Analysis of Circulating Immune Subsets in Primary Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Flow Cytometry Data Analysis
2.3. Construction of a Diagnostic Model
2.4. Gene Expression Profile Collection and Processing
2.5. xCell Algorithm
2.6. Differential Expression Analysis
2.7. Gene Ontology Enrichment Analysis
2.8. Correlation Analysis
2.9. Statistical Analysis
3. Results
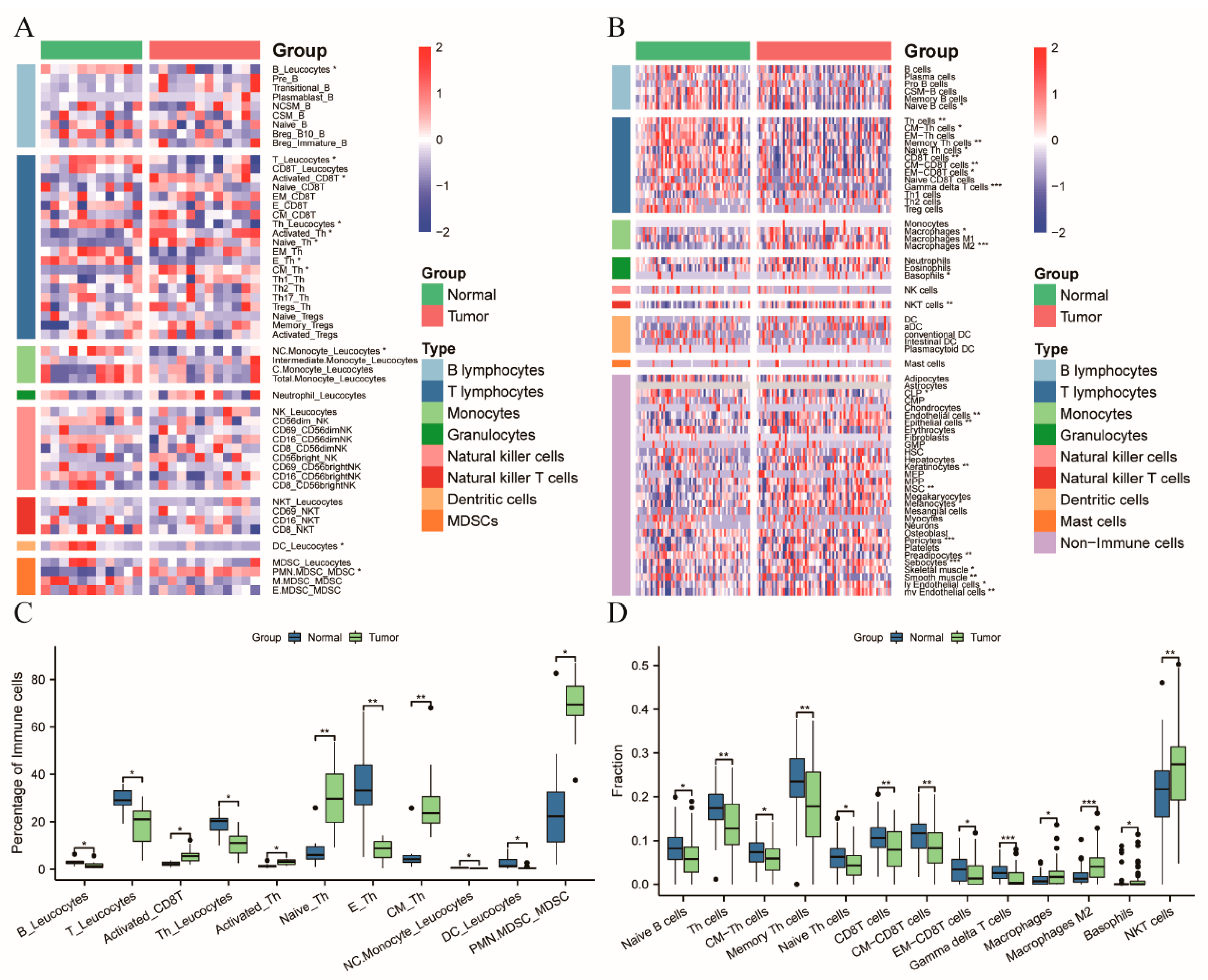
3.1. Patients with CRC Exhibiting a Systemic Immune Suppression
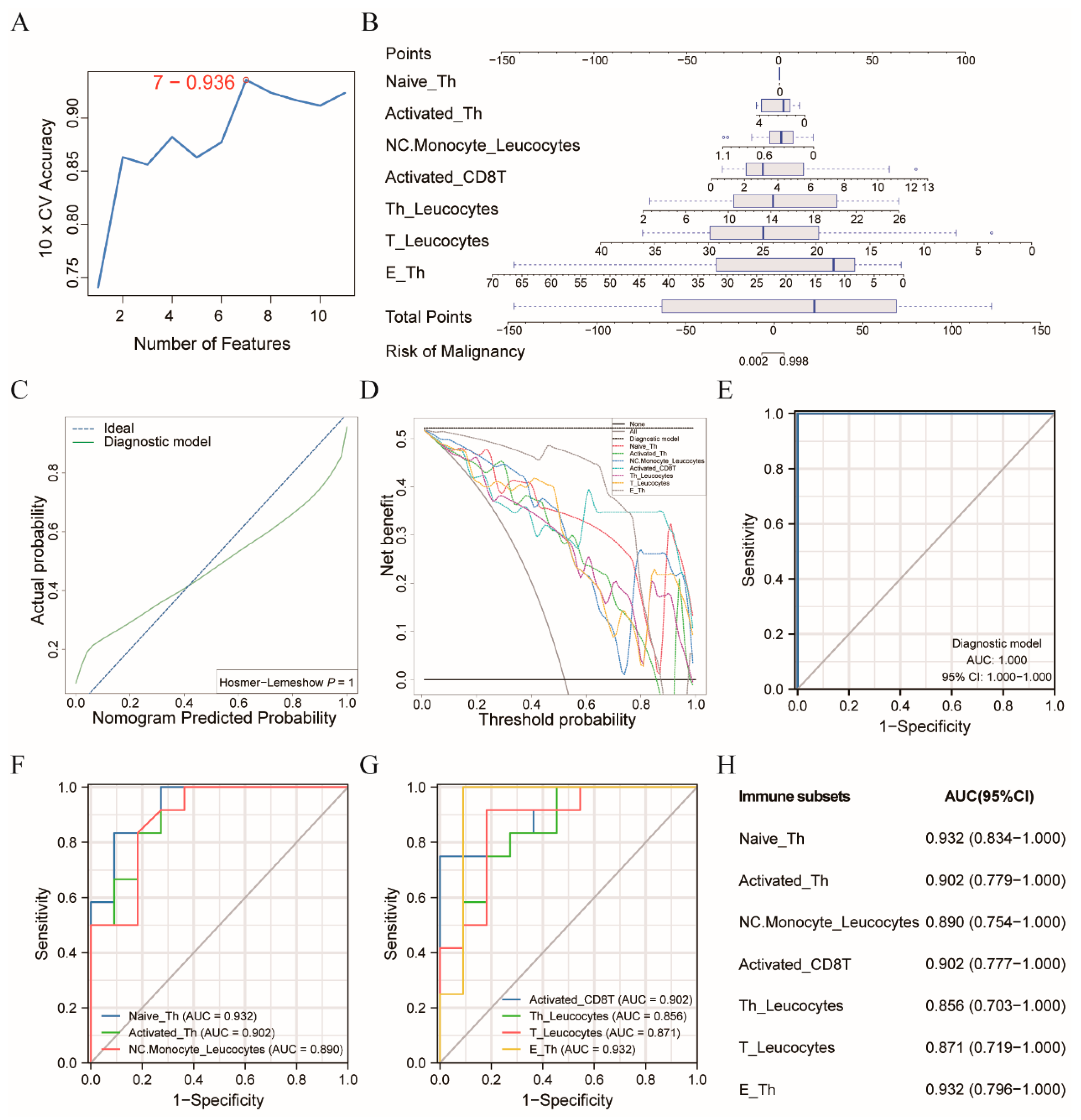
3.2. Diagnostic Model Allowed for the Differentiation of the CRC Patients from the Healthy Controls
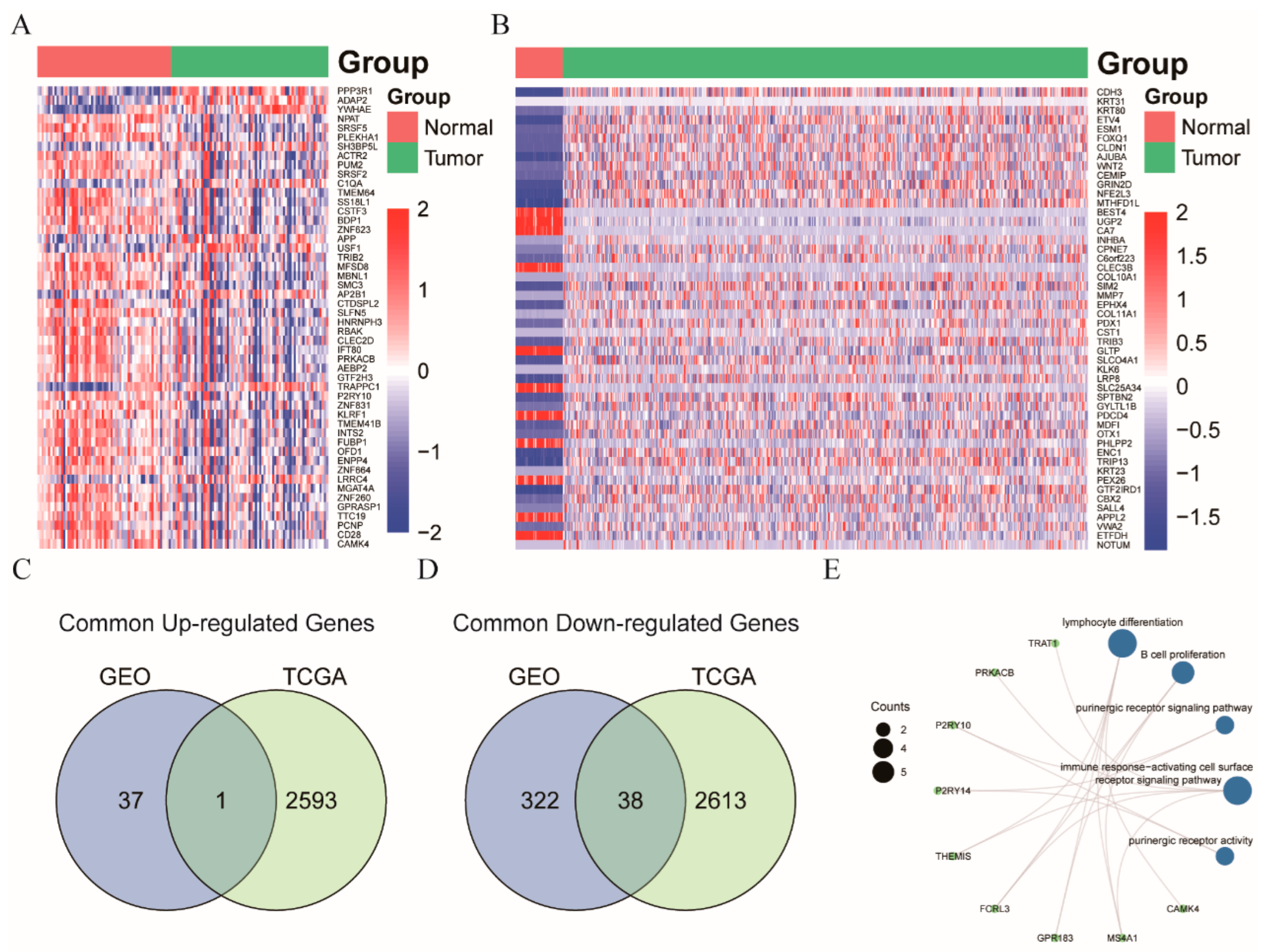
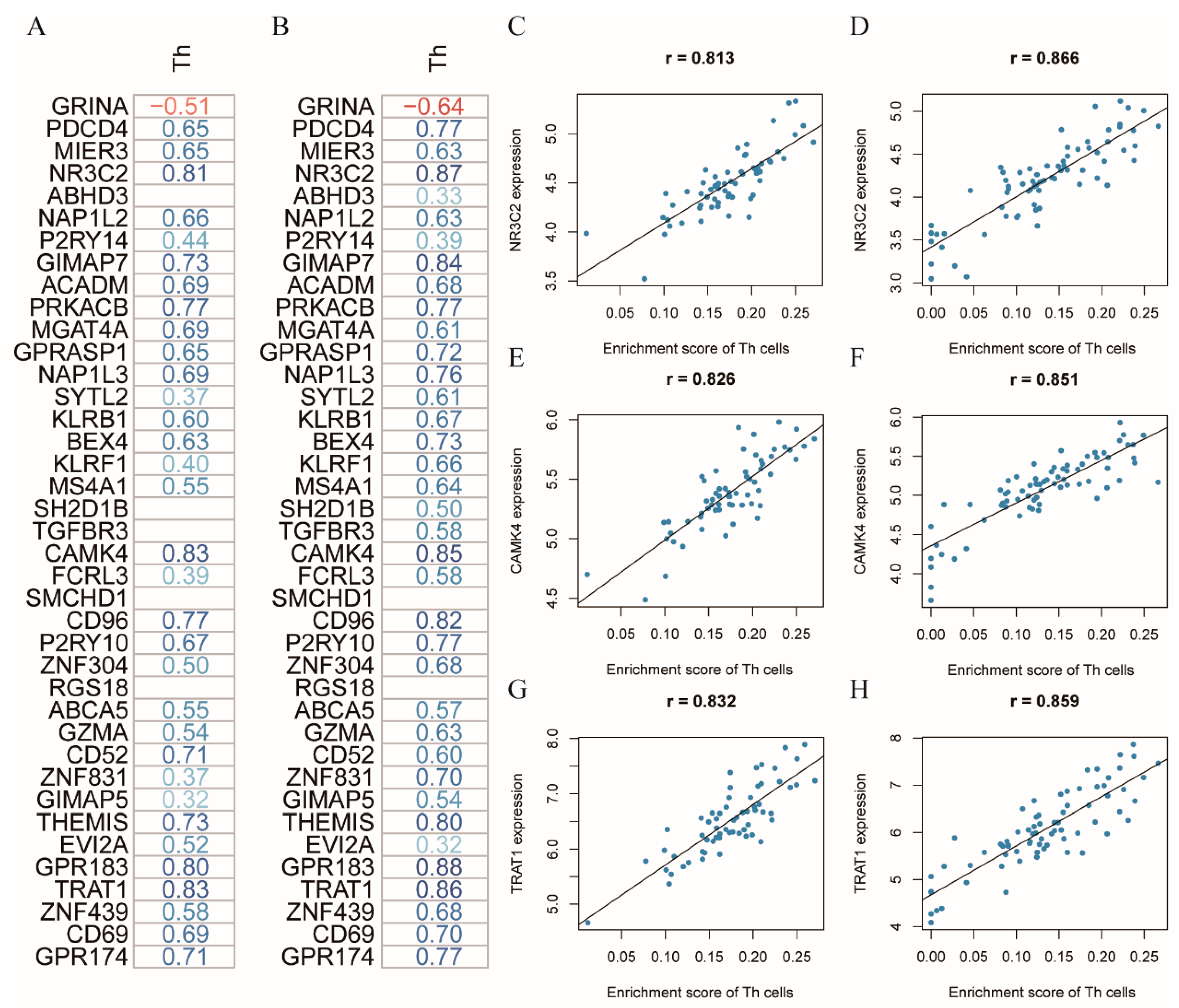
3.3. NR3C2, CAMK4, and TRAT1 Associated with the Composition of the Th Cells
3.4. Correlation of the Clinical Test Parameters with the Immune Subsets in the CRC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii1–iii9. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Markman, J.L.; Shiao, S.L. Impact of the immune system and immunotherapy in colorectal cancer. J. Gastrointest. Oncol. 2015, 6, 208–223. [Google Scholar] [CrossRef]
- Ferrone, C.; Dranoff, G. Dual roles for immunity in gastrointestinal cancers. J. Clin. Oncol. 2010, 28, 4045–4051. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, R.; Wang, Y.-J.; Schoen, R.E.; Finn, O.J.; Yu, J.; Zhang, L. Colorectal cancer prevention: Immune modulation taking the stage. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 138–148. [Google Scholar] [CrossRef]
- Xie, Y.; Xie, F.; Zhang, L.; Zhou, X.; Huang, J.; Wang, F.; Jin, J.; Zhang, L.; Zeng, L.; Zhou, F. Targeted Anti-Tumor Immunotherapy Using Tumor Infiltrating Cells. Adv. Sci. 2021, 8, e2101672. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Tosolini, M.; Kirilovsky, A.; Mlecnik, B.; Fredriksen, T.; Mauger, S.; Bindea, G.; Berger, A.; Bruneval, P.; Fridman, W.-H.; Pagès, F.; et al. Clinical Impact of Different Classes of Infiltrating T Cytotoxic and Helper Cells (Th1, Th2, Treg, Th17) in Patients with Colorectal Cancer. Cancer Res. 2011, 71, 1263–1271. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pagès, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Duan, Y.; Cheng, X.; Chen, X.; Xie, W.; Long, H.; Lin, Z.; Zhu, B. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem. Biophys. Res. Commun. 2011, 407, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Kinugasa, T.; Miyoshi, H.; Sato, K.; Yuge, K.; Ohchi, T.; Fujino, S.; Shiraiwa, S.; Katagiri, M.; Akagi, Y.; et al. A High RORγT/CD3 Ratio is a Strong Prognostic Factor for Postoperative Survival in Advanced Colorectal Cancer: Analysis of Helper T Cell Lymphocytes (Th1, Th2, Th17 and Regulatory T Cells). Ann. Surg. Oncol. 2016, 23, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olingy, C.E.; Dinh, H.Q.; Hedrick, C.C. Monocyte heterogeneity and functions in cancer. J. Leukoc. Biol. 2019, 106, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Ma, T.; Renz, B.W.; Ilmer, M.; Koch, D.; Yang, Y.; Werner, J.; Bazhin, A.V. Myeloid-Derived Suppressor Cells in Solid Tumors. Cells 2022, 11, 310. [Google Scholar] [CrossRef]
- Maecker, H.T.; Trotter, J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytom. Part A 2006, 69A, 1037–1042. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 3 January 2022).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R Package Version 1.7-9. 2021. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 3 January 2022).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 3 January 2022).
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Pages, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Anitei, M.G.; Zeitoun, G.; Mlecnik, B.; Marliot, F.; Haicheur, N.; Todosi, A.M.; Kirilovsky, A.; Lagorce, C.; Bindea, G.; Ferariu, D.; et al. Prognostic and predictive values of the immunoscore in patients with rectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 1891–1899. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Maeng, H.G.; Lee, S.J.; Kim, Y.J.; Kim, D.W.; Lee, H.N.; Namgung, J.H.; Oh, H.M.; Kim, T.J.; Jeong, J.E.; et al. Diagnostic value of peripheral blood immune profiling in colorectal cancer. Ann. Surg. Treat. Res. 2018, 94, 312–321. [Google Scholar] [CrossRef]
- Waidhauser, J.; Nerlinger, P.; Arndt, T.T.; Schiele, S.; Sommer, F.; Wolf, S.; Löhr, P.; Eser, S.; Müller, G.; Claus, R.; et al. Alterations of circulating lymphocyte subsets in patients with colorectal carcinoma. Cancer Immunol. Immunother. CII 2021, 71, 1937–1947. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Schlosser, H.A.; Gryschok, L.; Malcher, J.; Wennhold, K.; Garcia-Marquez, M.; Herbold, T.; Neuhaus, L.S.; Becker, H.J.; Fiedler, A.; et al. Characterization of tumor-associated B-cell subsets in patients with colorectal cancer. Oncotarget 2014, 5, 4651–4664. [Google Scholar] [CrossRef]
- Krijgsman, D.; de Vries, N.L.; Skovbo, A.; Andersen, M.N.; Swets, M.; Bastiaannet, E.; Vahrmeijer, A.L.; van de Velde, C.J.H.; Heemskerk, M.H.M.; Hokland, M.; et al. Characterization of circulating T-, NK-, and NKT cell subsets in patients with colorectal cancer: The peripheral blood immune cell profile. Cancer Immunol. Immunother. CII 2019, 68, 1011–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dylag-Trojanowska, K.; Rogala, J.; Pach, R.; Siedlar, M.; Baran, J.; Sierzega, M.; Zybaczynska, J.; Lenart, M.; Rutkowska-Zapala, M.; Szczepanik, A.M. T Regulatory CD4(+)CD25(+)FoxP3(+) Lymphocytes in the Peripheral Blood of Left-Sided Colorectal Cancer Patients. Medicina 2019, 55, 307. [Google Scholar] [CrossRef] [Green Version]
- Krijgsman, D.; De Vries, N.L.; Andersen, M.N.; Skovbo, A.; Tollenaar, R.A.E.M.; Møller, H.J.; Hokland, M.; Kuppen, P.J.K. CD163 as a Biomarker in Colorectal Cancer: The Expression on Circulating Monocytes and Tumor-Associated Macrophages, and the Soluble Form in the Blood. Int. J. Mol. Sci. 2020, 21, 5925. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wang, Z.; Wu, L.; Zhang, M.; Li, W.; Ding, J.; Zhu, J.; Wei, H.; Zhao, K. Circulating and tumor-infiltrating myeloid-derived suppressor cells in patients with colorectal carcinoma. PLoS ONE 2013, 8, e57114. [Google Scholar] [CrossRef] [Green Version]
- Solito, S.; Falisi, E.; Diaz-Montero, C.M.; Doni, A.; Pinton, L.; Rosato, A.; Francescato, S.; Basso, G.; Zanovello, P.; Onicescu, G.; et al. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood 2011, 118, 2254–2265. [Google Scholar] [CrossRef] [Green Version]
- Borrego, F.; Robertson, M.J.; Ritz, J.; Peña, J.; Solana, R. CD69 is a stimulatory receptor for natural killer cell and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology 1999, 97, 159–165. [Google Scholar] [CrossRef]
- Krijgsman, D.; De Vries, N.L.; Andersen, M.N.; Skovbo, A.; Tollenaar, R.A.E.M.; Bastiaannet, E.; Kuppen, P.J.K.; Hokland, M. The effects of tumor resection and adjuvant therapy on the peripheral blood immune cell profile in patients with colon carcinoma. Cancer Immunol. Immunother. 2020, 69, 2009–2020. [Google Scholar] [CrossRef]
- Bene, N.C.; Alcaide, P.; Wortis, H.H.; Jaffe, I.Z. Mineralocorticoid receptors in immune cells: Emerging role in cardiovascular disease. Steroids 2014, 91, 38–45. [Google Scholar] [CrossRef]
- Racioppi, L.; Means, A.R. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: Novel routes for an ancient traveller. Trends Immunol. 2008, 29, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Koga, T.; Hedrich, C.M.; Mizui, M.; Yoshida, N.; Otomo, K.; Lieberman, L.A.; Rauen, T.; Crispín, J.C.; Tsokos, G.C. CaMK4-dependent activation of AKT/mTOR and CREM-α underlies autoimmunity-associated Th17 imbalance. J. Clin. Investig. 2014, 124, 2234–2245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koga, T.; Kawakami, A. The role of CaMK4 in immune responses. Mod. Rheumatol. 2018, 28, 211–214. [Google Scholar] [CrossRef]
- Kirchgessner, H.; Dietrich, J.; Scherer, J.; Isomäki, P.; Korinek, V.; Hilgert, I.; Bruyns, E.; Leo, A.; Cope, A.P.; Schraven, B. The transmembrane adaptor protein TRIM regulates T cell receptor (TCR) expression and TCR-mediated signaling via an association with the TCR zeta chain. J. Exp. Med. 2001, 193, 1269–1284. [Google Scholar] [CrossRef]
- Valk, E.; Leung, R.; Kang, H.; Kaneko, K.; Rudd, C.E.; Schneider, H. T cell receptor-interacting molecule acts as a chaperone to modulate surface expression of the CTLA-4 coreceptor. Immunity 2006, 25, 807–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacchi, E.; Burrello, J.; Burrello, A.; Bolis, S.; Monticone, S.; Barile, L.; Kaelin-Lang, A.; Melli, G. Profiling Inflammatory Extracellular Vesicles in Plasma and Cerebrospinal Fluid: An Optimized Diagnostic Model for Parkinson’s Disease. Biomedicines 2021, 9, 230. [Google Scholar] [CrossRef]
- Vacchi, E.; Burrello, J.; Di Silvestre, D.; Burrello, A.; Bolis, S.; Mauri, P.; Vassalli, G.; Cereda, C.W.; Farina, C.; Barile, L.; et al. Immune profiling of plasma-derived extracellular vesicles identifies Parkinson disease. Neurol Neuroimmunol. Neuroinflamm. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Song, X.; Zhang, X.; Shankar, V.; Wang, S.; Yang, Y.; Chen, S.; Zhang, L.; Ni, Y.; Zare, R.N.; et al. In situ DESI-MSI lipidomic profiles of mucosal margin of oral squamous cell carcinoma. EBioMedicine 2021, 70, 103529. [Google Scholar] [CrossRef]


| Variables | CRC (n = 12) | Healthy Control (n = 11) | p-Value |
|---|---|---|---|
| Age, year * | 75.0 (69.0, 78.0) | 58.0 (53.5, 68.0) | 0.0600 a |
| Gender | 0.6800 b | ||
| Female | 6 (50.0%) | 4 (36.4%) | |
| Male | 6 (50.0%) | 7 (63.6%) | |
| Sidedness | |||
| Left side | 6 (50.0%) | ||
| Right side | 6 (50.0%) | ||
| Elective surgery | |||
| Yes | 12 (100.0%) | ||
| Surgery Type | |||
| Open surgery | 9 (75.0%) | ||
| Laparoscopic surgery | 2 (16.7%) | ||
| Robot-assisted surgery | 1 (8.3%) | ||
| T (AJCC 7th) | |||
| T1 | 1 (8.3%) | ||
| T2 | 6 (50.0%) | ||
| T3 | 3 (25.0%) | ||
| T4a | 2 (16.7%) | ||
| N (AJCC 7th) | |||
| N0 | 11 (91.7%) | ||
| N1b | 1 (8.3%) | ||
| M (AJCC 7th) | |||
| M0 | 12 (100.0%) | ||
| Tumor stage (AJCC 7th) | |||
| I | 7 (58.4%) | ||
| II | 4 (33.3%) | ||
| III | 1 (8.3%) | ||
| Residual tumor classification | |||
| R0 | 12 (100.0%) | ||
| MSI | |||
| No c | 8 (66.7%) | ||
| Yes d | 4 (33.3%) | ||
| Bethesda | |||
| No | 12 (100.0%) |
| Clinical Parameters | Immune Cells | Coefficient | p-Value |
|---|---|---|---|
| Gamma-glutamyltransferase | Dendritic cells | 0.96 | 1.31 × 10−6 |
| AST | Plasmablasts | 0.95 | 1.55 × 10−6 |
| AST | Activated CD8T cells | 0.91 | 4.91 × 10−5 |
| AST | Effector CD8T cells | 0.87 | 2.60 × 10−4 |
| ALT | T lymphocytes | 0.84 | 6.60 × 10−4 |
| AST | CSM-B cells | 0.83 | 7.29 × 10−4 |
| ALT | Memory Treg cells | 0.82 | 1.19 × 10−3 |
| AST | CD8T cells | 0.81 | 1.34 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Schardey, J.; Wirth, U.; von Ehrlich-Treuenstätt, V.; Neumann, J.; Gießen-Jung, C.; Werner, J.; Bazhin, A.V.; Kühn, F. Analysis of Circulating Immune Subsets in Primary Colorectal Cancer. Cancers 2022, 14, 6105. https://doi.org/10.3390/cancers14246105
Lu C, Schardey J, Wirth U, von Ehrlich-Treuenstätt V, Neumann J, Gießen-Jung C, Werner J, Bazhin AV, Kühn F. Analysis of Circulating Immune Subsets in Primary Colorectal Cancer. Cancers. 2022; 14(24):6105. https://doi.org/10.3390/cancers14246105
Chicago/Turabian StyleLu, Can, Josefine Schardey, Ulrich Wirth, Viktor von Ehrlich-Treuenstätt, Jens Neumann, Clemens Gießen-Jung, Jens Werner, Alexandr V. Bazhin, and Florian Kühn. 2022. "Analysis of Circulating Immune Subsets in Primary Colorectal Cancer" Cancers 14, no. 24: 6105. https://doi.org/10.3390/cancers14246105





