A GNAS Gene Mutation’s Independent Expression in the Growth of Colorectal Cancer: A Systematic Review and Meta-Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search and Selection Criteria
2.2. Data Extraction and Quality Assessment
2.3. Data Synthesis and Analysis
3. Result
3.1. Search Results and Study Selection
3.2. Characteristics of the Eligible Studies
3.3. Prevalence of GNAS Mutations in CRC Patients
3.4. Prevalence of GNAS Gene Mutation in Colorectal Cancer Stratified by Study Location and Period of Study
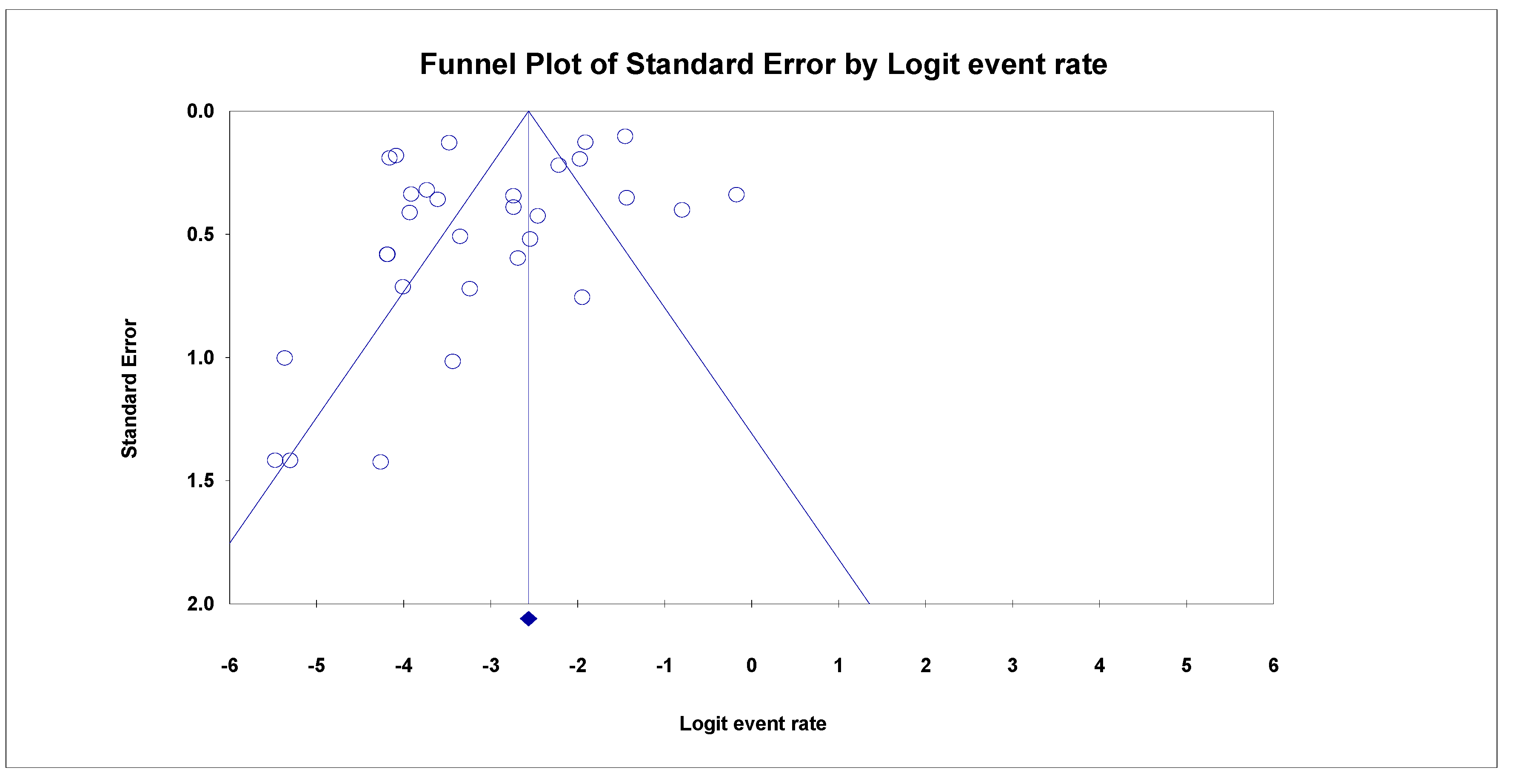
3.5. Analyses of Sensitivity and Publication Bias
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zauber, P.; Marotta, S.P.; Sabbath-Solitare, M. GNAS Gene Mutation May Be Present Only Transiently during Colorectal Tumorigenesis. Int. J. Mol. Epidemiol. Genet. 2016, 7, 24–31. [Google Scholar] [PubMed]
- Fadaka, A.O.; Bakare, O.O.; Pretorius, A.; Klein, A. Genomic Profiling of MicroRNA Target Genes in Colorectal Cancer. Tumor Biol. 2020, 42. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, X.; Luo, J.; Cui, B.; Lei, S.; Si, Z.; Shen, L.; Yao, H. GNA 13 Promotes Tumor Growth and Angiogenesis by Upregulating CXC Chemokines via the NF-κB Signaling Pathway in Colorectal Cancer Cells. Cancer Med. 2018, 7, 5611–5620. [Google Scholar] [CrossRef] [PubMed]
- Steffen, D.J.; Amornphimoltham, P.; Valera, J.L.C.; Taylor, S.; Hunter, T.; Tamayo, P.; Gutkind, J.S. GNAS-PKA Oncosignaling Network in Colorectal Cancer. FASEB J. 2018, 32, 695–699. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Tomosugi, T.; Kimura, R.; Nakamura, S.; Miyasaka, Y.; Nakata, K.; Mori, Y.; Morita, M.; Torata, N.; Shindo, K. Clinical Assessment of the GNAS Mutation Status in Patients with Intraductal Papillary Mucinous Neoplasm of the Pancreas. Surg. Today 2019, 49, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Sekine, S.; Ogawa, R.; Taniguchi, H.; Kushima, R.; Tsuda, H.; Kanai, Y. Frequent Activating GNAS Mutations in Villous Adenoma of the Colorectum. J. Pathol. 2012, 228, 113–118. [Google Scholar] [CrossRef]
- Kim, D.-H.; Park, J.C.; Lee, J.-S. G Protein-Coupled Receptors (GPCRs) in Rotifers and Cladocerans: Potential Applications in Ecotoxicology, Ecophysiology, Comparative Endocrinology, and Pharmacology. Comp. Biochem. Physiol. Part. C Toxicol. Pharmacol. 2022, 256, 109297. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Syst. Rev. 2021, 10, 1–11. [Google Scholar] [CrossRef]
- O’Hayre, M.; Vázquez-Prado, J.; Kufareva, I.; Stawiski, E.W.; Handel, T.M.; Seshagiri, S.; Gutkind, J.S. The Emerging Mutational Landscape of G Proteins and G-Protein-Coupled Receptors in Cancer. Nat. Rev. Cancer 2013, 13, 412–424. [Google Scholar] [CrossRef]
- Afolabi, H.; Md Salleh, S.; Zakaria, Z.; Seng, C.E.; Mohd Nafil, S.N.B.; Abdul Aziz, A.A.B.; Wada, Y.; Irekeola, A. A Systematic Review and Meta-Analysis on the Occurrence of Biomarker Mutation in Colorectal Cancer among the Asian Population. Biomed. Res. Int. 2022, 2022, 5824183. [Google Scholar] [CrossRef]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Shaib, W.L.; Zakka, K.; Staley, C.; Roberts, A.; Akce, M.; Wu, C.; Alese, O.B.; El-Rayes, B.F. Blood-Based Next-Generation Sequencing Analysis of Appendiceal Cancers. Oncologist 2020, 25, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Kweon, S.-S.; Tanikawa, C.; Jia, W.-H.; Xiang, Y.-B.; Cai, Q.; Zeng, C.; Schmit, S.L.; Shin, A.; Matsuo, K.; et al. Large-Scale Genome-Wide Association Study of East Asians Identifies Loci Associated With Risk for Colorectal Cancer. Gastroenterology 2019, 156, 1455–1466. [Google Scholar] [CrossRef]
- Philipovskiy, A.; Ghafouri, R.; Dwivedi, A.K.; Alvarado, L.; McCallum, R.; Maegawa, F.; Konstantinidis, I.T.; Hakim, N.; Shurmur, S.; Awasthi, S.; et al. Association Between Tumor Mutation Profile and Clinical Outcomes Among Hispanic-Latino Patients With Metastatic Colorectal Cancer. Front. Oncol. 2022, 11, 772225. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef]
- Pfarr, N.; Penzel, R.; Klauschen, F.; Heim, D.; Brandt, R.; Kazdal, D.; Jesinghaus, M.; Herpel, E.; Schirmacher, P.; Warth, A.; et al. Copy Number Changes of Clinically Actionable Genes in Melanoma, Non-Small Cell Lung Cancer and Colorectal CancerA Survey across 822 Routine Diagnostic Cases. GENES Chromosom. CANCER 2016, 55, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Munn, Z.; Moola, S.; Lisy, K.; Riitano, D.; Tufanaru, C. Methodological Guidance for Systematic Reviews of Observational Epidemiological Studies Reporting Prevalence and Cumulative Incidence Data. Int. J. Evid. Based. Healthc. 2015, 13, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Viechtbauer, W. Conducting Meta-Analyses in R with the Metafor Package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- Wang, C.K.; Sandhu, J.; Fakih, M. Mucinous Histology Is Associated with Resistance to Anti-EGFR Therapy in Patients with Left-Sided RAS/BRAF Wild-Type Metastatic Colorectal Cancer. Oncologist 2022, 27, 104–109. [Google Scholar] [CrossRef]
- Borazanci, E.; Millis, S.Z.; Kimbrough, J.; Doll, N.; von Hoff, D.; Ramanathan, R.K. Potential Actionable Targets in Appendiceal Cancer Detected by Immunohistochemistry, Fluorescent in Situ Hybridization, and Mutational Analysis. J. Gastrointest. Oncol. 2017, 8, 164–172. [Google Scholar] [CrossRef]
- Stein, M.K.; Williard, F.W.; Xiu, J.; Tsao, M.W.; Martin, M.G.; Deschner, B.W.; Dickson, P.V.; Glazer, E.S.; Yakoub, D.; Shibata, D.; et al. Comprehensive Tumor Profiling Reveals Unique Molecular Differences between Peritoneal Metastases and Primary Colorectal Adenocarcinoma. J. Surg. Oncol. 2020, 121, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.M. 642P—Frequent BRAF, GNAS and SMAD4 Mutations Identified in Colorectal Mucinous Carcinomas. Ann. Oncol. 2019, 30, v242. [Google Scholar] [CrossRef]
- Tokunaga, R.; Xiu, J.; Johnston, C.; Goldberg, R.M.; Philip, P.A.; Seeber, A.; Naseem, M.; Lo, J.H.; Arai, H.; Battaglin, F. Molecular Profiling of Appendiceal Adenocarcinoma and Comparison with Right-Sided and Left-Sided Colorectal CancerMolecular Profiling of Appendiceal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 3096–3103. [Google Scholar] [CrossRef]
- Ang, C.S.-P.; Shen, J.P.; Hardy-Abeloos, C.J.; Huang, J.K.; Ross, J.S.; Miller, V.A.; Jacobs, M.T.; Chen, I.L.; Xu, D.; Ali, S.M.; et al. Genomic Landscape of Appendiceal Neoplasms. JCO Precis. Oncol. 2018, 2, 1–18. [Google Scholar] [CrossRef]
- Parish, A.J.; Nguyen, V.; Goodman, A.M.; Murugesan, K.; Frampton, G.M.; Kurzrock, R. GNAS, GNAQ, and GNA11 Alterations in Patients with Diverse Cancers. Cancer 2018, 124, 4080–4089. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, T.; Liebmann-Reindl, S.; Glueck, O.; Starlinger, P.; Laengle, J.; Birner, P.; Klepetko, W.; Pils, D.; Streubel, B.; Hoetzenecker, K. Mutational Profile of Colorectal Cancer Lung Metastases and Paired Primary Tumors by Targeted next Generation Sequencing: Implications on Clinical Outcome after Surgery. J. Thorac. Dis. 2018, 10, 6147–6157. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Loree, J.M.; Advani, S.M.; Ning, J.; Li, W.; Pereira, A.A.L.L.; Lam, M.; Raghav, K.; Morris, V.K.; Broaddus, R.; et al. Prognostic Implications of Mucinous Differentiation in Metastatic Colorectal Carcinoma Can Be Explained by Distinct Molecular and Clinicopathologic Characteristics. Clin. Colorectal Cancer 2018, 17, e699–e709. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; La, B.M.; Hwang, I.; Kang, Y.N.; Choi, I.J.; Lee, J.H. Absence of GNAS Mutation in Colorectal Carcinogenesis. Tumori 2017, 103, 209–211. [Google Scholar] [CrossRef]
- Chang, P.Y.; Chen, J.S.; Chang, S.C.; Wang, M.C.; Chang, N.C.; Wen, Y.H.; Tsai, W.S.; Liu, W.H.; Liu, H.L.; Lu, J.J. Acquired Somatic TP53 or PIK3CA Mutations Are Potential Predictors of When Polyps Evolve into Colorectal Cancer. Oncotarget 2017, 8, 72352–72362. [Google Scholar] [CrossRef]
- Loree, J.M.; Pereira, A.A.L.; Lam, M.; Willauer, A.N.; Raghav, K.; Dasari, A.; Morris, V.; Advani, S.; Menter, D.G.; Eng, C. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular SubtypesmCRC Profile by Location. Clin. Cancer Res. 2018, 24, 1062–1072. [Google Scholar] [CrossRef]
- Liu, C.; McKeone, D.M.; Walker, N.I.; Bettington, M.L.; Leggett, B.A.; Whitehall, V.L.J. GNAS Mutations Are Present in Colorectal Traditional Serrated Adenomas, Serrated Tubulovillous Adenomas and Serrated Adenocarcinomas with Adverse Prognostic Features. Histopathology 2017, 70, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Jung, S.H.; Kim, T.-M.; Rhee, J.-K.; Park, H.-C.; Kim, M.S.; Kim, S.S.; An, C.H.; Lee, S.H.; Chung, Y.-J. Whole-Exome Sequencing Identified Mutational Profiles of High-Grade Colon Adenomas. Oncotarget 2017, 8, 6579–6588. [Google Scholar] [CrossRef] [PubMed]
- Jauhri, M.; Bhatnagar, A.; Gupta, S.; Shokeen, Y.; Minhas, S.; Aggarwal, S. Targeted Molecular Profiling of Rare Genetic Alterations in Colorectal Cancer Using Next-Generation Sequencing. Med. Oncol. 2016, 33. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.S.; Cates, J.M.M.M.; Washington, M.K.; Beauchamp, R.D.; Coffey, R.J.; Shi, C.J. Adenoma-like Adenocarcinoma: A Subtype of Colorectal Carcinoma with Good Prognosis, Deceptive Appearance on Biopsy and Frequent KRAS Mutation. Histopathology 2016, 68, 183–190. [Google Scholar] [CrossRef]
- Crumley, S.M.; Pepper, K.L.; Phan, A.T.; Olsen, R.J.; Schwartz, M.R.; Portier, B.P. Next-Generation Sequencing of Matched Primary and Metastatic Rectal Adenocarcinomas Demonstrates Minimal Mutation Gain and Concordance to Colonic Adenocarcinomas. Arch. Pathol. Lab. Med. 2016, 140, 529–535. [Google Scholar] [CrossRef]
- Stachler, M.D.; Rinehart, E.; Lindeman, N.; Odze, R.; Srivastava, A. Novel Molecular Insights from Routine Genotyping of Colorectal Carcinomas. Hum. Pathol. 2015, 46, 507–513. [Google Scholar] [CrossRef]
- Alakus, H.; Babicky, M.L.; Ghosh, P.; Yost, S.; Jepsen, K.; Dai, Y.; Arias, A.; Samuels, M.L.; Mose, E.S.; Schwab, R.B.; et al. Genome-Wide Mutational Landscape of Mucinous Carcinomatosis Peritonei of Appendiceal Origin. Genome Med. 2014, 6. [Google Scholar] [CrossRef]
- Fecteau, R.E.; Lutterbaugh, J.; Markowitz, S.D.; Willis, J.; Guda, K. GNAS Mutations Identify a Set of Right-Sided, RAS Mutant, Villous Colon Cancers. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Walther, B.M.; Walther, I.; Chen, Y.; Petersen, I. GNAS1 Mutation Analysis in Gastrointestinal Tumors. FOLIA Histochem. Cytobiol. 2014, 52, 90–95. [Google Scholar] [CrossRef]
- Wiland IV, H.O.; Shadrach, B.; Allende, D.; Carver, P.; Goldblum, J.R.; Liu, X.; Patil, D.T.; Rybicki, L.A.; Pai, R.K. Morphologic and Molecular Characterization of Traditional Serrated Adenomas of the Distal Colon and Rectum. Am. J. Surg. Pathol. 2014, 38, 1290–1297. [Google Scholar] [CrossRef]
- Abdul-Jalil, K.I.; Sheehan, K.M.; Toomey, S.; Schmid, J.; Prehn, J.; O’Grady, A.; Cummins, R.; O’Neill, B.; McNamara, D.A.; Deasy, J.; et al. The Frequencies and Clinical Implications of Mutations in 33 Kinase-Related Genes in Locally Advanced Rectal Cancer: A Pilot Study. Ann. Surg. Oncol. 2014, 21, 2642–2649. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, G.; Sekine, S.; Ogawa, R.; Matsubara, A.; Mori, T.; Taniguchi, H.; Kushima, R.; Hiraoka, N.; Tsuta, K.; Tsuda, H.; et al. Frequent GNAS Mutations in Low-Grade Appendiceal Mucinous Neoplasms. Br. J. Cancer 2013, 108, 951–958. [Google Scholar] [CrossRef]
- Idziaszczyk, S.; Wilson, C.H.; Smith, C.G.; Adams, D.J.; Cheadle, J.P. Analysis of the Frequency of GNAS Codon 201 Mutations in Advanced Colorectal Cancer. Cancer Genet. Cytogenet. 2010, 202, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Lamba, S.; Felicioni, L.; Buttitta, F.; Bleeker, F.E.; Malatesta, S.; Corbo, V.; Scarpa, A.; Rodolfo, M.; Knowles, M.; Frattini, M.; et al. Mutational Profile of GNAQ(Q209) in Human Tumors. PLoS ONE 2009, 4, e6833. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Xiao, H.J.; Wu, H.T.; Yao, C.; He, H.; Wang, C.; Li, W. G Protein Subunit Alpha q Regulates Gastric Cancer Growth via the P53/P21 and MEK/ERK Pathways. Oncol. Rep. 2017, 37, 1998–2006. [Google Scholar] [CrossRef]
- Domingo, E.; Camps, C.; Kaisaki, P.J.; Parsons, M.J.; Mouradov, D.; Pentony, M.M.; Makino, S.; Palmieri, M.; Ward, R.L.; Hawkins, N.J.; et al. Mutation Burden and Other Molecular Markers of Prognosis in Colorectal Cancer Treated with Curative Intent: Results from the QUASAR 2 Clinical Trial and an Australian Community-Based Series. Lancet Gastroenterol. Hepatol. 2018, 3, 635–643. [Google Scholar] [CrossRef]
- Tsai, J.-H.; Yuan, R.-H.; Chen, Y.-L.; Liau, J.-Y.; Jeng, Y.-M. GNAS Is Frequently Mutated in a Specific Subgroup of Intraductal Papillary Neoplasms of the Bile Duct. Am. J. Surg. Pathol. 2013, 37, 1862–1870. [Google Scholar] [CrossRef]
- Testa, U.; Castelli, G.; Pelosi, E. Genetic Alterations of Metastatic Colorectal Cancer. Biomedicines 2020, 8, 414. [Google Scholar] [CrossRef]
- Fu, X.; Lin, H.; Fan, X.; Zhu, Y.; Wang, C.; Chen, Z.; Tan, X.; Huang, J.; Cai, Y.; Huang, Y. The Spectrum, Tendency and Predictive Value of PIK3CA Mutation in Chinese Colorectal Cancer Patients. Front. Oncol. 2021, 11, 595675. [Google Scholar] [CrossRef]
- Ferlizza, E.; Solmi, R.; Miglio, R.; Nardi, E.; Mattei, G.; Sgarzi, M.; Lauriola, M. Colorectal Cancer Screening: Assessment of CEACAM6, LGALS4, TSPAN8 and COL1A2 as Blood Markers in Faecal Immunochemical Test Negative Subjects. J. Adv. Res. 2020, 24, 99–107. [Google Scholar] [CrossRef]
- Cueto-López, N.; García-Ordás, M.T.; Dávila-Batista, V.; Moreno, V.; Aragonés, N.; Alaiz-Rodríguez, R. A Comparative Study on Feature Selection for a Risk Prediction Model for Colorectal Cancer. Comput. Methods Programs Biomed. 2019, 177, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Raskin, L.; Dakubo, J.C.B.; Palaski, N.; Greenson, J.K.; Gruber, S.B. Distinct Molecular Features of Colorectal Cancer in Ghana. Cancer Epidemiol. 2013, 37, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Osasan, S.A. The pathological features of colorectal carcinoma in ile-ife:-a ten-year descriptive retrospective study. Fac. Pathol. 2007. [Google Scholar]
- Palos-Paz, F.; Perez-Guerra, O.; Cameselle-Teijeiro, J.; Rueda-Chimeno, C.; Barreiro-Morandeira, F.; Lado-Abeal, J.; Araujo Vilar, D.; Argueso, R.; Barca, O.; Botana, M. Prevalence of Mutations in TSHR, GNAS, PRKAR1A and RAS Genes in a Large Series of Toxic Thyroid Adenomas from Galicia, an Iodine-Deficient Area in NW Spain. Eur. J. Endocrinol. 2008, 159, 623. [Google Scholar] [CrossRef] [PubMed]
- Jauhri, M.; Bhatnagar, A.; Gupta, S.; Bp, M.; Minhas, S.; Shokeen, Y.; Aggarwal, S. Prevalence and Coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC Mutations in Indian Colorectal Cancer Patients: Next-Generation Sequencing–Based Cohort Study. Tumor Biol. 2017, 39, 1010428317692265. [Google Scholar] [CrossRef]
- Sekine, S.; Ogawa, R.; Oshiro, T.; Kanemitsu, Y.; Taniguchi, H.; Kushima, R.; Kanai, Y. Frequent Lack of GNAS Mutations in Colorectal Adenocarcinoma Associated with GNAS-Mutated Villous Adenoma. Genes Chromosom. Cancer 2014, 53, 366–372. [Google Scholar] [CrossRef]
- Al-Shamsi, H.O.; Jones, J.; Fahmawi, Y.; Dahbour, I.; Tabash, A.; Abdel-Wahab, R.; Abousamra, A.O.S.; Shaw, K.R.; Xiao, L.; Hassan, M.M. Molecular Spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC Somatic Gene Mutations in Arab Patients with Colorectal Cancer: Determination of Frequency and Distribution Pattern. J. Gastrointest. Oncol. 2016, 7, 882. [Google Scholar] [CrossRef]
- Tezcan, G.; Tunca, B.; Ak, S.; Cecener, G.; Egeli, U. Molecular Approach to Genetic and Epigenetic Pathogenesis of Early-Onset Colorectal Cancer. World, J. Gastrointest. Oncol. 2016, 8, 83. [Google Scholar] [CrossRef]
- Schult, A.L.; Botteri, E.; Hoff, G.; Randel, K.R.; Dalén, E.; Eskeland, S.L.; Holme, Ø.; de Lange, T. Detection of Cancers and Advanced Adenomas in Asymptomatic Participants in Colorectal Cancer Screening: A Cross-Sectional Study. BMJ Open 2021, 11, e048183. [Google Scholar] [CrossRef]
- Heshmat-Ghahdarijani, K.; Najafian, J.; Vafaei, Z.; Mostafavi, S.; Mohammadifard, N.; Mansourian, M.; Ashrafi, F.; Sharifi, M.; Khosravifarsani, M.; Darakhshandeh, A.; et al. Rational, Design and Preliminary Results of a Cohort Study on Breast and Colorectal Cancer to Develop a Risk Assessment Model to Predict Future Cardiovascular Events. “Cardio Vascular Events in Breast and Colorectal Cancers (CIBC) Study”. Curr. Probl. Cardiol. 2021, 47, 100958. [Google Scholar] [CrossRef]
- Dolin, T.G.; Mikkelsen, M.; Jakobsen, H.L.; Nordentoft, T.; Pedersen, T.S.; Vinther, A.; Zerahn, B.; Vistisen, K.K.; Suetta, C.; Nielsen, D. Geriatric Assessment and Intervention in Older Vulnerable Patients Undergoing Surgery for Colorectal Cancer: A Protocol for a Randomised Controlled Trial (GEPOC Trial). BMC Geriatr. 2021, 21, 88. [Google Scholar] [CrossRef] [PubMed]
- White, A.; Ironmonger, L.; Steele, R.J.C.; Ormiston-Smith, N.; Crawford, C.; Seims, A. A Review of Sex-Related Differences in Colorectal Cancer Incidence, Screening Uptake, Routes to Diagnosis, Cancer Stage and Survival in the UK. BMC Cancer 2018, 18, 906. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R. An Examination of Colorectal Cancer Burden by Socioeconomic Status: Evidence from GLOBOCAN 2018. EPMA J. 2020, 11, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Scherer, S.; Jansen, L.; Boakye, D.; Hoffmeister, M.; Brenner, H. Changes in Health-Related Outcomes among Colorectal Cancer Patients Undergoing Inpatient Rehabilitation Therapy: A Systematic Review of Observational and Interventional Studies. Acta Oncol. 2021, 60, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Thurmaier, J.; Heinemann, V.; Engel, J.; Schubert-Fritschle, G.; Wiedemann, M.; Nüssler, N.C.; Ruppert, R.; Kleeff, J.; Schepp, W.; Löhe, F. Patients with Colorectal Cancer and Brain Metastasis: The Relevance of Extracranial Metastatic Patterns Predicting Time Intervals to First Occurrence of Intracranial Metastasis and Survival. Int. J. Cancer 2021, 148, 1919–1927. [Google Scholar] [CrossRef]
- Kawamata, H.; Yamashita, K.; Kojo, K.; Ushiku, H.; Ooki, A.; Watanabe, M. Discrepancies between the K-Ras Mutational Status of Primary Colorectal Cancers and Corresponding Liver Metastases Are Found in Codon 13. Genomics 2015, 106, 71–75. [Google Scholar] [CrossRef]
- Koi, M.; Garcia, M.; Choi, C.; Kim, H.-R.; Koike, J.; Hemmi, H.; Nagasaka, T.; Okugawa, Y.; Toiyama, Y.; Kitajima, T. Microsatellite Alterations with Allelic Loss at 9p24. 2 Signify Less-Aggressive Colorectal Cancer Metastasis. Gastroenterology 2016, 150, 944–955. [Google Scholar] [CrossRef]
- Nakajima, Y.; Okamura, T.; Horiguchi, K.; Gohko, T.; Miyamoto, T.; Satoh, T.; Ozawa, A.; Ishii, S.; Yamada, E.; Hashimoto, K.; et al. GNAS Mutations in Adrenal Aldosterone-Producing Adenomas. Endocr. J. 2016, 63, 199–204. [Google Scholar]
- More, A.; Ito, I.; Haridas, V.; Chowdhury, S.; Gu, Y.; Dickson, P.; Fowlkes, N.; Shen, J.P. Oncogene Addiction to GNAS in GNASR201 Mutant Tumors. Oncogene 2022, 41, 4159–4168. [Google Scholar] [CrossRef]
- Nomura, R.; Saito, T.; Mitomi, H.; Hidaka, Y.; Lee, S.; Watanabe, S.; Yao, T. GNAS Mutation as an Alternative Mechanism of Activation of the Wnt/β-Catenin Signaling Pathway in Gastric Adenocarcinoma of the Fundic Gland Type. Hum. Pathol. 2014, 45, 2488–2496. [Google Scholar] [CrossRef]
- Schmoll, H.J.; Van Cutsem, E.; Stein, A.; Valentini, V.; Glimelius, B.; Haustermans, K.; Nordlinger, B.; Van de Velde, C.J.; Balmana, J.; Regula, J. ESMO Consensus Guidelines for Management of Patients with Colon and Rectal Cancer. a Personalized Approach to Clinical Decision Making. Ann. Oncol. 2012, 23, 2479–2516. [Google Scholar] [CrossRef] [PubMed]

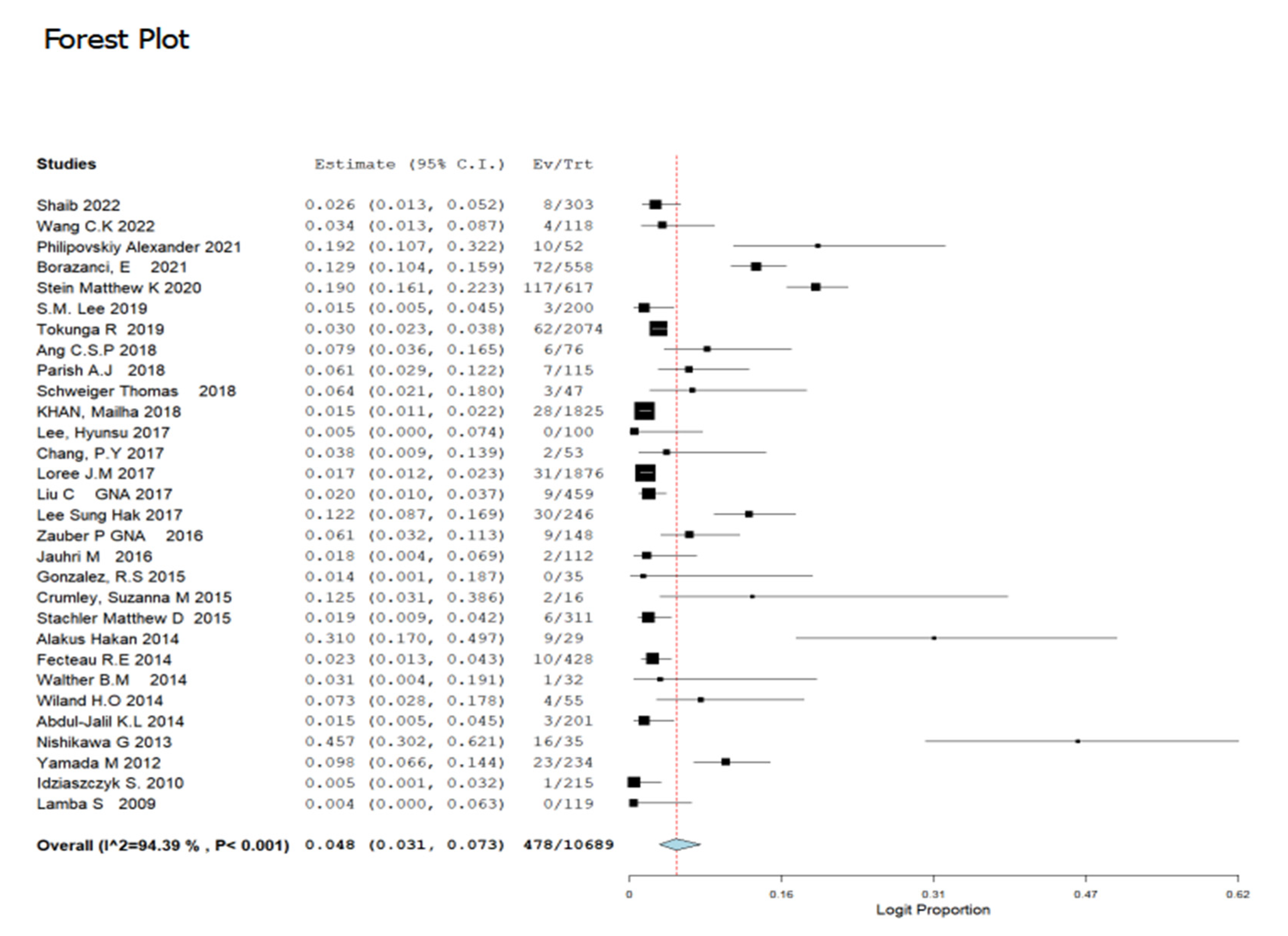
| Nr. | Author | Year | Location | Male n (%) | Age * | Sample size | Tumour Stage (Early Stage 1&2) | Tumour Stage (Late-Stage 3&4) | Tumour Location (Colon) | Tumour Location (Rectum) | Tumour Grade (Poor) | Tumour Grade (Moderate) | Tumour Grade (Well) | Method | Total GNAS Mutation (%) | GNAQ (%) | GNA11 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Shaib et al. [12]. | 2022 | USA | 44 | 56.8 (54–83) | 303 | NR | NR | NR | NR | NR | NR | NR | NGS-sequencing | 2.6 | NR | NR |
| 2 | Wang et al. [19] | 2022 | USA | 64.4 | 52 (19–88) | 118 | 22 | 78 | NR | NR | NR | NR | NR | Sanger sequencing | 3.4 | NR | NR |
| 3 | Philipovskiy et al. [14] | 2021 | USA | 69.2 | 58.67 ± 10.64 | 52 | NR | 52 | NR | NR | NR | NR | NR | sequencing | 19.2 | NR | NR |
| 4 | Borazanci et al. [20] | 2021 | Norway | 40.5 | 56 (20–88) | 558 | NR | NR | NR | NR | NR | NR | NR | NGS-sequencing | 12.9 | NR | NR |
| 5 | Stein et al. [21] | 2020 | USA | 55 | 59 (16–91) | 617 | NR | NR | 421 | 147 | 7.9 | 49 | 25 | sequencing | 19 | NR | NR |
| 6 | Lee, S.M. et al. [22] | 2019 | South Korea | 54.1 | 58 (20–80) | 200 | NR | NR | NR | NR | NR | NR | NR | sequencing | 1.5 | NR | NR |
| 7 | Tokunaga et al. [23] | 2019 | USA | 1435 | NR | 2074 | NR | NR | NR | NR | NR | NR | NR | NGS-sequencing | 3 | NR | NR |
| 8 | Ang et al. [24] | 2018 | USA | 40.8 | 53.4 (23.6–82.8) | 76 | 8 | 92 | NR | NR | 44.7 | 11.8 | NR | sequencing | 7.9 | NR | NR |
| 9 | Parish et al. [25] | 2018 | USA | 60.9 | 56.1 (1.0–95.1) | 115 | NR | NR | NR | NR | NR | NR | NR | sequencing | 6.1 | NR | NR |
| 10 | Schweiger et al. [26] | 2018 | Austria | 55.3 | 63(44–83) | 47 | 34 | 61.7 | 59.6 | 40.4 | NR | NR | NR | sequencing | 6.4 | 11 | 19.1 |
| 11 | Khan et al. [27] | 2018 | USA | 56.7 | 55.2 (19.1–91.8) | 1825 | 35.4 | 64.6 | NR | NR | NR | NR | NR | sequencing | 1.5 | NR | NR |
| 12 | Lee, H. et al. [28] | 2017 | South Korea | 60 | NR | 100 | NR | NR | NR | NR | NR | NR | NR | sequencing | 0.5 | NR | NR |
| 13 | Chang et al. [29] | 2017 | Taiwan | 75 | 58 (26–75) | 53 | NR | NR | 81 | 19 | NR | NR | NR | sequencing | 3.8 | NR | NR |
| 14 | Loree et al. [30] | 2017 | USA | 56 | 55(46–63) | 1876 | 78.2 | 3 | 77.5 | 22.5 | NR | NR | NR | NGS-sequencing | 1.7 | NR | NR |
| 15 | Liu, C. et al. [31] | 2017 | Australia | 53.8 | 68.3 ± 13.5 | 459 | NR | NR | NR | NR | NR | NR | NR | Sanger sequencing | 2 | NR | NR |
| 16 | Lee, S.H. et al. [32] | 2017 | South Korea | 150 | NR | 246 | NR | NR | NR | NR | NR | NR | NR | W.E. Sequencing | 12.2 | 0.9 | NR |
| 17 | Zauber, M. et al. [1] | 2016 | USA | 30 | 69 (24–95) | 148 | 52 | 44.6 | NR | NR | NR | NR | NR | sequencing | 6.1 | NR | NR |
| 18 | Jauhri et al. [33] | 2016 | India | 70 | NR | 112 | NR | NR | NR | NR | NR | NR | NR | NGS-sequencing | 1 | 0.9 | NR |
| 19 | Gonzalez et al. [34] | 2015 | USA | 40 | 69 (27–89) | 35 | 31 | 69 | 69 | 19 | 3 | 17 | NR | Sanger sequencing | 1.4 | NR | NR |
| 20 | Crumley et al. [35] | 2015 | USA | 56.3 | 57 (21–85) | 16 | 0 | 100 | 0 | 100 | 19 | 0 | NR | NGS-sequencing | 12.5 | NR | NR |
| 21 | Stachler et al. [36] | 2015 | USA | 61 | 56.9 (21–89) | 311 | 18 | 68.5 | 72 | 27.6 | 30.2 | 0 | 3.9 | sequencing | 1.9 | NR | NR |
| 22 | H Alakus et al. [37] | 2014 | USA | 20 | 54 (22–90) | 29 | NR | NR | NR | NR | NR | NR | NR | sequencing | 3.1 | NR | NR |
| 23 | Fecteau et al. [38] | 2014 | USA | 49.5 | NR | 428 | 34.3 | 65.6 | NR | NR | NR | NR | NR | pyrosequencing | 2.3 | NR | NR |
| 24 | B M Walther et al. [39] | 2014 | Germany | 20 | 77 (58–85) | 32 | NR | NR | NR | NR | NR | NR | NR | sequencing | 3.1 | NR | NR |
| 25 | Wiland IV et al. [40] | 2014 | USA | 47 | 60 (38–82) | 55 | NR | NR | 45.5 | 55.5 | NR | NR | NR | sequencing | 7.3 | NR | NR |
| 26 | Abdul-Jalil et al. [41] | 2014 | Ireland | 70 | 63 (38–80) | 201 | 9 | 87 | NR | NR | 18 | 12 | 4 | NGS-sequencing | 1.5 | NR | NR |
| 27 | Nishikawa et al. [42] | 2013 | Japan | 20 | 56 (18–80) | 35 | NR | NR | NR | NR | NR | NR | NR | Sequencing | 45.7 | NR | NR |
| 28 | M Yamada et al. [6] | 2012 | Japan | 140 | NR | 234 | NR | NR | NR | NR | NR | NR | NR | Sequencing | 9.8 | NR | NR |
| 29 | Idziaszczyk W et al. [43] | 2010 | UK | 130 | NR | 215 | NR | NR | NR | NR | NR | NR | NR | sequencing | 0.5 | NR | NR |
| 30 | S Lamba et al. [44] | 2009 | Italy | 70 | NR | 119 | NR | NR | NR | NR | NR | NR | NR | NGS-sequencing | 0.4 | NR | NR |
| Subgroup | No of Studies | Prevalence (%) | 95% CI | I2 (%) | Q | Heterogeneity Test | |
|---|---|---|---|---|---|---|---|
| DF | p | ||||||
| Study Location | |||||||
| USA | 16 | 4.8 | 0.033–0.062 | 90.74 | 161.95 | 15 | <0.001 |
| Norway | 1 | 12.9 | 0.101–0.157 | NA | NA | NA | NA |
| South Korea | 3 | 4.2 | 0.002–0.086 | 92.93 | 28.31 | 2 | <0.001 |
| Austria | 1 | 6.4 | 0.006–0.134 | NA | NA | NA | NA |
| Taiwan | 1 | 3.8 | 0.014–0.089 | 77.04 | NA | NA | NA |
| Australia | 1 | 2 | 0.007–0.032 | NA | NA | NA | NA |
| India | 1 | 1.8 | 0.007–0.042 | NA | NA | NA | NA |
| Germany | 1 | 3.1 | 0.029–0.092 | NA | NA | NA | NA |
| Ireland | 1 | 1.5 | 0.002–0.032 | NA | NA | NA | NA |
| Japan | 2 | 26.8 | 0.083–0.620 | 94.2 | 17.24 | 1 | <0.001 |
| United Kingdom | 1 | 0.5 | 0.007–0.016 | NA | NA | NA | NA |
| Italy | 1 | 0.4 | 0.176–0.306 | NA | NA | NA | NA |
| Overall | 30 | 4.5 | 0.034–0.056 | 90.82 | 315.89 | 29 | <0.001 |
| GNAS Subgroup by Gender of Study Conduct | |||||||
| Male gender | 20 | 56.9 | 0.482–0.595 | 94.84 | 367.997 | 19 | <0.001 |
| Female gender | 20 | 43.4 | 0.378–0.492 | 95.08 | 386.062 | 29 | <0.001 |
| GNAS Subgroup by Tumour Stage | |||||||
| Early Tumour Stage (1) | 11 | 27.3 | 0.152–0.441 | 98.99 | 987.069 | 10 | <0.001 |
| Late Tumour Stage (2) | 11 | 67.9 | 0.497–0.843 | 98.87 | 974.316 | 10 | <0.001 |
| GNAS Subgroup by Tumour Location | |||||||
| Colon | 8 | 50.5 | 0.332–0.676 | 97.93 | 338.303 | 7 | <0.001 |
| Rectum | 8 | 21 | 0.150–0.287 | 93.52 | 108.081 | 7 | <0.001 |
| GNAS Subgroup by Tumour Grading | |||||||
| Poor | 6 | 18.3 | 0.091–0.334 | 95.09 | 101.748 | 5 | <0.001 |
| Moderate | 6 | 10.7 | 0.033–0.296 | 95.99 | 124.645 | 5 | <0.001 |
| Well | 6 | 57.5 | 0.324–0.792 | 98.1 | 263.622 | 5 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afolabi, H.A.; Salleh, S.M.; Zakaria, Z.; Ch’ng, E.S.; Mohd Nafi, S.N.; Abdul Aziz, A.A.B.; Irekeola, A.A.; Wada, Y.; Al-Mhanna, S.B. A GNAS Gene Mutation’s Independent Expression in the Growth of Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 5480. https://doi.org/10.3390/cancers14225480
Afolabi HA, Salleh SM, Zakaria Z, Ch’ng ES, Mohd Nafi SN, Abdul Aziz AAB, Irekeola AA, Wada Y, Al-Mhanna SB. A GNAS Gene Mutation’s Independent Expression in the Growth of Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers. 2022; 14(22):5480. https://doi.org/10.3390/cancers14225480
Chicago/Turabian StyleAfolabi, Hafeez Abiola, Salzihan Md Salleh, Zaidi Zakaria, Ewe Seng Ch’ng, Siti Norasikin Mohd Nafi, Ahmad Aizat Bin Abdul Aziz, Ahmad Adebayo Irekeola, Yusuf Wada, and Sameer Badri Al-Mhanna. 2022. "A GNAS Gene Mutation’s Independent Expression in the Growth of Colorectal Cancer: A Systematic Review and Meta-Analysis" Cancers 14, no. 22: 5480. https://doi.org/10.3390/cancers14225480
APA StyleAfolabi, H. A., Salleh, S. M., Zakaria, Z., Ch’ng, E. S., Mohd Nafi, S. N., Abdul Aziz, A. A. B., Irekeola, A. A., Wada, Y., & Al-Mhanna, S. B. (2022). A GNAS Gene Mutation’s Independent Expression in the Growth of Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers, 14(22), 5480. https://doi.org/10.3390/cancers14225480








