Panel Sequencing of Primary Cutaneous B-Cell Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue
2.2. Targeted Sequencing
2.3. DNA Extraction
2.4. Hybridization-Based Panel Sequencing
2.5. Sanger Sequencing
2.6. FISH Analysis
2.7. Bioinformatical Data Analysis
3. Results
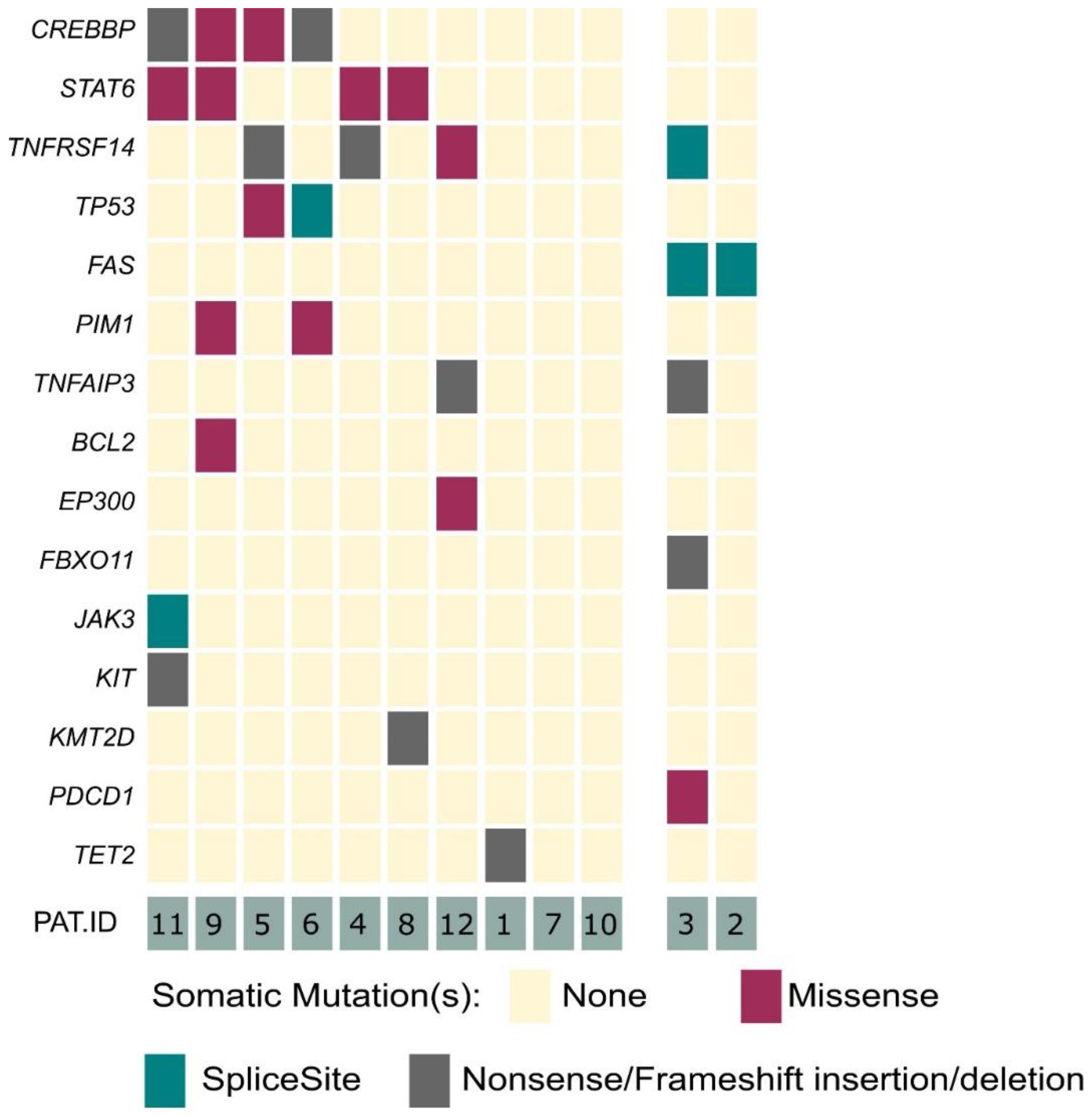
3.1. Recurrent Somatic Mutations Are Present in PCFBCL
3.2. Alterations of the FAS Gene May Serve as An Adjunctive Molecular Tool for Subtype Classification of Indolent B-Cell Lymphomas of the Skin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willemze, R.; Cerroni, L.; Kempf, W.; Berti, E.; Facchetti, F.; Swerdlow, S.H.; Jaffe, E.S. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019, 133, 1703–1714. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, J.P.; Wobser, M. Cutaneous B-cell lymphomas—Pathogenesis, diagnostic workup, and therapy. J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG 2016, 14, 1207–1224. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Willemze, R.; Pimpinelli, N.; Whittaker, S.; Olsen, E.A.; Ranki, A.; Dummer, R.; Hoppe, R.T. TNM classification system for primary cutaneous lymphomas other than mycosis fungoides and Sezary syndrome: A proposal of the International Society for Cutaneous Lymphomas (ISCL) and the Cutaneous Lymphoma Task Force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007, 110, 479–484. [Google Scholar] [CrossRef]
- Senff, N.J.; Willemze, R. The applicability and prognostic value of the new TNM classification system for primary cutaneous lymphomas other than mycosis fungoides and Sézary syndrome: Results on a large cohort of primary cutaneous B-cell lymphomas and comparison with the system used by the Dutch Cutaneous Lymphoma Group. Br. J. Dermatol. 2007, 157, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Senff, N.J.; Hoefnagel, J.J.; Jansen, P.M.; Vermeer, M.H.; van Baarlen, J.; Blokx, W.A.; Canninga-van Dijk, M.R.; Geerts, M.L.; Hebeda, K.M.; Kluin, P.M.; et al. Reclassification of 300 primary cutaneous B-Cell lymphomas according to the new WHO-EORTC classification for cutaneous lymphomas: Comparison with previous classifications and identification of prognostic markers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 1581–1587. [Google Scholar] [CrossRef] [PubMed]
- Wobser, M.; Goebeler, M. Cutaneous lymphomas: Clinical presentation—Diagnosis—Treatment. Hautarzt Z. Dermatol. Venerol. Verwandte Geb. 2019, 70, 815–830. [Google Scholar] [CrossRef]
- Wobser, M. Treatment of indolent cutaneous B-cell lymphoma. Hautarzt Z. Dermatol. Venerol. Verwandte Geb. 2017, 68, 721–726. [Google Scholar] [CrossRef]
- Dippel, E.; Assaf, C.; Becker, J.C.; von Bergwelt-Baildon, M.; Beyer, M.; Cozzio, A.; Eich, H.T.; Follmann, M.; Grabbe, S.; Hillen, U.; et al. S2k Guidelines—Cutaneous Lymphomas Update 2016-Part 2: Treatment and Follow-up (ICD10 C82–C86). J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG 2018, 16, 112–122. [Google Scholar] [CrossRef]
- Laban, É.; Beylot-Barry, M.; Ortonne, N.; Battistella, M.; Carlotti, A.; de Muret, A.; Wechsler, J.; Balme, B.; Petrella, T.; Lamant, L.; et al. Cutaneous lymphoproliferations: Proposal for the use of diagnostic algorithms based on 2760 cases of cutaneous lymphoproliferations taken from the INCa networks (LYMPHOPATH and GFELC) over a two-year period. Ann. Pathol. 2015, 35, 131–147. [Google Scholar] [CrossRef]
- Massone, C.; Fink-Puches, R.; Laimer, M.; Rütten, A.; Vale, E.; Cerroni, L. Miliary and agminated-type primary cutaneous follicle center lymphoma: Report of 18 cases. J. Am. Acad. Dermatol. 2011, 65, 749–755. [Google Scholar] [CrossRef]
- Cassisa, A.; Colpani, F.; Rinaldi, R.; Cima, L. Primary Cutaneous Follicle Center Lymphoma Clear Cell Variant: Expanding the Spectrum of Cutaneous Clear Cell Neoplasms. Am. J. Dermatopathol. 2018, 40, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Oschlies, I.; Kohler, C.W.; Szczepanowski, M.; Koch, K.; Gontarewicz, A.; Metze, D.; Hillen, U.; Richter, J.; Spang, R.; Klapper, W. Spindle-Cell Variants of Primary Cutaneous Follicle Center B-Cell Lymphomas Are Germinal Center B-Cell Lymphomas by Gene Expression Profiling Using a Formalin-Fixed Paraffin-Embedded Specimen. J. Investig. Dermatol. 2017, 137, 2450–2453. [Google Scholar] [CrossRef] [PubMed]
- Dilly, M.; Ben-Rejeb, H.; Vergier, B.; Feldis, M.; Toty, L.; Nohra, O.; Beylot-Barry, M.; Gros, A.; Merlio, J.P.; Parrens, M. Primary cutaneous follicle center lymphoma with Hodgkin and Reed-Sternberg-like cells: A new histopathologic variant. J. Cutan. Pathol. 2014, 41, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Hoefnagel, J.J.; Dijkman, R.; Basso, K.; Jansen, P.M.; Hallermann, C.; Willemze, R.; Tensen, C.P.; Vermeer, M.H. Distinct types of primary cutaneous large B-cell lymphoma identified by gene expression profiling. Blood 2005, 105, 3671–3678. [Google Scholar] [CrossRef]
- Mareschal, S.; Pham-Ledard, A.; Viailly, P.J.; Dubois, S.; Bertrand, P.; Maingonnat, C.; Fontanilles, M.; Bohers, E.; Ruminy, P.; Tournier, I.; et al. Identification of Somatic Mutations in Primary Cutaneous Diffuse Large B-Cell Lymphoma, Leg Type by Massive Parallel Sequencing. J. Investig. Dermatol. 2017, 137, 1984–1994. [Google Scholar] [CrossRef]
- Koens, L.; Zoutman, W.H.; Ngarmlertsirichai, P.; Przybylski, G.K.; Grabarczyk, P.; Vermeer, M.H.; Willemze, R.; Jansen, P.M.; Schmidt, C.A.; Tensen, C.P. Nuclear factor-κB pathway-activating gene aberrancies in primary cutaneous large B-cell lymphoma, leg type. J. Investig. Dermatol. 2014, 134, 290–292. [Google Scholar] [CrossRef]
- Menguy, S.; Beylot-Barry, M.; Parrens, M.; Ledard, A.P.; Frison, E.; Comoz, F.; Battistella, M.; Szablewski, V.; Balme, B.; Croue, A.; et al. Primary cutaneous large B-cell lymphomas: Relevance of the 2017 World Health Organization classification: Clinicopathological and molecular analyses of 64 cases. Histopathology 2019, 74, 1067–1080. [Google Scholar] [CrossRef]
- Hallermann, C.; Kaune, K.M.; Siebert, R.; Vermeer, M.H.; Tensen, C.P.; Willemze, R.; Gunawan, B.; Bertsch, H.P.; Neumann, C. Chromosomal aberration patterns differ in subtypes of primary cutaneous B cell lymphomas. J. Investig. Dermatol. 2004, 122, 1495–1502. [Google Scholar] [CrossRef]
- Vela, V.; Juskevicius, D.; Dirnhofer, S.; Menter, T.; Tzankov, A. Mutational landscape of marginal zone B-cell lymphomas of various origin: Organotypic alterations and diagnostic potential for assignment of organ origin. Virchows Arch. 2021, 480, 403–413. [Google Scholar] [CrossRef]
- Maurus, K.; Appenzeller, S.; Roth, S.; Kuper, J.; Rost, S.; Meierjohann, S.; Arampatzi, P.; Goebeler, M.; Rosenwald, A.; Geissinger, E.; et al. Panel Sequencing Shows Recurrent Genetic FAS Alterations in Primary Cutaneous Marginal Zone Lymphoma. J. Investig. Dermatol. 2018, 138, 1573–1581. [Google Scholar] [CrossRef]
- Ducharme, O.; Beylot-Barry, M.; Pham-Ledard, A.; Bohers, E.; Viailly, P.J.; Bandres, T.; Faur, N.; Frison, E.; Vergier, B.; Jardin, F.; et al. Mutations of the B-Cell Receptor Pathway Confer Chemoresistance in Primary Cutaneous Diffuse Large B-Cell Lymphoma Leg Type. J. Investig. Dermatol. 2019, 139, 2334–2342.e2338. [Google Scholar] [CrossRef] [PubMed]
- Pham-Ledard, A.; Beylot-Barry, M.; Barbe, C.; Leduc, M.; Petrella, T.; Vergier, B.; Martinez, F.; Cappellen, D.; Merlio, J.P.; Grange, F. High frequency and clinical prognostic value of MYD88 L265P mutation in primary cutaneous diffuse large B-cell lymphoma, leg-type. JAMA Dermatol. 2014, 150, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y. Genomics of diffuse large B cell lymphoma. Blood Res. 2021, 56, S75–S79. [Google Scholar] [CrossRef] [PubMed]
- Gángó, A.; Bátai, B.; Varga, M.; Kapczár, D.; Papp, G.; Marschalkó, M.; Kuroli, E.; Schneider, T.; Csomor, J.; Matolcsy, A.; et al. Concomitant 1p36 deletion and TNFRSF14 mutations in primary cutaneous follicle center lymphoma frequently expressing high levels of EZH2 protein. Virchows Arch. 2018, 473, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Barasch, N.J.K.; Liu, Y.C.; Ho, J.; Bailey, N.; Aggarwal, N.; Cook, J.R.; Swerdlow, S.H. The molecular landscape and other distinctive features of primary cutaneous follicle center lymphoma. Hum. Pathol. 2020, 106, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.A.; Yang, J.; Ringbloom, K.G.; Martinez-Escala, M.E.; Stevenson, K.E.; Wenzel, A.T.; Fantini, D.; Martin, H.K.; Moy, A.P.; Morgan, E.A.; et al. Genomic landscape of cutaneous follicular lymphomas reveals 2 subgroups with clinically predictive molecular features. Blood Adv. 2021, 5, 649–661. [Google Scholar] [CrossRef]
- Senff, N.J.; Noordijk, E.M.; Kim, Y.H.; Bagot, M.; Berti, E.; Cerroni, L.; Dummer, R.; Duvic, M.; Hoppe, R.T.; Pimpinelli, N.; et al. European Organization for Research and Treatment of Cancer and International Society for Cutaneous Lymphoma consensus recommendations for the management of cutaneous B-cell lymphomas. Blood 2008, 112, 1600–1609. [Google Scholar] [CrossRef]
- Willemze, R.; Meijer, C.J. Classification of cutaneous T-cell lymphoma: From Alibert to WHO-EORTC. J. Cutan. Pathol. 2006, 33, 18–26. [Google Scholar] [CrossRef]
- Dippel, E.; Assaf, C.; Becker, J.C.; von Bergwelt-Baildon, M.; Beyer, M.; Cozzio, A.; Eich, H.T.; Follmann, M.; Grabbe, S.; Hillen, U.; et al. S2k Guidelines—Cutaneous Lymphomas Update 2016-Part 1: Classification and Diagnosis (ICD10 C82–C86). J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG 2017, 15, 1266–1273. [Google Scholar] [CrossRef]
- van Dongen, J.J.; Langerak, A.W.; Brüggemann, M.; Evans, P.A.; Hummel, M.; Lavender, F.L.; Delabesse, E.; Davi, F.; Schuuring, E.; García-Sanz, R.; et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003, 17, 2257–2317. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Bergmann, E.A.; Arora, K.; Vacic, V.; Zody, M.C.; Iossifov, I.; O’Rawe, J.A.; Wu, Y.; Jimenez Barron, L.T.; Rosenbaum, J.; et al. Indel variant analysis of short-read sequencing data with Scalpel. Nat. Protoc. 2016, 11, 2529–2548. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Šedý, J.R.; Ramezani-Rad, P. HVEM network signaling in cancer. Adv. Cancer Res. 2019, 142, 145–186. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T. The Tumor Microenvironment in Follicular Lymphoma: Its Pro-Malignancy Role with Therapeutic Potential. Int. J. Mol. Sci. 2021, 22, 5352. [Google Scholar] [CrossRef]
- Hoefnagel, J.J.; Vermeer, M.H.; Jansen, P.M.; Fleuren, G.J.; Meijer, C.J.; Willemze, R. Bcl-2, Bcl-6 and CD10 expression in cutaneous B-cell lymphoma: Further support for a follicle centre cell origin and differential diagnostic significance. Br. J. Dermatol. 2003, 149, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Verdanet, E.; Dereure, O.; René, C.; Tempier, A.; Benammar-Hafidi, A.; Gallo, M.; Frouin, E.; Durand, L.; Gazagne, I.; Costes-Martineau, V.; et al. Diagnostic value of STMN1, LMO2, HGAL, AID expression and 1p36 chromosomal abnormalities in primary cutaneous B cell lymphomas. Histopathology 2017, 71, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Szablewski, V.; Ingen-Housz-Oro, S.; Baia, M.; Delfau-Larue, M.H.; Copie-Bergman, C.; Ortonne, N. Primary Cutaneous Follicle Center Lymphomas Expressing BCL2 Protein Frequently Harbor BCL2 Gene Break and May Present 1p36 Deletion: A Study of 20 Cases. Am. J. Surg. Pathol. 2016, 40, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Pastore, A.; Jurinovic, V.; Kridel, R.; Hoster, E.; Staiger, A.M.; Szczepanowski, M.; Pott, C.; Kopp, N.; Murakami, M.; Horn, H.; et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: A retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015, 16, 1111–1122. [Google Scholar] [CrossRef]
- Green, M.R. Chromatin modifying gene mutations in follicular lymphoma. Blood 2018, 131, 595–604. [Google Scholar] [CrossRef]
- Okosun, J.; Bödör, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Fernandez-Flores, A.; Smucler-Simonovich, A.; Escalante, F.; Manjon, J.A. The differential diagnosis between primary cutaneous large B-cell lymphoma and cutaneous follicular lymphoma: Prognostic and therapeutic implications. Am. J. Dermatopathol. 2011, 33, 819–826. [Google Scholar] [CrossRef]
- Felcht, M.; Klemke, C.D.; Nicolay, J.P.; Weiss, C.; Assaf, C.; Wobser, M.; Schlaak, M.; Hillen, U.; Moritz, R.; Tantcheva-Poor, I.; et al. Primary cutaneous diffuse large B-cell lymphoma, NOS and leg type: Clinical, morphologic and prognostic differences. J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG 2019, 17, 275–285. [Google Scholar] [CrossRef]
- Beltzung, F.; Ortonne, N.; Pelletier, L.; Beylot-Barry, M.; Ingen-Housz-Oro, S.; Franck, F.; Pereira, B.; Godfraind, C.; Delfau, M.H.; D’Incan, M.; et al. Primary Cutaneous CD4+ Small/Medium T-Cell Lymphoproliferative Disorders: A Clinical, Pathologic, and Molecular Study of 60 Cases Presenting With a Single Lesion: A Multicenter Study of the French Cutaneous Lymphoma Study Group. Am. J. Surg. Pathol. 2020, 44, 862–872. [Google Scholar] [CrossRef] [PubMed]

| Pat ID | Sex | Age at Primary Diagnosis [Years] | Tumor Stage at Primary Diagnosis | Tumor Stage at Date Last Seen | Cutaneous Relapses | Number of Cutaneous Relapses | Systemic Dissemination | Treatment | Overall Survival Since Primary Diagnosis [Months] | Final Status | Clonality | bcl-2 Expression Immunohistochemistry | bcl2 FISH Analysis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | male | 39 | T2 N0 M0 | T2 N0 M0 | yes | 1 | no | rituximab | 117 | alive | polyclonal | negative | negative |
| 2 | male | 60 | T2c N0 M0 | T2c N0 M0 | no | no | rituximab | 75 | dead | polyclonal | positive | negative | |
| 3 | female | 70 | T2 N0 M0 | T2 N0 M0 | no | no | rituximab | 131 | alive | oligoclonal/polyclonal | positive | negative | |
| 4 | male | 54 | T1 N0 M0 | T2 N0 M0 | yes | 3 | no | excision, radiation | 200 | alive | oligoclonal/polyclonal | negative | negative |
| 5 | female | 70 | T2 N0 M0 | T2b N0 M0 | yes | >5 | no | excision, radiation, topical steroids, rituximab | 117 | alive | monoclonal | positive | negative |
| 6 | female | 66 | T1 N0 M0 | T1 N0 M0 | no | no | watch-and-wait | 66 | alive | not done | negative | negative | |
| 7 | female | 89 | T1b N0 M0 | T1b N0 M0 | no | no | radiation | 47 | alive | not done | negative | negative | |
| 8 | male | 45 | T1a N0 M0 | T1a N0 M0 | no | no | excision | 23 | alive | polyclonal | positive | negative | |
| 9 | male | 50 | T1a N0 M0 | T1a N0 M0 | no | no | radiation | 21 | alive | not done | positive | positive | |
| 10 | male | 50 | T1 Nx Mx | T1 Nx Mx | no | no | watch-and-wait | 57 | alive | monoclonal | negative | negative | |
| 11 | female | 53 | T2 N0 M0 | T2 N0 M0 | yes | 2 | no | radiation, topical steroids | 35 | alive | not done | negative | negative |
| 12 | male | 43 | T2 N0 M0 | T2 N0 M0 | no | no | rituximab | 107 | alive | not done | negative | negative |
| Lymphoma Subtype | Characteristic Clinical Findings | Histology | Common Immunphenotype | Common Treatment Modalities | Prognosis |
|---|---|---|---|---|---|
| Primary cutaneous follicular B cell lymphoma (PCFBCL) | slowly growing erythematous plaques, papules, tumors; often at the head | Diffuse or follicular dermal B-cell infiltrates, mostly centrocytes | CD20+, bcl2 - (+), bcl-6 +, MUM-1 -, CD21 + remnants of follicular dendritic cells networks | watch and wait, excision, radiation, rituximab | Favorable. 5-year survival rate: >95% |
| Primary cutaneous marginal zone lymphoma (PCMZL) | slowly growing erythematous plaques, papules, tumors; often at trunk and extremities | Diffuse dermal B-cell infiltrates with germinal centers and plasma cells | CD20+, bcl2 +, bcl-6 -, MUM-1 -, mostly IgG4 + | watch and wait, excision, radiation, rituximab | Favorable. 5-year survival rate: >95% |
| Primary cutaneous diffuse large B-cell lymphoma (PCLBCL) | rapidly growing ulcerated tumors; mostly at the legs | Diffuse dermal sheet-like B-cell infiltrates, mostly centroblasts, immunoblasts | CD20+, bcl2 +, bcl-6 -/+, MUM-1 +, IgM +, CD21 lack of follicular dendritic cells networks | rituximab-CHOP +/- radiation | Poor. 5-year survival rate: 20–60% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wobser, M.; Schummer, P.; Appenzeller, S.; Kneitz, H.; Roth, S.; Goebeler, M.; Geissinger, E.; Rosenwald, A.; Maurus, K. Panel Sequencing of Primary Cutaneous B-Cell Lymphoma. Cancers 2022, 14, 5274. https://doi.org/10.3390/cancers14215274
Wobser M, Schummer P, Appenzeller S, Kneitz H, Roth S, Goebeler M, Geissinger E, Rosenwald A, Maurus K. Panel Sequencing of Primary Cutaneous B-Cell Lymphoma. Cancers. 2022; 14(21):5274. https://doi.org/10.3390/cancers14215274
Chicago/Turabian StyleWobser, Marion, Patrick Schummer, Silke Appenzeller, Hermann Kneitz, Sabine Roth, Matthias Goebeler, Eva Geissinger, Andreas Rosenwald, and Katja Maurus. 2022. "Panel Sequencing of Primary Cutaneous B-Cell Lymphoma" Cancers 14, no. 21: 5274. https://doi.org/10.3390/cancers14215274
APA StyleWobser, M., Schummer, P., Appenzeller, S., Kneitz, H., Roth, S., Goebeler, M., Geissinger, E., Rosenwald, A., & Maurus, K. (2022). Panel Sequencing of Primary Cutaneous B-Cell Lymphoma. Cancers, 14(21), 5274. https://doi.org/10.3390/cancers14215274







