Integrated Biomarker Analysis Reveals L1CAM as a Potential Stratification Marker for No Specific Molecular Profile High-Risk Endometrial Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Samples
2.2. Immunohistochemistry
2.3. DNA Extraction and Next-Generation Sequencing
2.4. Statistics
3. Results
3.1. Molecular Classification of High-Risk EC Patients Is Associated with Clinicopathological Characteristics
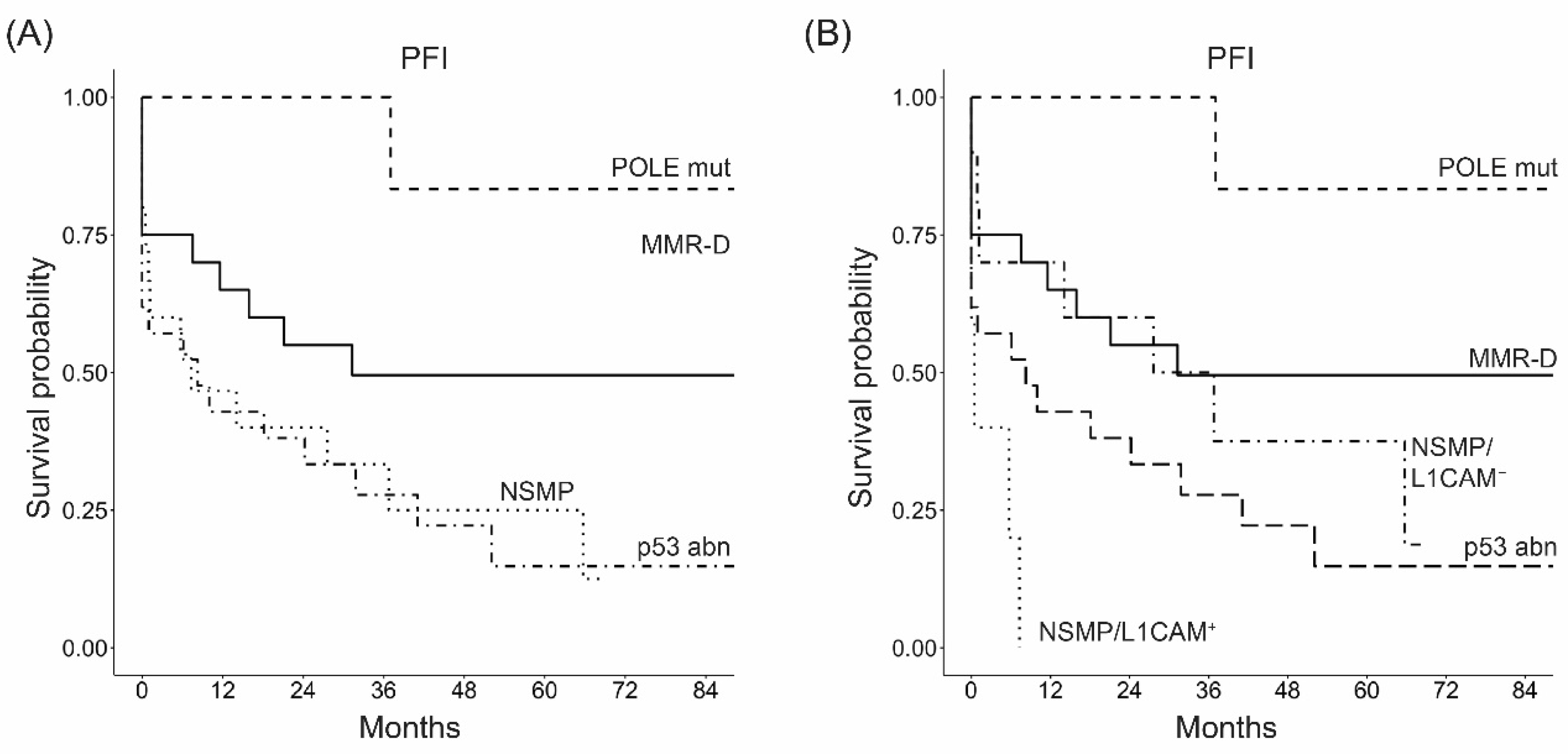
3.2. Molecular Classification Is an Independent Prognostic Factor in High-Risk ECs
3.3. Additional Biomarkers Are Significantly Associated with the Molecular Classification
3.4. L1CAM Is a Predictor of Worse Prognosis in the Whole EC Patients’ Cohort and in the NSMP Subgroup
3.5. L1CAM Correlates with the Time to Recurrence after Platinum-Based Chemotherapy in NSMP High-Risk ECs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Brescia Cohort | ENITEC Cohort | |
|---|---|---|
| Clinical Annotations | No. of Patients | No. of Patients |
| Age | ||
| Median (range) years | 65 (31–90) | 65 (37–83) |
| Ethnic group | ||
| Caucasian | 94 (100%) | NA |
| Histological type | ||
| Endometrioid | 69 (73.4%) | 27 (57.4%) |
| Clear cell | 5 (5.3%) | 5 (10.6%) |
| Serous | 11 (11.7%) | 10 (21.3%) |
| Mixed | 7 (7.4%) | 4 (8.5%) |
| Other | 2 (2.2) | 1 (2.1%) |
| FIGO Stage | ||
| I | 28 (29.8%) | 13 (27.7%) |
| II | 11 (11.7%) | 8 (17.0%) |
| III | 43 (45.7%) | 25 (53.2%) |
| IV | 12 (12.8%) | 1 (2.1%) |
| Tumor Grade | ||
| G1 | 0 | 4 (8.5%) |
| G2 | 17 (18.1%) | 10 (21.3%) |
| G3 | 77 (81.9%) | 33 (70.2%) |
| Myometrial invasion (M) | ||
| M1 | 19 (20.2%) | 13 (27.7%) |
| M2 | 75 (79.8%) | 34 (72.3%) |
| LVSI | ||
| No | 9 (9.6%) | 22 (46.8%) |
| Yes | 85 (90.4%) | 25 (53.2%) |
| Lymph node Metastasis | ||
| Absent | 46 (59.7%) | 25 (69.4%) |
| Present | 31 (40.3%) | 11 (30.6%) |
| Unknown | 17 | 11 |
| Risk Group (ESGO/ESTRO/ESP 2020) * | ||
| High–intermediate | 32 (34.0%) | 8 (17.0%) |
| High | 45 (47.9%) | 39 (83.0%) |
| Advanced Metastatic | 17 (18.1%) | 0 |
| Treatment | ||
| Radiotherapy | 29 (30.9%) | 33 (70.2%) |
| Chemotherapy | 30 (31.9%) | 2 (4.3%) |
| Chemo + Radio | 34 (36.2%) | 9 (19.1%) |
| None | 1 (1.1%) | 3 (6.4%) |
| Time to recurrence after CT | ||
| Late-relapsing | 37 (57.8%) | 10 (90.9%) |
| Early-relapsing | 27 (42.2%) | 1 (9.1%) |
| Status | ||
| Alive | 51 (54.3%) | 35 (74.5%) |
| DOD | 36 (38.3%) | 12 (25.5%) |
| DID | 7 (7.4%) | 0 |
| Clinical Annotations | N. of Patients | DSS | PFS | ||||
|---|---|---|---|---|---|---|---|
| HR | 95%CI | p Value | HR | 95%CI | p Value | ||
| Age | |||||||
| Year | 94 | 1.03 | 1.00–1.07 | 0.038 | 1.04 | 1.01–1.07 | 0.013 |
| Histological type | |||||||
| Endometrioid | 69 | 1 | - | - | 1 | - | - |
| Non-endometrioid | 24 | 3.49 | 1.81–6.74 | <0.001 | 2.29 | 1.29–4.07 | 0.005 |
| FIGO Stage | |||||||
| I–II | 39 | 1 | - | - | 1 | - | - |
| III–IV | 55 | 4.79 | 1.99–11.5 | <0.001 | 4.13 | 2.11–8.10 | <0.001 |
| Tumor Grade | |||||||
| G2 | 17 | 1 | - | - | 1 | - | - |
| G3 | 77 | 0.99 | 0.43–2.26 | 0.983 | 1.38 | 0.65–2.9 | 0.404 |
| Myometrial invasion | |||||||
| M1 (<50%) | 19 | 1 | - | - | 1 | - | - |
| M2 (≥50%) | 75 | 1.77 | 0.69–4.56 | 0.235 | 2.02 | 0.91–4.49 | 0.086 |
| Lymph node metastasis | |||||||
| Absent | 46 | 1 | - | - | 1 | - | - |
| Present | 31 | 2.69 | 1.19–6.09 | 0.018 | 1.99 | 1.06–3.74 | 0.033 |
| LVSI | |||||||
| Absent | 9 | 1 | - | - | 1 | - | - |
| Present | 85 | 0.21 | 0.03–1.56 | 0.128 | 0.63 | 0.23–1.76 | 0.378 |
| Risk Group (ESGO /ESTRO /ESP 2020) | |||||||
| High–intermediate | 46 | 1 | - | - | 1 | - | - |
| High | 31 | 3.59 | 1.21–10.6 | 0.021 | 3.25 | 1.47–7.16 | 0.004 |
| Advanced Metastatic | 17 | 21.8 | 6.95–68.1 | <0.001 | 9.65 | 4.01–23.2 | <0.001 |
| Time to recurrence after CT | |||||||
| Late-relapsing | 37 | 1 | - | - | 1 | - | - |
| Early-relapsing | 27 | 27.5 | 9.11–82.7 | <0.001 | 12.6 | 5.94–26.8 | <0.001 |
| Adjuvant therapy | |||||||
| RT | 29 | 1 | - | - | 1 | - | - |
| CT or CT+RT | 64 | 4.71 | 1.66–13.3 | 0.004 | 2.35 | 1.14–4.84 | 0.021 |
| Clinical Annotations | ER (% of Positive Cells) | PR (% of Positive Cells) | Ki67 (% of Positive Cells) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N. | Mean (SD) | Median (Q1–Q3) | p Value | Mean (SD) | Median (Q1–Q3) | p Value | Mean (SD) | Median (Q1–Q3) | p Value | |
| Histological type | 0.002 | <0.001 | 0.839 | |||||||
| Endometrioid | 69 | 43 (37) | 40 (5–80) | 32 (33) | 20 (0–60) | 42 (23) | 40 (25–65) | |||
| Non-endometrioid | 24 | 18 (29) | 0 (0–25) | 9 (19) | 0 (0–4) | 43 (24) | 40 (20–70) | |||
| FIGO Stage | 0.089 | 0.094 | 0.091 | |||||||
| I–II | 39 | 43 (37) | 30 (5–75) | 28 (30) | 15 (3–56) | 48 (24) | 40 (30–65) | |||
| III–IV | 55 | 32 (36) | 15 (0–70) | 24 (33) | 3 (0–40) | 39 (23) | 35 (20–60) | |||
| Tumor Grade | 0.735 | 0.168 | 0.022 | |||||||
| G2 | 17 | 37 (36) | 30 (5–80) | 33 (31) | 30 (5–60) | 32 (25) | 20 (10–45) | |||
| G3 | 77 | 36 (37) | 20 (0–70) | 24 (32) | 5 (0–40) | 45 (23) | 40 (28–65) | |||
| Myometrial invasion | 0.541 | 0.821 | 0.400 | |||||||
| M1 (<50%) | 19 | 41 837) | 30 (0–80) | 27 (31) | 10 (0–56) | 39 (22) | 30 (20–65) | |||
| M2 (≥50%) | 75 | 35 (37) | 15 (0–70) | 25 (32) | 10 (0–40) | 43 (24) | 40 (25–65) | |||
| Lymph node metastasis | 0.035 | 0.216 | 0.002 | |||||||
| Absent | 46 | 44 (40) | 35 (5–85) | 29 (33) | 15 (0–56) | 49 (23) | 48 (30–70) | |||
| Present | 31 | 27 (32) | 15 (0–40) | 25 (34) | 3 (0–50) | 31 (20) | 28 (15–45) | |||
| LVSI | 0.984 | 0.727 | 0.711 | |||||||
| Absent | 9 | 34 (41) | 15 (3–75) | 22 (34) | 3 (0–20) | 44 (17) | 40 (30–53) | |||
| Present | 85 | 37 (37) | 25 (0–70) | 26 (31) | 10 (0–40) | 43 (24) | 40 (20–65) | |||
| Risk Group (ESGO/ESMO/ESP 2020) | 0.006 | 0.002 | 0.461 | |||||||
| High–intermediate | 32 | 45 (38) | 58 (5–80) | 29 (31) | 15 (4–50) | 47 (24) | 43 (30–65) | |||
| High | 45 | 38 (37) | 30 (0–80) | 31 (34) | 20 (0–56) | 40 (23) | 40 (20–58) | |||
| Advanced Metastatic | 17 | 15 (29) | 0 (0–5) | 5 (14) | 0 (0–3) | 43 (24) | 35 (20–65) | |||
| p53 | 0.002 | <0.001 | 0.338 | |||||||
| Wt | 67 | 45 (38) | 40 (3–80) | 33 (33) | 30 (0–70) | 41 (23) | 40 (20–65) | |||
| Abn | 27 | 16 (25) | 5 (0–25) | 6 (13) | 0 (0–5) | 46 (25) | 40 (25–70) | |||
| Time to recurrence after CT | 0.018 | 0.002 | 0.802 | |||||||
| Late-relapsing | 37 | 40 (37) | 25 (5–80) | 32 (35) | 15 (0–70) | 38 (21) | 38 (23–58) | |||
| Early-relapsing | 27 | 23 (33) | 0 (0–40) | 10 (22) | 0 (0–5) | 41 (23) | 35 (20–65) | |||
| Clinical Annotations | L1CAM | CTNNB1 | ARID1A | DEGREE OF INFLAMMATION | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤10% (n. 66) | >10% (n. 28) | p Value | Wild Type (n. 82) | Mutated (n.12) | p Value | Present (n.62) | Loss (n.32) | p Value | Absent/ Weak (n.52) | Moderate/ Prominent (n.42) | p Value | |
| Age (years) | 0.110 | 0.200 | 0.004 | 0.200 | ||||||||
| Mean (SD) | 64 (11) | 68 (10) | 66 (11) | 62 (9) | 68 (10) | 60 (11) | 67 (9) | 63 (13) | ||||
| Median (Q1–Q3) | 64 (57–72) | 70 (62–74) | 66 (60–74) | 62 (58–68) | 68 (63–74) | 62 (52–69) | 66 (61–74) | 64 (55–70) | ||||
| Histological type | <0.001 | 0.032 | 0.140 | 0.200 | ||||||||
| Endometrioid | 61 (94%) | 8 (29%) | 57 (70%) | 12 (100%) | 42 (69%) | 27 (84%) | 36 (69%) | 33 (80%) | ||||
| Non-endometrioid | 4 (6%) | 20 (71%) | 24 (30%) | 0 (0%) | 19 (31%) | 5 (16%) | 16 (31%) | 8 (20%) | ||||
| Unknown | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | ||||
| FIGO Stage | 0.041 | 0.110 | 0.700 | 0.007 | ||||||||
| I–II | 32 (48%) | 7 (25%) | 37 (45%) | 2 (17%) | 27 (44%) | 12 (38%) | 15 (29%) | 24 (57%) | ||||
| III–IV | 34 (52%) | 21 (75%) | 45 (55%) | 10 (83%) | 35 (56%) | 20 (62%) | 37 (71%) | 18 (43%) | ||||
| Tumor Grade | 0.018 | >0.9 | >0.9 | >0.9 | ||||||||
| G2 | 16 (24%) | 1 (4%) | 15 (18%) | 2 (17%) | 11 (18%) | 6 (19%) | 9 (17%) | 8 (19%) | ||||
| G3 | 50 (76%) | 27 (96%) | 67 (82%) | 10 (83%) | 51 (82%) | 26 (81%) | 43 (83%) | 34 (81%) | ||||
| Myometrial invasion | 0.300 | 0.120 | 0.300 | 0.078 | ||||||||
| M1 (<50%) | 11 (17%) | 8 (29%) | 19 (23%) | 0 (0%) | 15 (24%) | 4 (12%) | 7 (13%) | 12 (29%) | ||||
| M2 (≥50%) | 55 (83%) | 20 (71%) | 63 (77%) | 12 (100%) | 47 (76%) | 28 (88%) | 45 (87%) | 30 (71%) | ||||
| Lymph node metastasis | 0.077 | 0.110 | >0.9 | 0.500 | ||||||||
| Absent | 36 (67%) | 10 (43%) | 42 (64%) | 4 (36%) | 30 (60%) | 16 (59%) | 22 (55%) | 24 (65%) | ||||
| Present | 18 (33%) | 13 (57%) | 24 (36%) | 7 (64%) | 20 (40%) | 11 (41%) | 18 (45%) | 13 (35%) | ||||
| Unknown | 12 | 5 | 16 | 1 | 12 | 5 | 12 | 5 | ||||
| LVSI | 0.400 | 0.600 | 0.700 | 0.500 | ||||||||
| Absent | 5 (8%) | 4 (14%) | 9 (11%) | 0 (0%) | 7 (11%) | 2 (6%) | 4 (8%) | 5 (12%) | ||||
| Present | 61 (92%) | 24 (86%) | 73 (89%) | 12 (100%) | 55 (89%) | 30 (94%) | 48 (92%) | 37 (88%) | ||||
| Risk Group ESMO/ESGO/ESP | 0.002 | 0.120 | 0.300 | 0.012 | ||||||||
| High–intermediate | 30 (45%) | 2 (7%) | 30 (37%) | 2 (17%) | 23 (37%) | 9 (28%) | 12 (23%) | 20 (48%) | ||||
| High | 27 (41%) | 18 (64%) | 36 (44%) | 9 (75%) | 26 (42%) | 19 (59%) | 26 (50%) | 19 (45%) | ||||
| Advanced Metastatic | 9 (14%) | 8 (29%) | 16 (20%) | 1 (8%) | 13 (21%) | 4 (12%) | 14 (27%) | 3 (7%) | ||||
| L1CAM | - | 0.500 | 0.035 | >0.9 | ||||||||
| Negative (≤10%) | - | - | 56 (68%) | 10 (83%) | 39 (63%) | 27 (84%) | 36 (69%) | 30 (71%) | ||||
| Positive (>10%) | - | - | 26 (32%) | 2 (17%) | 23 (37%) | 5 (16%) | 16 (31%) | 12 (29%) | ||||
| CTNNB1 | 0.500 | - | 0.500 | >0.9 | ||||||||
| Wild type | 56 (85%) | 26 (93%) | - | - | 55 (89%) | 27 (84%) | 45 (87%) | 37 (88%) | ||||
| Mutated | 10 (15%) | 2 (7%) | - | - | 7 (11%) | 5 (16%) | 7 (13%) | 5 (12%) | ||||
| ARID1A | 0.035 | 0.500 | - | 0.300 | ||||||||
| Present | 39 (59%) | 23 (83%) | 55 (67%) | 7 (58%) | - | - | 37 (71%) | 25 (60%) | ||||
| Loss | 27 (41%) | 5 (18%) | 27 (33%) | 5 (42%) | - | - | 15 (29%) | 17 (40%) | ||||
| p53 | <0.001 | 0.017 | 0.016 | 0.400 | ||||||||
| Wt | 57 (86%) | 10 (36%) | 55 (67%) | 12 (100%) | 39 (63%) | 28 (88%) | 35 (67%) | 32 (76%) | ||||
| Abn | 9 (14%) | 18 (64%) | 27 (33%) | 0 (0%) | 23 (37%) | 4 (12%) | 17 (33%) | 10 (24%) | ||||
| Degree of inflammation | >0.9 | >0.9 | 0.300 | - | ||||||||
| Absent/weak | 36 (55%) | 16 (57%) | 45 (55%) | 7 (58%) | 37 (60%) | 15 (47%) | - | - | ||||
| Moderate/prominent | 30 (45%) | 12 (43%) | 37 (45%) | 5 (42%) | 25 (40%) | 17 (53%) | - | - | ||||
| ER (% of positive cells) | <0.001 | 0.200 | 0.700 | 0.800 | ||||||||
| Mean (SD) | 46 (37) | 15 (24) | 34 (36) | 51 (40) | 36 (37) | 37 (37) | 38 (39) | 35 (35) | ||||
| Median (Q1–Q3) | 40 (5–80) | 0 (0–18) | 18 (0–70) | 55 (11–81) | 25 (0–70) | 23 (2–78) | 23 (0–80) | 25 (1–70) | ||||
| PR (% of positive cells) | <0.001 | 0.037 | >0.9 | 0.300 | ||||||||
| Mean (SD) | 32 (33) | 10 (20) | 22 (29) | 48 (41) | 28 (34) | 21 (25) | 23 (32) | 28 (31) | ||||
| Median (Q1–Q3) | 20 (3–64) | 0 (0–10) | 5 (0–35) | 40 (4–86) | 8 (0–60) | 10 (0–35) | 5 (0–36) | 13 (0–55) | ||||
| Ki67 (% of positive cells) | 0.900 | 0.400 | 0.500 | 0.091 | ||||||||
| Mean (SD) | 43 (24) | 43 (24) | 43 (24) | 38 (24) | 42 (24) | 45 (23) | 39 (23) | 47 (24) | ||||
| Median (Q1–Q3) | 40 (21–65) | 35 (25–68) | 40 (25–65) | 28 (24–58) | 35 (20–65) | 48 (29–65) | 35 (20–60) | 48 (30–65) | ||||
| Time to recurrence after CT | 0.120 | 0.700 | 0.067 | 0.002 | ||||||||
| Late-relapsing | 26 (67%) | 11 (44%) | 31 (56%) | 6 (67%) | 20 (49%) | 17 (74%) | 17 (42% | 20 (83%) | ||||
| Early-relapsing | 13 (33%) | 14 (56%) | 24 (44%) | 3 (33%) | 21 (51%) | 6 (26%) | 23 (57%) | 4 (17%) | ||||
| Unknown | 27 | 3 | 27 | 3 | 21 | 9 | 12 | 18 | ||||
Appendix B
Appendix B.1. Immunohistochemistry
Appendix B.2. Next-Generation Sequencing
Appendix C

References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International Patterns and Trends in Endometrial Cancer Incidence, 1978–2013. J. Natl. Cancer Inst. 2018, 110, 354–361. [Google Scholar] [CrossRef] [PubMed]
- van den Heerik, A.S.V.M.; Horeweg, N.; de Boer, S.M.; Bosse, T.; Creutzberg, C.L. Adjuvant therapy for endometrial cancer in the era of molecular classification: Radiotherapy, chemoradiation and novel targets for therapy. Int. J. Gynecol. Cancer 2021, 31, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Gilks, C.B.; Oliva, E.; Soslow, R.A. Poor interobserver reproducibility in the diagnosis of high-grade endometrial carcinoma. Am. J. Surg. Pathol. 2013, 37, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jürgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Britton, H.; Kommoss, F.; Grevenkamp, F.; Karnezis, A.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann. Oncol. 2018, 29, 1180–1188. [Google Scholar] [CrossRef]
- Vermij, L.; Horeweg, N.; Leon-Castillo, A.; Rutten, T.A.; Mileshkin, L.R.; Mackay, H.J.; Leary, A.; Powell, M.E.; Singh, N.; Crosbie, E.J.; et al. HER2 Status in High-Risk Endometrial Cancers (PORTEC-3): Relationship with Histotype, Molecular Classification, and Clinical Outcomes. Cancers 2020, 13, 44. [Google Scholar] [CrossRef]
- León-Castillo, A.; de Boer, S.M.; Powell, M.E.; Mileshkin, L.R.; Mackay, H.J.; Leary, A.; Nijman, H.W.; Singh, N.; Pollock, P.M.; Bessette, P.; et al. TransPORTEC consortium. Molecular Classification of the PORTEC-3 Trial for High-Risk Endometrial Cancer: Impact on Prognosis and Benefit From Adjuvant Therapy. J. Clin. Oncol. 2020, 38, 3388–3397. [Google Scholar] [CrossRef]
- Auguste, A.; Genestie, C.; De Bruyn, M.; Adam, J.; Le Formal, A.; Drusch, F.; Pautier, P.; Crosbie, E.J.; MacKay, H.; Kitchener, H.C.; et al. Refinement of high-risk endometrial cancer classification using DNA damage response biomarkers: A TransPORTEC initiative. Mod. Pathol. 2018, 31, 1851–1861. [Google Scholar] [CrossRef]
- Köbel, M.; Atenafu, E.G.; Rambau, P.F.; Ferguson, S.E.; Nelson, G.S.; Ho, T.C.; Panzarella, T.; McAlpine, J.N.; Gilks, C.B.; Clarke, B.A.; et al. Progesterone receptor expression is associated with longer overall survival within high-grade histotypes of endometrial carcinoma: A Canadian high risk endometrial cancer consortium (CHREC) study. Gynecol. Oncol. 2016, 141, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Karnezis, A.N.; Leung, S.; Magrill, J.; McConechy, M.K.; Yang, W.; Chow, C.; Kobel, M.; Lee, C.H.; Huntsman, D.G.; Talhouk, A.; et al. Evaluation of endometrial carcinoma prognostic immunohistochemistry markers in the context of molecular classification. J. Pathol. Clin. Res. 2017, 3, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, I.C.; Stelloo, E.; Nout, R.A.; Nijman, H.W.; Edmondson, R.J.; Church, D.N.; MacKay, H.J.; Leary, A.; Powell, M.E.; Mileshkin, L.; et al. Prognostic significance of L1CAM expression and its association with mutant p53 expression in high-risk endometrial cancer. Mod. Pathol. 2016, 29, 174–181. [Google Scholar] [CrossRef]
- Kommoss, F.; Kommoss, F.; Grevenkamp, F.; Bunz, A.K.; Taran, F.A.; Fend, F.; Brucker, S.Y.; Wallwiener, D.; Schönfisch, B.; Greif, K.; et al. L1CAM: Amending the "low-risk" category in endometrial carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 255–262. [Google Scholar] [CrossRef]
- Guo, M.; Gong, H.; Nie, D.; Li, Z. High L1CAM expression predicts poor prognosis of patients with endometrial cancer: A systematic review and meta-analysis. Medicine 2021, 100, e25330. [Google Scholar] [CrossRef] [PubMed]
- Kurnit, K.C.; Kim, G.N.; Fellman, B.M.; Urbauer, D.L.; Mills, G.B.; Zhang, W.; Broaddus, R.R. CTNNB1 (beta-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Mod. Pathol. 2017, 30, 1032–1041. [Google Scholar] [CrossRef]
- Allo, G.; Bernardini, M.Q.; Wu, R.C.; Shih, I.e.M.; Kalloger, S.; Pollett, A.; Gilks, C.B.; Clarke, B.A. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod. Pathol. 2014, 27, 255–261. [Google Scholar] [CrossRef]
- Kato, M.K.; Yoshida, H.; Tanase, Y.; Uno, M.; Ishikawa, M.; Kato, T. Loss of ARID1A Expression as a Favorable Prognostic Factor in Early-Stage Grade 3 Endometrioid Endometrial Carcinoma Patients. Pathol. Oncol. Res. 2021, 27, 598550. [Google Scholar] [CrossRef]
- Kitson, S.; Sivalingam, V.N.; Bolton, J.; McVey, R.; Nickkho-Amiry, M.; Powell, M.E.; Leary, A.; Nijman, H.W.; Nout, R.A.; Bosse, T.; et al. Ki-67 in endometrial cancer: Scoring optimization and prognostic relevance for window studies. Mod. Pathol. 2017, 30, 459–468. [Google Scholar] [CrossRef]
- Guo, F.; Dong, Y.; Tan, Q.; Kong, J.; Yu, B. Tissue Infiltrating Immune Cells as Prognostic Biomarkers in Endometrial Cancer: A Meta-Analysis. Dis. Markers. 2020, 2020, 1805764. [Google Scholar] [CrossRef]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef] [PubMed]
- van der Putten, L.J.; Visser, N.C.; van de Vijver, K.; Santacana, M.; Bronsert, P.; Bulten, J.; Hirschfeld, M.; Colas, E.; Gil-Moreno, A.; Garcia, A.; et al. L1CAM expression in endometrial carcinomas: An ENITEC collaboration study. Br. J. Cancer 2016, 115, 716–724. [Google Scholar] [CrossRef] [PubMed]
- León-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; Rau, T.T.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J. Pathol. 2020, 250, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.; Borsboom, G.; Houwelingen, H.; Eijkemans, M.; Habbema, J. Validation and updating of predictive logistic regression models: A study on sample size and shrinkage. Stat. Med. 2004, 23, 2567–2586. [Google Scholar] [CrossRef]
- Newson, R. Confidence intervals for rank statistics: Somers’ D and extensions. Stata J. 2006, 6, 309–334. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 20 June 2022).
- León-Castillo, A.; Gilvazquez, E.; Nout, R.; Smit, V.T.; McAlpine, J.N.; McConechy, M.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; et al. Clinicopathological and molecular characterisation of ’multiple-classifier’ endometrial carcinomas. J. Pathol. 2020, 250, 312–322. [Google Scholar] [CrossRef]
- Kim, G.; Kurnit, K.C.; Djordjevic, B.; Singh, C.; Munsell, M.F.; Wang, W.L.; Lazar, A.J.; Zhang, W.; Broaddus, R. Nuclear β-catenin localization and mutation of the CTNNB1 gene: A context-dependent association. Mod. Pathol. 2018, 31, 1553–1559. [Google Scholar] [CrossRef]
- Liu, Y.; Patel, L.; Mills, G.B.; Lu, K.H.; Sood, A.K.; Ding, L.; Kucherlapati, R.; Mardis, E.R.; Levine, D.A.; Shmulevich, I.; et al. Clinical significance of CTNNB1 mutation and Wnt pathway activation in endometrioid endometrial carcinoma. J. Natl. Cancer Inst. 2014, 106, dju245. [Google Scholar] [CrossRef]
- Leon-Castillo, A.; Horeweg, N.; Peters, E.E.M.; Rutten, T.; Ter Haar, N.; Smit, V.T.H.B.M.; Kroon, C.D.; Boennelycke, M.; Hogdall, E.; Hogdall, C.; et al. Prognostic relevance of the molecular classification in high-grade endometrial cancer for patients staged by lymphadenectomy and without adjuvant treatment. Gynecol. Oncol. 2022, 164, 577–586. [Google Scholar] [CrossRef]
- van den Heerik, A.S.V.M.; Horeweg, N.; Nout, R.A.; Lutgens, L.C.H.W.; van der Steen-Banasik, E.M.; Westerveld, G.H.; van den Berg, H.A.; Slot, A.; Koppe, F.L.A.; Kommoss, S.; et al. PORTEC-4a: International randomized trial of molecular profile-based adjuvant treatment for women with high-intermediate risk endometrial cancer. Int. J. Gynecol. Cancer 2020, 30, 2002–2007. [Google Scholar] [CrossRef]
- Cavaliere, A.F.; Perelli, F.; Zaami, S.; D’Indinosante, M.; Turrini, I.; Giusti, M.; Gullo, G.; Vizzielli, G.; Mattei, A.; Scambia, G.; et al. Fertility Sparing Treatments in Endometrial Cancer Patients: The Potential Role of the New Molecular Classification. Int. J. Mol. Sci. 2021, 22, 12248. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, F.K.; Karnezis, A.N.; Kommoss, F.; Talhouk, A.; Taran, F.A.; Staebler, A.; Gilks, C.B.; Huntsman, D.G.; Krämer, B.; Brucker, S.Y.; et al. L1CAM further stratifies endometrial carcinoma patients with no specific molecular risk profile. Br. J. Cancer 2018, 119, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Romani, C.; Capoferri, D.; Reijnen, C.; Lonardi, S.; Ravaggi, A.; Ratti, M.; Bugatti, M.; Zanotti, L.; Tognon, G.; Sartori, E.; et al. L1CAM expression as a predictor of platinum response in high-risk endometrial carcinoma. Int. J. Cancer 2022, 151, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Mileshkin, L.; Edmondson, R.; O’Connell, R.L.; Sjoquist, K.M.; Andrews, J.; Jyothirmayi, R.; Beale, P.; Bonaventura, T.; Goh, J.; Hall, M.; et al. Phase 2 study of anastrozole in recurrent estrogen (ER)/progesterone (PR) positive endometrial cancer: The PARAGON trial—ANZGOG 0903. Gynecol. Oncol. 2019, 154, 29–37. [Google Scholar] [CrossRef]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef]
- Okamura, R.; Kato, S.; Lee, S.; Jimenez, R.E.; Sicklick, J.K.; Kurzrock, R. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J. Immunother. Cancer 2020, 8, e000438. [Google Scholar] [CrossRef]
- Doberstein, K.; Harter, P.N.; Haberkorn, U.; Bretz, N.P.; Arnold, B.; Carretero, R.; Moldenhauer, G.; Mittelbronn, M.; Altevogt, P. Antibody therapy to human L1CAM in a transgenic mouse model blocks local tumor growth but induces EMT. Int. J. Cancer 2015, 136, E326–E339. [Google Scholar] [CrossRef]
- Schäfer, H.; Dieckmann, C.; Korniienko, O.; Moldenhauer, G.; Kiefel, H.; Salnikov, A.; Krüger, A.; Altevogt, P.; Sebens, S. Combined treatment of L1CAM antibodies and cytostatic drugs improve the therapeutic response of pancreatic and ovarian carcinoma. Cancer Lett. 2012, 319, 66–82. [Google Scholar] [CrossRef]
- Cho, S.; Lee, T.S.; Song, I.H.; Kim, A.R.; Lee, Y.J.; Kim, H.; Hwang, H.; Jeong, M.S.; Kang, S.G.; Hong, H.J. Combination of anti-L1 cell adhesion molecule antibody and gemcitabine or cisplatin improves the therapeutic response of intrahepatic cholangiocarcinoma. PLoS ONE 2017, 12, e0170078. [Google Scholar] [CrossRef]

| Clinical Annotations | POLE mut | MMR-D | NSMP | p53abn | Total | p Value |
|---|---|---|---|---|---|---|
| n. 15 (16%) | n. 34 (36%) | n. 20 (21%) | n. 25 (27%) | n. 94 (100%) | ||
| Age (years) | 0.300 1 | |||||
| Mean (SD) | 62 (16) | 63 (11) | 66 (11) | 68 (7) | 65 (11) | |
| Median (Q1–Q3) | 64 (54–74) | 64 (55–71) | 64 (60–75) | 70 (64–72) | 65 (59–73) | |
| Histological type | <0.001 2 | |||||
| Endometrioid | 12 (80%) | 33 (97%) | 17 (85%) | 7 (29%) | 69 (73%) | |
| Non-endometrioid | 3 (20%) | 1 (3%) | 3 (15%) | 17 (71%) | 24 (27%) | |
| Unknown | 0 | 0 | 0 | 1 | 1 | |
| FIGO Stage | 0.015 2 | |||||
| I–II | 11 (73%) | 15 (44%) | 4 (20%) | 9 (36%) | 39 (41%) | |
| III–IV | 4 (27%) | 19 (56%) | 16 (80%) | 16 (64%) | 55 (59%) | |
| Tumor Grade | 0.024 2 | |||||
| G2 | 3 (20%) | 5 (15%) | 8 (40%) | 1 (4%) | 17 (18%) | |
| G3 | 12 (80%) | 29 (85%) | 12 (60%) | 24 (96%) | 77 (82%) | |
| Myometrial invasion | 0.110 2 | |||||
| M1 (<50%) | 5 (33%) | 6 (18%) | 1 (5%) | 7 (28%) | 19 (20%) | |
| M2 (≥50%) | 10 (67%) | 28 (82%) | 19 (95%) | 18 (72%) | 75 (80%) | |
| Lymph node metastasis | 0.006 2 | |||||
| Absent | 10 (91%) | 19 (63%) | 4 (25%) | 13 (65%) | 46 (60%) | |
| Present | 1 (9%) | 11 (37%) | 12 (75%) | 7 (35%) | 31 (40%) | |
| Unknown | 4 | 4 | 4 | 5 | 17 | |
| LVSI | 0.110 2 | |||||
| Absent | 2 (13%) | 1 (3%) | 1 (5%) | 5 (20%) | 9 (10%) | |
| Present | 13 (87%) | 33 (97%) | 19 (95%) | 20 (80%) | 85 (90%) | |
| Risk Group | 0.048 2 | |||||
| High–intermediate | 9 (60%) | 15 (44%) | 4 (20%) | 4 (16%) | 32 (34%) | |
| High | 4 (27%) | 14 (41%) | 13 (65%) | 14 (56%) | 45 (48%) | |
| Advanced Metastatic | 2 (13%) | 5 (15%) | 3 (15%) | 7 (28%) | 17 (18%) | |
| Adjuvant therapy | 0.051 2 | |||||
| None | 1 (7%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) | |
| CT | 5 (33%) | 9 (26%) | 6 (30%) | 10 (40%) | 30 (32%) | |
| RT | 7 (47%) | 14 (41%) | 5 (25%) | 3 (12%) | 29 (31%) | |
| CT+ RT | 2 (13%) | 11 (32%) | 9 (45%) | 12 (48%) | 34 (36%) | |
| Time to recurrence after CT | 0.140 2 | |||||
| Late-relapsing | 6 (86%) | 14 (70%) | 7 (47%) | 10 (45%) | 37 (58%) | |
| Early-relapsing | 1 (14%) | 6 (30%) | 8 (53%) | 12 (55%) | 27 (42%) | |
| Unknown | 8 | 14 | 5 | 3 | 30 | |
| Variable | DSS | PFS | ||||||
|---|---|---|---|---|---|---|---|---|
| n. Events/n. Patients | HR | 95%CI | p Value | n. Events/ n. Patients | HR | 95%CI | p Value | |
| Univariable analysis | ||||||||
| Molecular Groups | ||||||||
| MMR-D | 8/34 | 1 | - | - | 14/33 | 1 | - | - |
| POLE mut | 0/15 | 0.10 | 0.00–0.78 | 0.023 | 1/15 | 0.11 | 0.01–0.83 | 0.032 |
| NSMP | 11/20 | 2.56 | 1.06–6.47 | 0.037 | 16/20 | 2.41 | 1.17–4.98 | 0.017 |
| p53abn | 17/25 | 3.62 | 1.63–8.70 | 0.001 | 19/25 | 2.3 | 1.15–4.61 | 0.018 |
| Multivariable analysis | ||||||||
| Molecular Groups | ||||||||
| MMR-D | 8/34 | 1 | - | - | 14/33 | 1 | - | - |
| POLE mut | 0/15 | 0.10 | 0.00–0.78 | 0.024 | 1/15 | 0.10 | 0.01–0.76 | 0.026 |
| NSMP | 11/20 | 1.98 | 0.79–5.14 | 0.142 | 16/20 | 2.43 | 1.13–5.22 | 0.023 |
| p53abn | 17/25 | 3.54 | 1.57–8.67 | 0.002 | 19/25 | 2.24 | 1.11–4.54 | 0.025 |
| Age (year) | 36/94 | 1.05 | 1.01–1.10 | 0.013 | 50/93 | 1.04 | 1.01–1.07 | 0.010 |
| Grade (G3 vs. G2) | 36/94 | 1.04 | 0.45–2.67 | 0.929 | 50/93 | 2.19 | 0.95–5.05 | 0.065 |
| Stage (III-IV vs. I-II) | 36/94 | 4.01 | 1.75–10.6 | <0.001 | 50/93 | 3.76 | 1.86–7.58 | <0.001 |
| Clinical Annotations | POLE mut n. 15 (16%) | MMR-D n. 34 (36%) | NSMP n. 20 (21%) | p53abn n. 25 (27%) | Total n. 94 (100%) | p Value |
|---|---|---|---|---|---|---|
| L1CAM | <0.001 1 | |||||
| Negative (≤10%) | 12 (80%) | 31 (91%) | 15 (75%) | 8 (32%) | 66 (70%) | |
| Positive (>10%) | 3 (20%) | 3 (9%) | 5 (25%) | 17 (68%) | 28 (30%) | |
| CTNNB1 | 0.003 1 | |||||
| Wild type | 15 (100%) | 28 (82%) | 14 (70%) | 25 (100%) | 82 (87%) | |
| Mutated | 0 (0%) | 6 (18%) | 6 (30%) | 0 (0%) | 12 (13%) | |
| ER (% of positive cells) | 0.023 2 | |||||
| Mean (SD) | 37 (37) | 51 (38) | 35 (38) | 18 (26) | 37 (4) | |
| Median (Q1–Q3) | 30 (5–70) | 62 (6–84) | 22 (0–80) | 5 (0–25) | 25 (0–75) | |
| PR (% of positive cells) | 0.002 2 | |||||
| Mean (SD) | 24 (27) | 36 (35) | 33 (37) | 6 (13) | 3 (3) | |
| Median (Q1–Q3) | 15 (5–35) | 30 (3–69) | 15 (0–65) | 0 (0–5) | 10 (0–40) | |
| Ki67 (% of positive cells) | 0.012 2 | |||||
| Mean (SD) | 50 (20) | 46 (22) | 29 (22) | 46 (26) | 4 (2) | |
| Median (Q1–Q3) | 55 (38–65) | 42 (30–65) | 20 (10–40) | 38 (25–71) | 40 (23–65) | |
| ARID1A | 0.008 1 | |||||
| Present | 8 (53%) | 19 (56%) | 12 (60%) | 23 (92%) | 62 (66%) | |
| Loss | 7 (47%) | 15 (44%) | 8 (40%) | 2 (8%) | 32 (34%) | |
| Degree of inflammation Absent/weak Moderate/prominent | 2 (13%) 13 (87%) | 19 (56%) 15 (44%) | 15 (75%) 5 (25%) | 16 (64%) 9 (36%) | 52 (56%) 41 (44%) | 0.001 1 |
| Variables | N. of Patients | DSS | PFS | ||||
|---|---|---|---|---|---|---|---|
| HR | 95%CI | p Value | HR | 95%CI | p Value | ||
| L1CAM | |||||||
| Negative (≤10%) | 66 | 1 | - | - | 1 | - | - |
| Positive (>10%) | 28 | 2.689 | 1.391–5.200 | 0.003 | 1.672 | 0.955–2.929 | 0.072 |
| CTNNB1 | |||||||
| Wild type | 82 | 1 | - | - | 1 | - | - |
| Mutated | 12 | 1.023 | 0.361–2.899 | 0.966 | 1.950 | 0.909–4.183 | 0.086 |
| Estrogen receptor | |||||||
| % of positive cells | 94 | 0.987 | 0.976–0.997 | 0.015 | 0.996 | 0.988–1.004 | 0.301 |
| Progesterone receptor | |||||||
| % of positive cells | 94 | 0.977 | 0.960–0.993 | 0.006 | 0.995 | 0.985–1.005 | 0.298 |
| Ki67 | |||||||
| % of positive cells | 94 | 0.994 | 0.980–1.009 | 0.454 | 0.991 | 0.978–1.004 | 0.177 |
| ARID1A | |||||||
| Present | 62 | 1 | - | - | 1 | - | - |
| Loss | 32 | 0.429 | 0.194–0.945 | 0.036 | 0.578 | 0.306–1.092 | 0.091 |
| Inflammation | |||||||
| Absent/weak | 53 | 1 | - | - | 1 | - | - |
| Moderate/prominent | 41 | 0.429 | 0.213–0.862 | 0.018 | 0.342 | 0.186–0.631 | 0.001 |
| DSS | PFS | |||||||
|---|---|---|---|---|---|---|---|---|
| Variables | n. Events/n. Patients | HR | 95%CI | p Value | n. Events/n. Patients | HR | 95%CI | p Value |
| Univariable analysis | ||||||||
| Molecular Groups | ||||||||
| MMR-D | 8/34 | 1 | - | - | 14/33 | 1 | - | - |
| POLE mut | 0/15 | 0.10 | 0.00–0.77 | 0.023 | 1/15 | 0.11 | 0.01–0.84 | 0.034 |
| NSMP/L1CAM − | 7/15 | 2.04 | 0.74–5.52 | 0.163 | 11/15 | 2.01 | 0.91–4.45 | 0.085 |
| NSMP/L1CAM + | 4/5 | 5.76 | 1.63–17.9 | 0.009 | 5/5 | 5.57 | 1.94–16.0 | 0.001 |
| p53abn | 17/25 | 3.65 | 1.64–8.75 | 0.001 | 19/25 | 2.39 | 1.19–4.78 | 0.014 |
| Multivariable analysis | ||||||||
| Molecular Groups | ||||||||
| MMR-D | 8/34 | 1 | - | - | 14/33 | 1 | - | - |
| POLE mut | 0/15 | 0.10 | 0.00–0.81 | 0.027 | 1/15 | 0.10 | 0.01–0.76 | 0.027 |
| NSMP/L1CAM − | 7/15 | 1.59 | 0.55–4.53 | 0.385 | 11/15 | 2.09 | 0.89–4.93 | 0.092 |
| NSMP/L1CAM + | 4/5 | 3.48 | 0.97–11.1 | 0.056 | 5/5 | 3.42 | 1.17–9.99 | 0.024 |
| p53abn | 17/25 | 3.56 | 1.58–8.67 | 0.002 | 19/25 | 2.26 | 1.11–4.57 | 0.024 |
| Age (year) | 36/94 | 1.05 | 1.01–1.09 | 0.018 | 50/93 | 1.04 | 1.01–1.07 | 0.011 |
| Grade (G3 vs. G2) | 36/94 | 0.93 | 0.39–2.43 | 0.870 | 50/93 | 1.97 | 0.82–4.71 | 0.130 |
| Stage (III-IV vs. I-II) | 36/94 | 3.82 | 1.65–10.2 | 0.001 | 50/93 | 3.65 | 1.80–7.42 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravaggi, A.; Capoferri, D.; Ardighieri, L.; Ghini, I.; Ferrari, F.; Romani, C.; Bugatti, M.; Zanotti, L.; Vrede, S.; Tognon, G.; et al. Integrated Biomarker Analysis Reveals L1CAM as a Potential Stratification Marker for No Specific Molecular Profile High-Risk Endometrial Carcinoma. Cancers 2022, 14, 5429. https://doi.org/10.3390/cancers14215429
Ravaggi A, Capoferri D, Ardighieri L, Ghini I, Ferrari F, Romani C, Bugatti M, Zanotti L, Vrede S, Tognon G, et al. Integrated Biomarker Analysis Reveals L1CAM as a Potential Stratification Marker for No Specific Molecular Profile High-Risk Endometrial Carcinoma. Cancers. 2022; 14(21):5429. https://doi.org/10.3390/cancers14215429
Chicago/Turabian StyleRavaggi, Antonella, Davide Capoferri, Laura Ardighieri, Iacopo Ghini, Federico Ferrari, Chiara Romani, Mattia Bugatti, Laura Zanotti, Stephanie Vrede, Germana Tognon, and et al. 2022. "Integrated Biomarker Analysis Reveals L1CAM as a Potential Stratification Marker for No Specific Molecular Profile High-Risk Endometrial Carcinoma" Cancers 14, no. 21: 5429. https://doi.org/10.3390/cancers14215429
APA StyleRavaggi, A., Capoferri, D., Ardighieri, L., Ghini, I., Ferrari, F., Romani, C., Bugatti, M., Zanotti, L., Vrede, S., Tognon, G., Pijnenborg, J. M. A., Sartori, E., Calza, S., Bignotti, E., & Odicino, F. (2022). Integrated Biomarker Analysis Reveals L1CAM as a Potential Stratification Marker for No Specific Molecular Profile High-Risk Endometrial Carcinoma. Cancers, 14(21), 5429. https://doi.org/10.3390/cancers14215429








