [PubMed]
- LeVasseur, N.; Sun, J.; Gondara, L.; Diocee, R.; Speers, C.; Lohrisch, C.; Chia, S. Impact of pathologic complete response on survival after neoadjuvant chemotherapy in early-stage breast cancer: A population-based analysis. J. Cancer Res. Clin. Oncol. 2020, 146, 529–536. [Google Scholar] [CrossRef]
- Maas, M.; Nelemans, P.J.; Valentini, V.; Das, P.; Rödel, C.; Kuo, L.J.; Calvo, F.A.; García-Aguilar, J.; Glynne-Jones, R.; Haustermans, K.; et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol. 2010, 11, 835–844. [Google Scholar] [CrossRef]
- Hospers, G.; Bahadoer, R.R.; Dijkstra, E.A.; Etten, B.V.; Marijnen, C.; Putter, H.; Kranenbarg, E.M.K.; Roodvoets, A.G.; Nagtegaal, I.D.; Beets-Tan, R.G.; et al. Short-course radiotherapy followed by chemotherapy before TME in locally advanced rectal cancer: The randomized RAPIDO trial. J. Clin. Oncol. 2020, 38, 4006. [Google Scholar] [CrossRef]
- Conroy, T.; Lamfichekh, N.; Etienne, P.-L.; Rio, E.; Francois, E.; Mesgouez-Nebout, N.; Vendrely, V.; Artignan, X.; Bouché, O.; Gargot, D.; et al. Total neoadjuvant therapy with mFOLFIRINOX versus preoperative chemoradiation in patients with locally advanced rectal cancer: Final results of PRODIGE 23 phase III trial, a UNICANCER GI trial. J. Clin. Oncol. 2020, 38, 4007. [Google Scholar] [CrossRef]
- Habr-Gama, A.; Perez, R.O.; Proscurshim, I.; Campos, F.G.; Nadalin, W.; Kiss, D.; Gama-Rodrigues, J. Patterns of failure and survival for nonoperative treatment of stage c0 distal rectal cancer following neoadjuvant chemoradiation therapy. J. Gastrointest. Surg. 2006, 10, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Van der Valk, M.J.M.; Hilling, D.E.; Bastiaannet, E.; Meershoek-Klein Kranenbarg, E.; Beets, G.L.; Figueiredo, N.L.; Habr-Gama, A.; Perez, R.O.; Renehan, A.G.; van de Velde, C.J.H.; et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): An international multicentre registry study. Lancet 2018, 391, 2537–2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beets, G.L.; Figueiredo, N.L.; Habr-Gama, A.; van de Velde, C.J. A new paradigm for rectal cancer: Organ preservation: Introducing the International Watch & Wait Database (IWWD). Eur. J. Surg. Oncol. 2015, 41, 1562–1564. [Google Scholar] [CrossRef]
- Feeney, G.; Sehgal, R.; Sheehan, M.; Hogan, A.; Regan, M.; Joyce, M.; Kerin, M. Neoadjuvant radiotherapy for rectal cancer management. World J. Gastroenterol. 2019, 25, 4850–4869. [Google Scholar] [CrossRef]
- Joye, I.; Haustermans, K. Early and late toxicity of radiotherapy for rectal cancer. Recent Results Cancer Res. 2014, 203, 189–201. [Google Scholar] [CrossRef]
- De Mattia, E.; Roncato, R.; Palazzari, E.; Toffoli, G.; Cecchin, E. Germline and Somatic Pharmacogenomics to Refine Rectal Cancer Patients Selection for Neo-Adjuvant Chemoradiotherapy. Front. Pharmacol. 2020, 11, 897. [Google Scholar] [CrossRef]
- Alves, A.; Panis, Y.; Mathieu, P.; Kwiatkowski, F.; Slim, K.; Mantion, G.; Association Francaise de, C. Mortality and morbidity after surgery of mid and low rectal cancer. Results of a French prospective multicentric study. Gastroenterol. Clin. Biol. 2005, 29, 509–514. [Google Scholar] [CrossRef]
- Vitti, E.T.; Parsons, J.L. The Radiobiological Effects of Proton Beam Therapy: Impact on DNA Damage and Repair. Cancers 2019, 11, 946. [Google Scholar] [CrossRef]
- Lühr, A.; von Neubeck, C.; Krause, M.; Troost, E.G.C. Relative biological effectiveness in proton beam therapy—Current knowledge and future challenges. Clin. Transl. Radiat. Oncol. 2018, 9, 35–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaios, E.J.; Wo, J.Y. Proton beam radiotherapy for anal and rectal cancers. J. Gastrointest. Oncol. 2020, 11, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Suckert, T.; Nexhipi, S.; Dietrich, A.; Koch, R.; Kunz-Schughart, L.A.; Bahn, E.; Beyreuther, E. Models for Translational Proton Radiobiology-From Bench to Bedside and Back. Cancers 2021, 13, 4216. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrials.gov. Preoperative Short-Course Radiation Therapy with PROtons Compared to Photons In High-Risk RECTal Cancer (PRORECT). 25 August 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04525989 (accessed on 1 February 2022).
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütte, M.; Risch, T.; Abdavi-Azar, N.; Boehnke, K.; Schumacher, D.; Keil, M.; Yildiriman, R.; Jandrasits, C.; Borodina, T.; Amstislavskiy, V.; et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 2017, 8, 14262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.; Kwon, J.; Kong, J.; Moon, S.M.; Cho, S.; Yang, K.Y.; Jang, W.I.; Kim, M.S.; Kim, Y.; Shin, U.S. A Patient-Derived Organoid-Based Radiosensitivity Model for the Prediction of Radiation Responses in Patients with Rectal Cancer. Cancers 2021, 13, 3760. [Google Scholar] [CrossRef]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauve, C.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell 2020, 26, 17–26. [Google Scholar] [CrossRef]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olshen, A.B.; Bengtsson, H.; Neuvial, P.; Spellman, P.T.; Olshen, R.A.; Seshan, V.E. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics 2011, 27, 2038–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Helmbrecht, S.; Baumann, M.; Enghardt, W.; Fiedler, F.; Krause, M.; Lühr, A. Design and implementation of a robust and cost-effective double-scattering system at a horizontal proton beamline. J. Instrum. 2016, 11, T11001. [Google Scholar] [CrossRef]
- Suckert, T.; Rassamegevanon, T.; Muller, J.; Dietrich, A.; Graja, A.; Reiche, M.; Lock, S.; Krause, M.; Beyreuther, E.; von Neubeck, C. Applying Tissue Slice Culture in Cancer Research-Insights from Preclinical Proton Radiotherapy. Cancers 2020, 12, 1589. [Google Scholar] [CrossRef]
- Slinker, B.K. The statistics of synergism. J. Mol. Cell. Cardiol. 1998, 30, 723–731. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, PO.17.00011. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Beyreuther, E.; Baumann, M.; Enghardt, W.; Helmbrecht, S.; Karsch, L.; Krause, M.; Pawelke, J.; Schreiner, L.; Schürer, M.; von Neubeck, C.; et al. Research Facility for Radiobiological Studies at the University Proton Therapy Dresden. Int. J. Part. Ther. 2018, 5, 172–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickson, I.; Zhao, Y.; Richardson, C.J.; Green, S.J.; Martin, N.M.; Orr, A.I.; Reaper, P.M.; Jackson, S.P.; Curtin, N.J.; Smith, G.C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004, 64, 9152–9159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlin, J.; Allen, J.; Ahmad, S.F.; Hughes, G.; Sheridan, V.; Odedra, R.; Farrington, P.; Cadogan, E.B.; Riches, L.C.; Garcia-Trinidad, A.; et al. Orally Bioavailable and Blood-Brain Barrier-Penetrating ATM Inhibitor (AZ32) Radiosensitizes Intracranial Gliomas in Mice. Mol. Cancer Ther. 2018, 17, 1637–1647. [Google Scholar] [CrossRef] [Green Version]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.M.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.J.G.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, L.D.; Parsons, D.W.; Jones, S.; Lin, J.; Sjöblom, T.; Leary, R.J.; Shen, D.; Boca, S.M.; Barber, T.; Ptak, J.; et al. The genomic landscapes of human breast and colorectal cancers. Science 2007, 318, 1108–1113. [Google Scholar] [CrossRef] [Green Version]
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis. Oncol. 2021, 5, 30. [Google Scholar] [CrossRef]
- Shinoto, M.; Ebner, D.K.; Yamada, S. Particle Radiation Therapy for Gastrointestinal Cancers. Curr. Oncol. Rep. 2016, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Tatsuzaki, H.; Urie, M.M.; Willett, C.G. 3-D comparative study of proton vs. x-ray radiation therapy for rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 1992, 22, 369–374. [Google Scholar] [CrossRef]
- Isacsson, U.; Montelius, A.; Jung, B.; Glimelius, B. Comparative treatment planning between proton and X-ray therapy in locally advanced rectal cancer. Radiother. Oncol. 1996, 41, 263–272. [Google Scholar] [CrossRef]
- Colaco, R.J.; Nichols, R.C.; Huh, S.; Getman, N.; Ho, M.W.; Li, Z.; Morris, C.G.; Mendenhall, W.M.; Mendenhall, N.P.; Hoppe, B.S. Protons offer reduced bone marrow, small bowel, and urinary bladder exposure for patients receiving neoadjuvant radiotherapy for resectable rectal cancer. J. Gastrointest. Oncol. 2014, 5, 3–8. [Google Scholar] [CrossRef]
- Palmer, M.; Mok, H.; Ciura, K.; Georges, R.; Nguyen, B.; Crawford, C.; Beddar, S.; Zhu, R.; Crane, C.; Das, P. Dose Reduction to Small Bowel and Other Relevant Structures in Rectal Carcinoma with Proton Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, S846. [Google Scholar] [CrossRef]
- Wolff, H.A.; Wagner, D.M.; Conradi, L.-C.; Hennies, S.; Ghadimi, M.; Hess, C.F.; Christiansen, H. Irradiation with protons for the individualized treatment of patients with locally advanced rectal cancer: A planning study with clinical implications. Radiother. Oncol. 2012, 102, 30–37. [Google Scholar] [CrossRef]
- Lühr, A.; von Neubeck, C.; Pawelke, J.; Seidlitz, A.; Peitzsch, C.; Bentzen, S.M.; Bortfeld, T.; Debus, J.; Deutsch, E.; Langendijk, J.A.; et al. “Radiobiology of Proton Therapy”: Results of an international expert workshop. Radiother. Oncol. 2018, 128, 56–67. [Google Scholar] [CrossRef]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.W. Cancer and radiation therapy: Current advances and future directions. Int. J. Med. Sci. 2012, 9, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Meneceur, S.; Löck, S.; Gudziol, V.; Hering, S.; Bütof, R.; Rehm, M.; Baumann, M.; Krause, M.; von Neubeck, C. Residual gammaH2AX foci in head and neck squamous cell carcinomas as predictors for tumour radiosensitivity: Evaluation in pre-clinical xenograft models and clinical specimens. Radiother. Oncol. 2019, 137, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Willers, H.; Gheorghiu, L.; Liu, Q.; Efstathiou, J.A.; Wirth, L.J.; Krause, M.; von Neubeck, C. DNA Damage Response Assessments in Human Tumor Samples Provide Functional Biomarkers of Radiosensitivity. Semin. Radiat. Oncol. 2015, 25, 237–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rassamegevanon, T.; Löck, S.; Baumann, M.; Krause, M.; von Neubeck, C. Comparable radiation response of ex vivo and in vivo irradiated tumor samples determined by residual γH2AX. Radiother. Oncol. 2019, 139, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Lieke, H.T.; Guchelaar, H.-J.; Gelderblom, H. Pharmacogenetics in chemotherapy of colorectal cancer. Best Pract. Res. Clin. Gastroenterol. 2009, 23, 257–273. [Google Scholar] [CrossRef]
- Wilson, G.D.; Bentzen, S.M.; Harari, P.M. Biologic basis for combining drugs with radiation. Semin. Radiat. Oncol. 2006, 16, 2–9. [Google Scholar] [CrossRef]
- Grem, J.L. 5-Fluorouracil: Forty-Plus and Still Ticking. A Review of its Preclinical and Clinical Development. Investig. New Drugs 2000, 18, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.L.; Luengas, J.; Rich, T.A.; Murray, D. Radiosensitization of cultured human colon adenocarcinoma cells by 5-fluorouracil: Effects on cell survival, DNA repair, and cell recovery. Int. J. Radiat. Oncol. Biol. Phys. 1992, 23, 983–991. [Google Scholar] [CrossRef]
- Bruso, C.E.; Shewach, D.S.; Lawrence, T.S. Fluorodeoxyuridine-induced radiosensitization and inhibition of DNA double strand break repair in human colon cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 1990, 19, 1411–1417. [Google Scholar] [CrossRef]
- Lempiäinen, H.; Halazonetis, T.D. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009, 28, 3067–3073. [Google Scholar] [CrossRef] [Green Version]
- Clinicaltrials.gov. First-in-human Study of M4076 in Advanced Solid Tumors (DDRiver Solid Tumors 410). 12 May 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04882917 (accessed on 12 March 2021).
- Clinicaltrials.gov. Study to Assess the Safety and Preliminary Efficacy of AZD0156 at Increasing Doses Alone or in Combination with Other Anti-cancer Treatment in Patients with Advanced Cancer (AToM). 27 October 2015. Available online: https://clinicaltrials.gov/ct2/show/NCT02588105 (accessed on 11 March 2022).
- Clinicaltrials.gov. A Study of ART0380 for the Treatment of Advanced or Metastatic Solid Tumors. 8 December 2020. Available online: https://clinicaltrials.gov/ct2/show/study/NCT04657068 (accessed on 11 March 2022).
- Ivanov, V.N.; Zhou, H.; Partridge, M.A.; Hei, T.K. Inhibition of Ataxia Telangiectasia Mutated Kinase Activity Enhances TRAIL-Mediated Apoptosis in Human Melanoma Cells. Cancer Res. 2009, 69, 3510–3519. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pape, K.; Lößner, A.J.; William, D.; Czempiel, T.; Beyreuther, E.; Klimova, A.; Lehmann, C.; Schmäche, T.; Merker, S.R.; Naumann, M.; et al. Sensitization of Patient-Derived Colorectal Cancer Organoids to Photon and Proton Radiation by Targeting DNA Damage Response Mechanisms. Cancers 2022, 14, 4984. https://doi.org/10.3390/cancers14204984
Pape K, Lößner AJ, William D, Czempiel T, Beyreuther E, Klimova A, Lehmann C, Schmäche T, Merker SR, Naumann M, et al. Sensitization of Patient-Derived Colorectal Cancer Organoids to Photon and Proton Radiation by Targeting DNA Damage Response Mechanisms. Cancers. 2022; 14(20):4984. https://doi.org/10.3390/cancers14204984
Chicago/Turabian StylePape, Kristin, Anna J. Lößner, Doreen William, Tabea Czempiel, Elke Beyreuther, Anna Klimova, Claudia Lehmann, Tim Schmäche, Sebastian R. Merker, Max Naumann, and et al. 2022. "Sensitization of Patient-Derived Colorectal Cancer Organoids to Photon and Proton Radiation by Targeting DNA Damage Response Mechanisms" Cancers 14, no. 20: 4984. https://doi.org/10.3390/cancers14204984
APA StylePape, K., Lößner, A. J., William, D., Czempiel, T., Beyreuther, E., Klimova, A., Lehmann, C., Schmäche, T., Merker, S. R., Naumann, M., Ada, A. -M., Baenke, F., Seidlitz, T., Bütof, R., Dietrich, A., Krause, M., Weitz, J., Klink, B., von Neubeck, C., & Stange, D. E. (2022). Sensitization of Patient-Derived Colorectal Cancer Organoids to Photon and Proton Radiation by Targeting DNA Damage Response Mechanisms. Cancers, 14(20), 4984. https://doi.org/10.3390/cancers14204984