Simple Summary
Cancer is a genetic disease that is caused by changes in genes controlling cell growth, migration, and differentiation. Usually, cancer cells hijack processes used by healthy cells during organism development. The Hippo pathway is a developmental signaling system with a critical role in tissue and organ size regulation, which is frequently deregulated in cancer. Indeed, the contribution of Hippo dysfunction to cancer development has been extensively reported in carcinomas, but it is increasingly recognized in sarcomas. Sarcomas are rare cancers that develop in the bones and soft tissues, encompassing a large variety of different subtypes. Here we review the relevance of the Hippo pathway in specific sarcoma subtypes, with a focus on both the genetic alterations in Hippo pathway genes as well as other molecular mechanisms involved in its deregulation.
Abstract
The Hippo pathway is an evolutionarily conserved modulator of developmental biology with a key role in tissue and organ size regulation under homeostatic conditions. Like other signaling pathways with a significant role in embryonic development, the deregulation of Hippo signaling contributes to oncogenesis. Central to the Hippo pathway is a conserved cascade of adaptor proteins and inhibitory kinases that converge and regulate the activity of the oncoproteins YAP and TAZ, the final transducers of the pathway. Elevated levels and aberrant activation of YAP and TAZ have been described in many cancers. Though most of the studies describe their pervasive activation in epithelial neoplasms, there is increasing evidence pointing out its relevance in mesenchymal malignancies as well. Interestingly, somatic or germline mutations in genes of the Hippo pathway are scarce compared to other signaling pathways that are frequently disrupted in cancer. However, in the case of sarcomas, several examples of genetic alteration of Hippo members, including gene fusions, have been described during the last few years. Here, we review the current knowledge of Hippo pathway implication in sarcoma, describing mechanistic hints recently reported in specific histological entities and how these alterations represent an opportunity for targeted therapy in this heterogeneous group of neoplasm.
1. Introduction
The Hippo pathway is an evolutionary and functionally conserved pathway that controls developmental processes, differentiation, and regeneration by regulating organ size and tissue homeostasis [1,2]. This pathway was initially discovered in Drosophila melanogaster due to tumor suppressor screens and was later revealed to be conserved in mammals. These studies identified Warts (Wts, LATS1/2 in humans) [3,4] and Hippo (Hpo, or STK4/3 encoding MST1/2 in humans) [5,6] genes, which encode the kinases that constitute the principal phosphorylation cascade to the signaling pathway. Likewise, in flies, Hippo mutants display phenotypes of extremely sized organs and apparently resemble a hippopotamus, naming this signaling pathway as it is currently known—the Hippo pathway [5].
In recent years, aberrations on the Hippo pathway have been increasingly associated with cancer development. Thus, many studies have experimentally established its tumor suppressor function. For example, Mst1/2 loss leads to uncontrolled cell proliferation and differentiation in a mouse liver [7], and Yap1/Taz overexpression, the transcriptional coactivators of the pathway, triggers tissue overgrowth and cancer [8,9]. Therefore, dysregulation of the hippo pathway has been reported in various cancer types, including sarcomas [7,10,11,12,13,14,15,16], and correlated with poor prognosis [17]. This review will focus on the genomic alterations disturbing the Hippo pathway and how these aberrations might be potential therapeutic targets in bone and soft tissue sarcomas.
2. The Hippo Signaling Pathway: Critical Components in Mammals and Basic Biology
The primary function of the Hippo pathway is to inhibit proliferation and promote apoptosis, thereby controlling organ growth [18]. This role is arbitrated by a cascade of kinases that transmit, from the plasma to the nucleus, various upstream mechanical, architectural, and metabolic signals.
The Hippo regulating plasma membrane proteins principally include E-cadherin (CHD1) [19], protocadherin FAT4 [20], wingless-related integration (WNT) [21,22,23], the Crumbs polarity complex [24], LIM domain-containing protein Ajuba (AJUBA) [25], the hyaluronic acid receptor CD44 [26], and G-protein coupled receptors (GPCR) [27]. These proteins control the members of the upstream intracellular pathway, which include neurofibromatosis type 2 (NF2), also known as merlin [28], kidney and brain protein (KIBRA or WWC1) [29], Ras-association domain family members (RASSF1–10) [30], TAO kinases (1–3) [31] and angiomotin (AMOT) [32]. All these upstream regulators play a vital role in initiating the cascade of phosphorylation of the core Hippo pathway members.
When the Hippo pathway is activated, the STE20-like kinase 1/2 (MST1/2) is phosphorylated on threonine 183/180, mainly by TAO kinases [33], although it has been described that the activation can be achieved by MST1/2 autophosphorylation itself [34]. Active MST1/2 then phosphorylates the large tumor suppressor kinase 1/2 (LATS1/2) protein [35], but LATS1/2 can also be directly activated by the upstream regulators NF2, AJUBA, and TAO kinases [28,36,37]. MST1/2 also phosphorylates the Salvador family WW domain-containing protein 1 (SAV1) and MOB kinase activator 1A and 1B (MOB1A/B), which are scaffold proteins that coordinate the phosphorylation of MST1/2 and LATS1/2 protein kinases [38,39]. In turn, active LATS1/2 phosphorylates the paralogous transcriptional cofactors Yes-associated protein 1 (YAP) (gene symbol, YAP1) and PDZ-binding motif (TAZ) (gene symbol, WWTR1) on the serine S127 and S89, respectively, which results in their inactivation through translocation from the nucleus to the cytosol, binding with 14-3-3 protein and proteasomal degradation [40,41]. Thus, the cofactors TAZ and YAP are negatively regulated by the Hippo pathway.
When the Hippo signaling pathway is inactivated, non-phosphorylated YAP or TAZ are stabilized and translocated into the nucleus. Because of the lack of DNA-binding domains of YAP/TAZ, they require to cooperate with DNA-binding transcription factors to induce the expression of genes involved in cell proliferation, migration, survival, tissue growth, and inhibition of apoptosis [42]. YAP and TAZ interact preferentially with transcriptional enhanced associate domain (TEAD) proteins (TEAD1–4) [43,44] but also with other transcription factors such as SMAD family members [45,46], Erb-B2 receptor tyrosine kinase 4 (ERBB4) [47], T-box transcription factor 5 (TBX5) [48,49], RUNX family transcription factor 1, 2 and 3 (RUNX1/2/3) [50,51], early-growth response 1 (EGR1) [52], hypoxia-inducible factor 1 alpha (HIF1Aα) [53], core-binding factor subunit beta (CBFB) (also called PEBP2) [54] and tumor protein p73 [55]. Depending on the binding of YAP to one of these DNA-binding transcription factors and, subsequently, associated promoters, diverse target genes are activated. For example, target genes of the YAP/TAZ-TEAD complex include CYR61, CTGF, AREG, or MYC; YAP-TBX5 complex induces the expression of transcriptional targets such as BCL2L1 and BIRC5; and YAP-ERBB4 regulates the expression of CTGF, CYR61, and ANKRD1 [42], involved in cell proliferation, growth, migration, and survival.
In addition to the central inhibitory kinase core, the regulation of YAP and TAZ activity is also controlled by multiple Hippo-independent mechanisms. There is extensive crosstalk with other pathways that influence YAP/TAZ activity beyond the canonical Hippo pathway, such as WNT signaling, TGFβ signaling, GPCR, Rho GTPases or tyrosine kinases-PI3K-AKT signaling [56,57]. Of note, the prominent role of YAP and TAZ integrating morphogenic signals in mechanotransduction processes is modulated both by Hippo-dependent and independent mechanisms. The organization of the actin cytoskeleton seems to be the main input of mechanical cues involving Rho-family GTPases and ROCK (Rho-associated protein kinase) proteins that control F-actine polymerization and ultimately affect YAP/TAZ activity in a LATS-dependent or independent manner [57,58]. Moreover, cell-substratum interaction mediated by integrins promotes the activation of YAP/TAZ by SRC kinase. Indeed, SRC and other SRC family kinases can activate YAP/TAZ through multiple mechanisms, including direct phosphorylation conferring protein stability, enhancing transcriptional activity, and/or interaction with other transcription factors. SRC-mediated activation of YAP/TAZ can also occur through repression of LATS or Hippo pathway-independent mechanisms [59,60].
3. Deregulation of the Hippo Signaling Pathway in Bone and Soft Tissue Sarcoma
Given the critical role of the Hippo pathway in regulating these multiple cellular processes, it is not surprising that aberrant activation of YAP/TAZ leads to uncontrolled cell proliferation and malignant transformation. Indeed, cancer cells commonly hijack the Hippo pathway to acquire malignant properties.
There is extensive evidence that increased expression of YAP/TAZ associates with tumor onset and progression in a large variety of cancers [17]. Actually, the Cancer Genome Atlas (TCGA) project that performed multi-omics profiling in a pan-cancer cohort of 9125 patients across 33 cancer types and characterization of 19 Hippo core genes indicated widespread deregulation of the Hippo pathway members in human cancers. Their main finding is that Hippo signaling is especially relevant in the pathogenesis of carcinomas with squamous cell differentiation. This was mainly attributed to the elevated proportion of cases with YAP1/WWTR1 genomic amplification and high expression heterogeneity of YAP/TAZ target gene signature, which correlated with decreased overall survival of patients with squamous cell cancers. With regard to sarcomas, attending to this report, they seem to be among the malignancies with less genomic alterations in Hippo-related genes and exhibited a poor correlation between YAP/TAZ target gene signature and overall survival. The somatic copy number alteration study showed a significant deletion peak in 17p in sarcomas, where TAOK1 resides [16]. However, it is important to bear in mind that the data analyzed corresponded to a small subset of sarcoma subtypes (leiomyosarcomas, dedifferentiated liposarcomas, and myxofibrosarcomas/undifferentiated pleomorphic sarcomas), which does not represent the enormous diversity of different entities. Besides, pooled analysis of different sarcoma entities may hinder specific features. Therefore, the functional relevance of Hippo signaling in different types of sarcomas should be evaluated in specific entities.
Sarcomas are a highly heterogeneous and complex group of mesenchymal malignancies, both in terms of morphology and pathobiology, that represent <1% of all malignant neoplasms in adults [61].The WHO classification of bone and soft tissue sarcoma listed approximately 100 different sarcomas and mesenchymal tumors of intermediate malignancy. From the genomic point of view, sarcomas can be broadly classified into two groups. Around 1/3 are translocation-associated sarcomas (t-sarcomas), mainly arising in children and young adults, and 2/3 are non-t sarcomas that display complex karyotypes with no specific genomic patterns. In the case of t-sarcomas, the translocation generates a specific fusion gene, which is the driver oncogene of the disease and is an important hallmark to differentiate between different neoplasms among the large variety of entities. In fact, t-sarcomas show an overall low mutational burden apart from gene fusion. Interestingly, several t-sarcomas exhibit recurrent translocations involving Hippo-related genes. Particularly, the genes YAP1 and WWTR1 are identified to be rearranged in certain subtypes of sarcomas and in other unrelated tumor types, such as supratentorial ependymoma (YAP1::MAMLD1, YAP1::FAM118B), cervical squamous cell carcinoma and endocervical adenocarcinoma (YAP1::SS18), poroma/porocarcinoma (YAP1::MAML2, YAP1::NUTM1), or NF2-wild type meningioma (YAP1::MAML2, YAP1::FAM118B, YAP1::PYGO1, YAP1::LMO1) [62]. Moreover, Hippo pathway deregulation mediates the oncogenic properties of other recurrent sarcoma gene fusions. Intriguingly, most of the reports describing the functional relevance of the Hippo pathway in sarcomas deal with t-sarcomas, despite the fact that they represent only 1/3 of the mesenchymal malignancies.
Several studies have demonstrated that the Hippo pathway is deregulated in sarcomas. For example, fusion genes involving WWTR1 and YAP1 are found in nearly all cases of epithelioid haemangioendothelioma [63,64]; YAP1 copy number gain has been described in embryonal rhabdomyosarcoma [65] and frequent hypermethylation of MST1, MST2 and RASSF1A has been shown in several subtypes of soft tissue sarcoma [66]. Furthermore, a study encompassing an immunohistochemistry (IHQ) assessment of TAZ and YAP in 159 sarcomas representing the most prevalent types showed that 50% and 66% of samples exhibit activation (or nuclear location) of YAP and TAZ, respectively [67]. A later study analyzed the expression levels of YAP and TAZ by IHQ in a cohort of 486 sarcoma tissues. Nuclear YAP and TAZ expression levels were detected in 53% and 33% to be moderate to intense, respectively [68]. Additionally, deregulation of the hippo pathway has been related to poor prognosis in several subtypes of sarcomas [67,69,70,71]. These pieces of evidence suggest that the Hippo pathway plays a crucial role in sarcoma tumorigenesis, progression, and outcome.
In this section, we will discuss alterations that affect Hippo pathway members in specific subtypes of sarcomas (Figure 1 and Table 1).
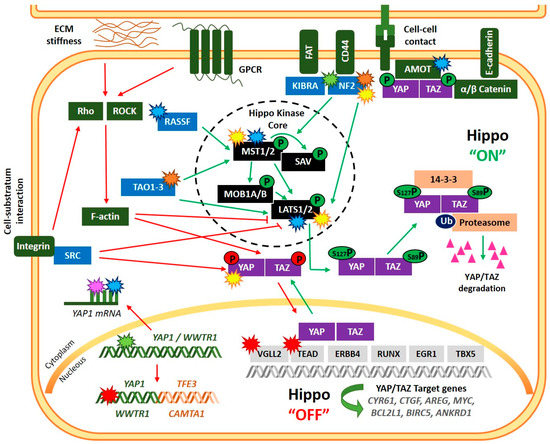
Figure 1.
Regulation of the Hippo signaling pathway and main alterations of Hippo-pathway members reported in sarcomas. Green arrow lines and phosphates indicate induction of Hippo “ON” status, while red arrow lines and phosphates indicate “OFF” status. Alterations of the Hippo-pathway members are displayed with start symbols with colors denoting: transcriptional (green); post-transcriptional (pink); post-translational (yellow); epigenetic (blue), mutation/copy number alteration (orange) and chromosomal rearrangement (red) aberrations. An example of chromosomal rearrangement involving YAP1 and WWTR1 is showed in the figure (YAP1::TFE3 and WWTR1::CAMTA1).

Table 1.
Deregulation mechanisms of Hippo pathway in sarcomas.
3.1. Osteosarcomas
Osteosarcoma (OS) is the most common primary malignancy of bone and one of the most common primary malignant tumors in children and adolescents. OS can occur in any bone, with 75% of all cases occurring in the distal femur and proximal tibia [127,128]. OS is characterized by heterogeneous genetic complexity, including complex genomic rearrangements as well as copy number alterations [129,130]. In addition, aberrations in the Hippo pathway have been extensively reported, and the deregulation of several members of this signaling pathway is described as tumorigenic factors in OS.
3.1.1. YAP
In 2013, Zhang et al. reported nuclear localization of YAP in OS patient tumor biopsies and that YAP1 knockdown inhibited the proliferation and invasion of OS cells by downregulation of the RUNX2 pathway [131]. The association between YAP nuclear localization and a poor prognosis in OS was reported by Bouvier et al., who suggested that the Hippo pathway could represent a therapeutic target in conventional OS [70]. Additionally, the transcription factor TEAD1 has been reported to be involved in YAP-driven OS development. Genetic silencing of TEAD1 suppresses several malignant phenotypes of OS cells, including cell proliferation, resistance to apoptosis, and invasiveness [132]. Interestingly, it has been shown that YAP and pSmad2 (a marker of active TGFβ signaling) have potential prognostic value in canine appendicular OS [133].
YAP1 can be upregulated by Hedgehog (Hh) pathway activation. Chan L.H. et al. have reported that YAP1 was overexpressed in both human and mouse tumor tissues and that YAP1 expression was reduced by targeting the Hh signaling pathway. They also showed that the upregulation of the Hh signaling significantly prompted osteoblastic OS cells in mature osteoblasts. In addition, they described the aberrant expression of the long noncoding RNA (lncRNA), H19, and proved that its regulation was Hh signaling and YAP expression-dependent [72].
YAP can also be upregulated by the human HLA-F adjacent transcript 10 (FAT10) protein, a member of the ubiquitin-like protein family. It has been reported that FAT10 plays an essential role in developing malignant tumors and stabilizes YAP expression by modifying its ubiquitination and degradation. Moreover, this study revealed that FAT10 is overexpressed in OS, and in vivo and in vitro assays proved that FAT10 silencing inhibited OS proliferation [79].
A functional connection between Rho-associated coiled-coil containing protein kinase 2 (ROCK2) and YAP in regulating OS cell migration and metastasis formation has been described by Zucchini et al. They reported that ROCK2 silencing induced a reduction in the nuclear expression and transcriptional activity of YAP and significantly reduced tumor growth, and eradicated the metastatic potential of OS cell lines [80]. In this context, ROCK2 has been reported to be significantly upregulated in OS tissues compared with adjacent normal tissues. The expression level is related to tumor size and patient prognosis [81,82].
HuR, an RNA-binding protein, can also control YAP1 expression. Thus, Li Z. et al. showed that the expression of HuR is meaningfully increased in OS tissues and positively correlates with OS progression. Moreover, the knockdown of HuR suppressed OS cell migration and invasion, the epithelial-mesenchymal transition (EMT) process, and the stemness of OS cells. Mechanistically, it was proved that HuR directly binds to YAP1 mRNA, stabilizing and increasing its transcriptional activity. Significantly, HuR and YAP1 expression was positively correlated in OS tissues [74]. A similar study by Xu, W., et al. revealed that the expression of the lncRNA, B4GALT1-AS1, was considerably increased in OS tissues. B4GALT1-AS1 was found to recruit HuR to enhance YAP1 mRNA stability and its transcriptional activity. B4GALT1-AS1 knockdown repressed proliferation, migration, and stemness of OS cells. Importantly, in vitro and in vivo assays of YAP1 overexpression rescued the inhibition of B4GALT1-AS1 knockdown on OS cell progression [75].
Liu G. et al. have observed significant upregulation of circFAT1, a circular RNA originating from exon two of the FAT1 gene, in human OS tissues and cell lines. In this study, the in vitro inhibition of circFAT1 efficiently prevented the migration, invasion, and tumorigenesis of OS cells and repressed in vivo OS growth. Mechanistic studies showed that circFAT1 could sponge microRNA-375 (miR-375), which was found to be downregulated in OS tissues and cell lines. Furthermore, they described that YAP1 3′-UTR mRNA is directly targeted by miR-375, revealing other potential regulatory properties of the circularized protein-coding exons or “sponging miRNAs” and providing a new therapeutic target for the OS treatment [76].
Luo Y. et al. described the upregulated expression of miR-624-5p in OS cells and tissues. A higher malignant phenotype of OS was observed when overexpressing miR-624-5p in in vitro and in vivo assays. In addition, they revealed that the expression of the protein tyrosine phosphatase receptor type B (PTPRB) was negatively correlated and identified the Hippo signaling pathway to be involved in the miR-624-5p/PTPRB axis, although precise mechanisms demand further research [134].
An opposite role to that described for miR-624-5p has been reported for miR-625. Luo Z. et al. revealed that miR-625 was markedly downregulated in OS tissues and cell lines. Mechanistically, they showed that miR-625 mimic attenuated the cell proliferation and invasion of OS cells by directly binding to YAP1 3′-UTR mRNA and suppressing YAP1 expression. Furthermore, YAP1 upregulation rescued the inhibitory properties of miR-625 on OS cell proliferation and invasion [77].
Cheng L. et al. have shown that Gankyrin, a regulatory subunit of the proteasome complex, is upregulated in OS and predicts disease progression and poor prognosis. Mechanistic studies revealed that gankyrin avoids YAP1 downregulation mediated by miR-200a through P53 and origins a positive feedback loop to regulate YAP signaling in OS cells. Furthermore, in vitro and in vivo studies showed that gankyrin interacts with YAP to induce OS tumorigenesis [78].
3.1.2. NF2
NF2 has also been described as playing a role in OS development. In human, germline or somatic mutations in one allele of NF2 results in the disease neurofibromatosis type 2, which is associated with schwannomas, meningiomas, and ependymomas. Nevertheless, heterozygous Nf2 mutant mice develop mainly osteomas and OS [83,84].
NF2 activity depends on specific interaction with the cytoplasmic tail of CD44, a transmembrane hyaluronate receptor that functions as an upstream regulator sensing the extracellular environment to modulate ERK, AKT, and Hippo pathways [85,86]. A study carried out by Gvozdenovic A. et al. revealed that CD44 silencing in OS cells reduces the number of proliferative cells and decreases the content of NF2 protein. However, in vivo studies showed that OS cells with reduced CD44 expression enhanced the malignant phenotype when compared to control cells. They suggested that the apparent discrepancy between in vitro and in vivo results highlights the critical impact of the tumor environment on OS progression [87]. A recent study has identified increased levels of total CD44 mRNA and membrane localization of CD44 in primary and metastatic OS compared to normal bone. In addition, they showed that CD44 promotes transendothelial migration of tumor OS cells [88].
Some studies have proven that Hippo signaling dysregulation is associated with SOX2 level in OS. Basilico et al. described that SOX2 maintains cancer stem cells (CSC) in OS and antagonizes the Hippo pathway by directly repressing two Hippo activators, NF2 and WWC1, leading to exaggerated YAP function. Moreover, this study showed the requirement of SOX2 for OS formation and survival of the tumor cells, proposing that disruption of these pathways initiated by SOX2 is an attractive strategy for the treatment of OS [89,90]. In addition, it has been described that YAP can regulate the expression of SOX2 by interacting with TEAD on two TEAD-binding DNA elements near the SOX2 gene. Thus, SOX2 and YAP reinforce each other’s expression to maintain stemness and tumorigenicity in OS [73]. The crucial role of SOX2 in OS was likewise described by Upal Basu-Roy et al., who reported that thiazolidinedione drugs (TZDs), a class of small-molecule activators of PPARγ, decrease the expression of target genes of YAP with a simultaneous reduction in SOX2 and YAP nuclear localization. They demonstrated that TZDs target the PPARγhigh-expressing CSC population and restores the tumor-suppressive Hippo signaling effects in OS [135].
3.1.3. LATS1/2
A recent study showed that the inhibition of Tankyrase 1 (TANK1), classified as a positive regulator of telomere length, by antisense oligodeoxynucleotides (TANK1-ASODN) decreased cell growth, migration, invasion, and EMT in OS cells. Mechanistically, the inhibition of TANK1 expression modulated the Hippo/YAP signaling, inducing significantly LATS1 expression and, subsequently, YAP phosphorylation [91].
Another study by Su X. et al. showed the overexpression of the miR-100HG in OS tissues and cell lines and the correlation with poor prognosis for OS patients. Inhibition of OS progression was observed after a miR-100HG knockdown by reducing cell proliferation, cell cycle distortion, and apoptosis resistance. Mechanism investigation revealed that miR-100HG exerted oncogenic function in OS by inactivating the Hippo signaling pathway. Concretely, RNA immunoprecipitation assay revealed the binding between miR-100HG and EZH2 in OS cells, suggesting that the expression of miR-100HG downstream targets is inhibited by epigenetic mechanisms involving EZH2. Further experiments revealed that both miR-100HG and EZH2 knockdown significantly upregulated the LATS1/2 expression in OS cells. Finally, ChIP assay results showed that EZH2 binding to the LATS1/2 promoter is inhibited by miR-100HG silencing, and consequently, a reduction of H3K27 trimethylation is displayed [92].
A more recent study developed by the same research group has reported that the deubiquitinase YOD1, which stabilizes ITCH (Itchy E3 Ubiquitin Protein Ligase) and facilitates ITCH-mediated LATS1/2 ubiquitination and degradation, was highly expressed in OS cells. They described that overexpression of miR-302b decreased the mRNA expression of YOD1 (direct target of miR-302b), ICTH, and YAP1. In contrast, LATS1 expression increased, suggesting that the YOD1-ICTH-LATS1-YAP axis is controlled by miR-302b [93].
Wu X. et al. described that the upregulation of the lysyl hydroxylase PLOD1 was correlated with the progression and worse survival probability of OS patients. Moreover, PLOD1 overexpression promoted OS tumorigenesis and metastasis in vitro and in vivo, and the mRNA levels of CTGF and CYR61 were significantly upregulated. In contrast, protein levels of p-LATS1 and p-YAP were decreased without disturbing p-MST1/2. Mechanistically, they proved that PLOD1 is directly regulated by miR-34c and PLOD1 mRNA, and miR-34c levels negatively correlated in OS samples [94].
3.1.4. RASSF
Three RASSFs (RASSF4, RASSF5, and RASSF10) proteins have been identified as tumor suppressors in OS. RASSF5 and RASSF10 have been reported to be epigenetically inactivated by hypermethylation of their CpG island promoters in OS. In vitro experiments in OS cell lines proved that overexpression of RASSF4 significantly inhibited proliferation, migration, and invasion as well as the EMT process [95], and RASSF5 overexpression markedly suppressed cell proliferation and invasion and induced cell apoptosis through activation of the MST1/LATS1 pathway [96].
3.1.5. TAZ
Interestingly, although there is not much data on the potential role of TAZ on OS tumorigenesis, some studies link TAZ and miRNAs to OS oncogenic behavior. Thus, Ma J. et al. demonstrated the upregulation of TAZ in OS tissues and cell lines, and OS cell migration, invasion, and proliferation could be induced by TAZ overexpression. The mechanistic study revealed that TAZ overexpression leads to miR-224 upregulation, which inhibits the tumor suppressor SMAD4 [136]. Similar findings were reported by Shen S. et al., which described that TAZ is upregulated in OS and modulates EMT. They demonstrated that TAZ induces miR-135b and suppresses the expression of LATS2, APC, and GSK-3β [137].
3.2. Ewing Sarcoma
Ewing sarcoma (EwS) is the second most frequent primary bone tumor and affects mainly children and young adolescents. EwS is characterized by gene fusions between EWSR1 and members of the ETS gene family (usually FLI1), which are considered the main oncogenic driver of the disease, but exhibit a low somatic mutation rate, and secondary genetic alterations are uncommon [61,138]. No recurrent genetic alterations in members of the Hippo pathway have been described in EwS. Instead, aberrant activation of TAZ and YAP has been observed in several studies, and we have shown that it associates with poor patient prognosis [67,69,98,139]. Moreover, TAZ and YAP suppression negatively affects proliferation and invasion capacity in EwS cell lines, and YAP could also mediate resistance to contact inhibition [69,140].
Interestingly, we described a transcriptional antagonism between the fusion EWSR1::FLI1 and YAP/TAZ [69], which may underlay the phenotypic plasticity of EwS cells. Franzetti G.A. et al. proposed that this plasticity relies on the expression levels of the fusion protein, with low levels favoring a migratory phenotype and, therefore, the dissemination of the disease in EwS [141]. Opposing gene expression signatures could result from interference between the fusion protein and YAP/TAZ/TEAD–AP1 complexes, as evidenced by Katschnig et al. [97], but direct or indirect transcriptional repression of TAZ by EWSR1::FLI1 could also contribute to this antagonism [69,98]. We have also speculated that Ewing sarcoma-associated transcript 1 (EWSAT1), a long noncoding RNA that mediates EWSR1::FLI1 gene repression by interacting with a heterogeneous nuclear ribonucleoprotein [142], might modulate the opposing gene signatures. We observed increased EWSAT1 mRNA expression upon YAP/TAZ silencing in the EwS cell line SK-N-MC [142].
Activation of YAP/TAZ in EwS could be mediated by epigenetic regulation of the RASSF1 locus [69]. RASSF1 encodes different isoforms, which affect the activity of the final Hippo effectors YAP/TAZ in opposite ways. The isoform RASSF1A contributes to the repression of YAP/TAZ by Hippo core kinases, whereas RASSF1C promotes the activation of YAP through functional interaction with SRC family kinases [143]. These two isoforms are differently regulated by the hypermethylation of the locus. Whereas RASSF1A is silenced, RASSF1C expression is induced from an alternative promoter. This may explain the correlation of DNA hypermethylation of RASSF genes with poor outcomes of EwS patients [99,100].
Activation of YAP by SRC has also been proposed as the mechanism mediating tenascin C (TNC) induction of Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long noncoding RNA with oncogenic properties [144]. Indeed, a feed-forward loop between TNC and SRC promotes cell metastatic behavior [145].
3.3. Epithelial Hemangioendothelioma
Epithelial Hemangioendothelioma (EHE) is a rare malignant vascular tumor that originates from vascular pre-endothelial or endothelial lineage cells, arising at a great variety of anatomic sites but mainly affecting lung, liver, and soft tissue with a variable clinical course [61,146]. In 2001, a chromosomal translocation t(1;3)(p36;q25) was identified in EHE [147], which was later on described as a genetic alteration generating the gene fusion WWTR1::CAMTA1 [63,148], present in >90% of the cases and hence considered a useful genetic hallmark for differential diagnosis [149,150]. A less frequent fusion gene, YAP1::TFE3, is present in <10% of EHE, and those cases display a different morphology [64]. Moreover, YAP1::TFE3 fusion seems to be associated with better patient prognosis than WWTR1::CAMTA1 positive patients [64,102]. Additional oncogenic alterations related to DNA damage response, cell cycle, and epigenetic pathways are present in at least 20% of cases [102]. However, pathognomonic gene fusion appears as the primary oncogenic driver in EHE.
Mechanistically, Tanas et al. have shown that WWTR1::CAMTA1 nuclear localization and TEAD-dependent transcriptional activity cannot be restrained by the Hippo pathway, and therefore the fusion oncoprotein is constitutively active [151]. Several fusion variants have been described, but all of them conserve the TEAD binding domain, 14-3-3 binding motif, and all or most of the WW domain of TAZ fused to the transactivation domain (TAD), TIG domain, ankyrin repeats, and IQ domains of CAMTA1 [101]. Besides, CAMTA1 also contributes to a non-canonical nuclear localization signal which translocates the fusion into the nucleus [151]. This results in the induction of a TAZ-like transcriptional program which promotes cellular transformation and adhesion-independent growth. Furthermore, it has been suggested that YAP/TAZ-induced transcriptome could contribute to the prominent fibrous stroma commonly observed in EHE [101].
3.4. Myxoid Liposarcoma
Myxoid liposarcoma (MLS), the second most common type of liposarcoma, is a malignant adipose tissue neoplasm that develops in deep soft tissues and is characterized by a chromosomal rearrangement between FUS and DDIT3 genes, producing a chimeric transcription factor [152]. This genetic hallmark is considered the primary oncogenic driver of the disease [153,154].
A recent report identified YAP1 in an RNA screen as an essential gene in FUS::DDIT3-expressing mesenchymal stem cells [103]. In addition, this study describes nuclear YAP expression in 96% of MLS human specimens and expression of the downstream targets FOXM1 and PLK1. Prevalent YAP expression in MLS is further confirmed in other immunohistochemical studies [67,68]. Functional assays indicated that the oncogenic properties of FUS::DDIT3 could be mainly mediated by YAP. FUS::DDIT3 not only induces YAP1 transcription but also promotes YAP nuclear localization and physically interacts with YAP in the nucleus, suggesting a cooperative function between both factors to modulate the transcriptional output in MLS cells [103]. It has been lately described that FUS::DDIT3 induces concurrent activation of IGF-IR/PI3K/AKT signaling and cooperates with YAP to regulate oncogenic gene sets in MLS and disrupt terminal adipogenic differentiation [104].
3.5. Sclerosing Epithelioid Fibrosarcoma and Low-Grade Fibromyxoid Sarcoma
Sclerosing epithelioid fibrosarcoma (SEF) is an aggressive sarcoma, classically composed of nests and cords of epithelioid cells within a dense collagenous matrix, with the presence of both large paucicellular fibrous zones and focal myxoid areas, features also seen in low-grade fibromyxoid sarcoma (LGFMS) [61,155]. LGFMS is a malignant, often late-metastasizing tumor with low to moderate cellularity and consists of bland spindle cells with small, angulated nuclei and scarce cytoplasm, typically showing an abrupt transition from myxoid to fibrous areas [61,156].
Conventional SEF and LGFMS are two closely related mesenchymal entities, with SEF harboring mostly EWSR1::CREB3L1 fusions and LGFMS exhibiting FUS::CREB3L2 fusions [157,158]. Both entities present the upregulation of MUC4, which is detectable at the protein level and used as a surrogate marker. However, a subset of cases negatives for MUC4 expression were reported to harbor complex rearrangements between YAP1 and lysine methyltransferase 2A (KMT2A) loci which exhibit unifying morphologic features slightly different from conventional cases and show an aggressive behavior [105,106,107,108,109]. For these reasons, the possibility of reclassifying YAP1::KMT2A tumors with SEF-like histologic features as a distinct entity related to SEF has been raised.
The most recent study by Massoth L.R. et al. [108] interrogated public genomic data from 14,680 sarcomas and found 33 patients with KMT2A rearrangements (0.2%), including 16 patients with tumors positive for YAP1::KMT2A fusion. Several cases were also reported to bear fusions between KMT2A and other partners, such as Vimentin (VIM). This study and the previous reports are coincident in reporting poor performance of FISH to detect the chromosomal aberration that could be due to the complex rearrangement with the configuration YAP1::KMT2A::YAP1 [108]. This configuration retains the CxxC-binding domain of KMT2A, which is functionally relevant in the pathogenesis of acute leukemias [110], and the TEAD-binding domain and PDZ-binding motif of YAP.
3.6. Rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma (STS) in children and adolescents. The WHO [61,156] recognizes four RMS subtypes, being the two most common subtypes the embryonal and alveolar RMS (ERMS and ARMS, respectively). The presence of the PAX3/7::FOXO1 fusion gene is detected in most ARMS cases, and it is considered the oncogenic driver of this entity. Less common fusion gene variants include the fusion of PAX3 to FOXO4, NCOA1 or INO80D, and FOXO1 to FGFR1. In contrast to ARMS, the oncogenic drivers in ERMS are still undefined. The two rarer RMS subtypes are pleomorphic RMS (PRMS) and spindle cell/sclerosing RMS (SRMS). Gene fusions involving VGLL2, SRF, TEAD1, NCOA2, CITED2, EWSR1, FUS, TFCP2, and MEIS genes have been identified in some subtypes of SRMS [61,156].
3.6.1. Alveolar RMS
Interestingly, PAX3::FOXO1 gene fusion has been found to suppress the Hippo pathway in ARMS [159]. Specifically, this study revealed that RASSF4 expression was highly increased in PAX3::FOXO1–positive ARMS, and its expression was necessary for ARMS cell proliferation, senescence evasion, and tumorigenesis. Mechanistically, it was evidenced that the gene fusion upregulates RASSF4, which associates with MST1 kinase to inhibit downstream signaling in PAX3::FOXO1–positive ARMS. In addition, they showed that YAP was upregulated in both ERMS (in part to the increased copy number of YAP1 locus) and ARMS subtypes, which suggests that Hippo pathway dysregulation is crucial for RMS tumorigenesis [65,159].
Similar studies have reported that the RASSF1 promoter is methylated in pediatric RMS but not adult RMS [66,111]. Thus, pediatric RMS becomes a potential candidate for epigenetic modifiers that can activate RASSF1. Indeed, Slemmons K.K. et al. have recently proved that treatment with a DNA methyltransferase inhibitor (DNMTi) can upregulate Hippo-activators RASSF1 and RASSF5 by promoter demethylation in RMS. Moreover, they reported that combined treatment with DNMTi and dasatinib ablates ARMS cell growth in vitro and trends towards decreased tumor growth in vivo [112].
3.6.2. Spindle Cell/Sclerosing RMS
A study of pediatric SRMS identified three different subsets with distinctive molecular features. A subset of pediatric SRMS presenting at birth or within one year of age exhibited recurrent gene fusions involving VGLL2, SRF, TEAD1, or NCOA2 and appeared to be associated with a better outcome [113]. Specifically, VGLL2 rearrangements were observed in 63% of cases of this subset (VGLL2::CITED2 in four patients and VGLL2::NCOA2 in two cases), and NCOA2 rearrangements were detected in the rest of the cases (TEAD1::NCOA2 in two cases, and SRF::NCOA2 in one case). Subsequently, another study identified six VGLL2::NCOA2 cases and one VGLL2::CITED2 case also occurring in very young children [109].
The NCOA2 gene rearrangements were reported in congenital/infantile SRMS in 2013, including a TEAD1::NCOA2 fusion in a case located in the chest wall of a 4-week-old child [114]. Afterward, several studies reported the TEAD1::NCOA2 gene rearrangement in a subset of pediatric SRMS, which followed a favorable clinical outcome compared to those with MYOD1 mutations [113,115,116]. Although the NCOA2::TEAD1 finding in pediatric SRMS has a prognostic value in clinical practice, the molecular significance of TEAD rearrangement and this involvement in the dysregulation of the hippo signaling is still unknown.
VGLL2 belongs to the Vestigial-like (VGLL) family, whose members have been shown to interact with TEADs in overlapping binding sites for YAP and TAZ. Thus, VGLL family members function as TEAD cofactors and are involved in tumor development in various types of neoplasms [117,118,119]. VGLL2 was identified as a VGLL1 homolog with expression limited to the skeletal muscle lineage. VGLL2, TEAD1, and SRF are transcriptional activators of muscle-specific genes [160,161], and VGLL2-fused tumors express muscle-related genes [109]. VGLL2, TEAD1, and SRF retain most of their functional domains as 5’ partners in the gene fusions. Still, the absence of overt rhabdomyoblastic differentiation in SRMS has led to speculation that the gene fusions could block skeletal muscle differentiation to maintain a primitive phenotype [113]. Interestingly, despite VGLL2-fused tumors expressing some muscle differentiation markers, they are not transcriptionally related to ERMS tumors [109]. Moreover, VGLL2::NCOA2 and VGLL2::CITED show some transcriptome heterogeneity, which may underlie histological differences. VGLL2::NCOA2 tumors present low cellularity and fibrous stroma, whereas VGLL2::CITED tumors exhibit an SRMS-like morphology [109].
3.7. Synovial Sarcoma
Synovial sarcoma (SS) is an aggressive mesenchymal tumor that usually occurs in soft tissues. SS constitutes 8–10% of all soft tissue sarcomas, mainly affecting adolescents and young adults [162]. SS is characterized by a pathognomonic translocation between chromosomes X and 18 that involves SS18 and SSX genes, commonly SS18::SSX1 and SS18::SSX2 [163].
Analysis of YAP/TAZ levels in different sarcoma cell lines and tumor samples showed that SS is one of the tumors with higher levels of nuclear YAP/TAZ proteins [68]. Similar to other sarcoma subtypes, the YAP/TAZ activity has been associated with the loss of Hippo kinases. In the previously cited study, Merrit et al. show that all SS-analyzed samples are negative for at least one of the kinases [122]. The presence of SS18::SSX translocation has also been described as a regulator of YAP/TAZ activity. In SS cell lines, the loss of SS18::SSX expression is associated with a reduction of YAP/TAZ-mediated transcriptional activity. In SS, the SS18::SSX-mediated dysregulation of YAP/TAZ has been linked to IGF-1R/PI3K/AKT activation, a pathway implicated in tumorigenesis in several types of cancer, through a decreased phosphorylation of LATS1 and MOB1. Because of the importance of the Hippo pathway in SS malignancy, SS cells and tumors show a high sensitivity to Verteportin, a suppressor of YAP/TAZ-TEAD binding [120].
3.8. Osteoblastoma
Osteoblastoma (OB) is an infrequent primary osseous tumor, locally aggressive and typically occurring in the medulla of long bones and the neural arch. A high proportion of cases present recurrent rearrangements in FOS or FOSB genes, but a subset of cases do not present these distinctive alterations [121,164]. Instead, they seem to be characterized by a homozygous deletion in chromosome band 22q12. Since the NF2 gene localizes at this region, the authors speculate that it may play a role in the pathogenesis of that subgroup of tumors [121]. Loss of NF2 expression could thus ultimately lead to YAP/TAZ activation, which is able to cooperate with the AP-1 transcriptional complex. As FOS is one of the main components of the AP-1 complex, the mechanisms underlying the pathogenesis of OB could be similar irrespective of the genetic alteration [121].
3.9. Undifferentiated Pleomorphic Sarcoma
Undifferentiated pleomorphic sarcoma (UPS), previously named malignant fibrous histiocytoma (MFH), is an aggressive adult sarcoma usually located in the extremities [165]. UPS is characterized by the presence of complex karyotypes, non-specific differentiation, and atypical anaplastic spindles and round cells [166]. Similar to other sarcoma subtypes, such as fibrosarcoma or liposarcoma, oncogenic driver mutations have not been described in this type of tumor [167].
Remarkably, YAP/TAZ stabilization has been described in UPS, and their expression has been correlated with decreased overall survival [67]. Mechanistically, deregulation of the Hippo pathway is associated with two different processes: the loss of Hippo kinases and the epigenetic repression of AMOT [123,124]. Because of the negative regulation of TAZ and YAP by the Hippo pathway, Merrit et al. hypothesize that the loss of Hippo kinases (MST1, MST2, LATS1, and LATS2) could be implicated in the activation of these proteins. In this study, 77% of UPS analyzed samples (20/26) were negative for at least one Hippo kinase. They also demonstrate that proteasomal degradation and epigenetic modifications, including deacetylated histones and hypermethylated promoters, are implicated in the negative regulation of Hippo kinases. These results suggest that proteasome or DNA methyltransferase/histone deacetylase inhibitors could be used in UPS patients with activation of YAP/TAZ [122].
Deregulation of the Hippo pathway in UPS promotes tumorigenesis through the modulation of the expression of different factors. Forkhead box M1 (FOXM1) is a YAP transcriptional target highly expressed in sarcomas. Downregulation of FOXM1 in in vitro and in vivo sarcoma models reduces cell proliferation and sarcomagenesis [168]. In UPS, FOXM1 expression has been associated with the development of metastases in mouse models [169]. FOXM1 also induces the expression of pluripotency-related genes. Similar to embryonic carcinoma or neuroblastoma, FOXM1 in UPS could maintain the characteristic undifferentiated state of this sarcoma [170]. Different strategies have been developed targeting FOXM1 that could be used for the treatment of UPS patients. Thiostrepton, a proteasome inhibitor, efficiently reduces the expression of FOXM1, suppressing tumor growth in fibrosarcoma models [168].
High levels of YAP in UPS tumors have also been associated with the upregulation of the NF-kB factor. NF-kB is expressed in normal myoblast, the most accepted cell-of-origin of UPS, promoting proliferation and an undifferentiated state [171,172]. Shuai Ye et al. described that YAP-related regulation of NF-kB depends on Ubiquitin Specific Peptidase 31 (USP31), a negative regulator of NF-kB expression repressed by YAP. In this study, repression of USP31 induced more NF-kB activity, promoting proliferation and reducing the differentiation capacity. They also show that the use of epigenetic modulators such as Vorinostat/SAHA and JQ1 reduces the expression of YAP and, in consequence, the pathogenic effects of the protein in UPS models [124]. Finally, the same authors discovered that YAP is implicated in UPS tumorigenesis blocking autophagy in NF-kB independent manner and repressing circadian clock activity through NF-kB upregulation. Circadian clock genes promote the expression of unfolded protein response (UPR) genes. Loss of UPR activity in UPS could be associated with the undifferentiated state of this tumor [173].
The interaction between the UPS cells and extracellular matrix components, such as hyaluronic acid (HA), has also been associated with tumorigenesis and metastatic capacity. The expression of the hyaluronan-mediated mobility receptor (HMMR) gene, which encodes HA surface receptor RHAMM, is activated by YAP and TGFβ signaling (upregulated in UPS). In addition, it has been reported that the loss of YAP/TGFβ activity in UPS animal models reduces the invasion and migration of tumor cells [123].
3.10. Chondrosarcoma
Chondrosarcomas (CS) are groups of locally aggressive or malignant neoplasms that produce a cartilaginous matrix and represent the second most common primary bone tumor [174]. A recent report describes the elevated expression of protein arginine methyltransferase 1 (PRMT1) and nuclear accumulation of YAP in CS specimens. Furthermore, PMRT1 and YAP were positively correlated and associated with high histologic grade and shorter overall survival, being YAP an independent prognostic marker of poor survival [125]. Accordingly, a previous report had also described higher frequencies of YAP and TAZ IHC expression in high-grade CS specimens [67]. PRMT1 is the predominant type I PRMT in mammalian cells, accounting for at least 85% of all arginine methylation in human cells, with implications in several types of cancer [175]. Functional assays in the study by Chen et al. revealed that PMRT1 promoted CS cell growth through suppression of apoptosis, and this could be mediated in part by activation of YAP. PMRT1-dependent activation of YAP was reported to involve LATS1 [125].
3.11. Ossifying Fibromyxoid Tumor
Ossifying fibromyxoid tumor (OFMT) is a rare soft tissue neoplasm of an uncertain line of differentiation and intermediate risk of malignancy. Up to 85% of OFMT present recurrent rearrangements mostly involving PHD finger protein 1 (PHF1), a Polycomb group protein, but also translocations of other genes related to histone modification functions as well [176,177]. A transcriptome sequencing study assessed the presence of alternate gene fusions in a subset of cases lacking those translocations [126]. Two novel gene fusions were identified, CREBBP::BCORL1 and KDM2A::WWTR1. KDM2A is a histone demethylase with a prominent role in the cell proliferation of mesenchymal stem cells. Interestingly, transcriptional profiling grouped OFMT cases with different gene fusions, except the case with KDM2A::WWTR1, which clustered with other tumor types [126].
4. Targeting the Hippo Pathway as a Therapeutic Approach for Sarcomas
The potential of the Hippo Signaling Pathway activation/inhibition as a prognostic indicator and its key role in CSC renewal, tumor growth, migration, and invasion in several types of cancers, including sarcomas, has led many research groups to develop diverse strategies targeting YAP/TAZ network for anti-cancer therapy. Furthermore, it has been described that YAP/TAZ upregulation is involved in mechanisms inducing drug resistance, and YAP levels might limit the clinical efficacy of RAF and MEK inhibitors in melanoma [178]. Likewise, Li et al. described the link between the Hippo pathway and CDK4/6 inhibitors resistance in breast cancer cells. Mechanistically, they revealed that the Hippo pathway is suppressed because of FAT1 loss, and subsequently, YAP and TAZ bind to the CDK6 promoter and upregulate its expression, promoting drug sensitivity [179]. In the same way, it has been suggested the potential combination strategy of CDK4/6 and IGF1R inhibitors for EwS, due to IGF-1R signaling activation, has been reported as a CDK4/6 drug resistance mechanism [180].
Interestingly, some small molecule inhibitors or drugs have been discovered to modulate Hippo pathway activity directly or indirectly at various levels. In this review, we will focus on those molecules that target the Hippo Signaling Pathway and are being tested in cancer clinical trials, particularly in sarcomas (Table 2).

Table 2.
List of Hippo pathway-regulators under clinical investigation for treating sarcomas. Source: ClinicalTrials.gov (accessed on 14 November 2022).
4.1. Inhibition of YAP-TEAD Interaction: Verteporfin
The most used molecule is verteporfin, a benzoporphyrin-derived compound that has been approved by the FDA for the photodynamic treatment of age-related neovascular macular degeneration [181]. Verteporfin is the only reported direct inhibitor of YAP/TAZ. It was described that verteporfin binds to YAP and changes its conformation, inhibiting the binding of YAP-TEAD [182]. Later, it was reported that verteporfin increases 14-3-3σ levels, which promotes the translocation of YAP from nuclear to cytoplasm, decreasing its transcriptional co-activation function [183]. A recent study has revealed a mechanism by which the function of YAP is inhibited by verteporfin by regulating YAP SUMOylation in endometrial cancer. They also described that Serine127 phosphorylation of YAP is important for YAP sumo modification [184]. At the transcriptional level, verteporfin has been described to reduce the expression of Hippo pathway targets genes, and in vitro and in vivo studies have proven that verteporfin decrease proliferation and migration, and invasion of certain cancer cells [182,185,186,187,188,189,190], including EwS and SS cells [98,120]. Furthermore, Visudyne, the FDA-approved liposomal formulation of verteporfin, is being tested in some clinical trials, such as the treatment of cutaneous metastases of breast cancer [191].
4.2. YAP/TAZ Cytoplasmic Retention: Dasatinib, Statins, Pazopanib, and Metformin
A small molecule screening carried out by Oku et al. in 2015 showed that dasatinib, statins, and pazopanib inhibited the nuclear localization of YAP/TAZ and TEAD-dependent transcription, and induced YAP/TAZ phosphorylation in breast cancer cell lines [192].
Dasatinib was originally described as an SRC kinase inhibitor and then shown to inhibit Bcr-Abl and other tyrosine kinases. It has been reported that dasatinib blocks cell migration and invasion in many diverse human sarcoma cell lines and induces apoptosis in the bone sarcoma subgroup through inhibition of SRC-mediated signaling [193]. Numerous studies have reported that YAP and TAZ can be activated and stabilized by SRC-family kinases -mediated phosphorylation [60]. Dasatinib has shown antitumor efficacy in several types of sarcomas, including alveolar soft part sarcoma (ASPS) [194], uterine leiomyosarcoma (LMS) [195], neuroblastoma, EwS [69,196], childhood RMS [112] and uterine sarcoma [197]. Indeed, dasatinib is being tested in several clinical trials in cancer, highlighting chronic myeloid leukemia [198,199,200], acute lymphoblastic leukemia in adults [201], metastatic breast carcinoma [202], lung cancer [203,204], and several types of sarcomas [205,206,207,208,209].
Statins are reductase-competitive inhibitors that are commonly used to treat hypercholesterolemia by inhibiting the mevalonate pathway. They function by suppressing hydroxymethylglutaryl-coenzyme A (HMG-CoA) reductases, the rate-limiting enzymes in the synthesis of a fatty acid intermediate named mevalonate [210]. Aberrant inactivation of the mevalonate pathway has been reported to promote tumor progression and has a marked negative effect on YAP/TAZ transcriptional activity, as YAP/TAZ actions need mevalonate, geranylgeranyl pyrophosphate (GGPP) and Rho GTPases [30]. Many studies have demonstrated that statin use could exhibit potential survival benefits for cancer patients and appeared to be very promising in combined therapies, as they have been shown to reduce the resistance of cancer cells to other anti-cancer drugs [210,211,212,213,214]. Statin antitumoral effects have also been demonstrated in fibrosarcoma and OS cell lines [215,216]. Accordingly, a protective role in breast-cancer-related mortality [214], an improvement in ovarian cancer survival and multiple myeloma [217,218], and a reduction of the risk of developing lethal prostate cancer [219] have been observed among statin users. In this same context, a strong association between preoperative statin therapy and reduced postoperative mortality following surgical resection for rectal cancer has been reported [220]. Furthermore, statin treatment in chronic obstructive pulmonary disease (COPD) may reduce the risk of lung cancer [221]. Consequently, statins are being tested in several cancer clinical trials, such as oesophageal adenocarcinoma [222] and rectal cancer [223].
Pazopanib is a c-KIT, FGFR, PDGFR, and VEGFR multi-kinase inhibitor, but it has also been proved that it induces proteasomal degradation of YAP and TAZ [192,224,225,226]. Pazopanib has shown potent antitumor activity in many cancer cells [224,227,228,229] and is being tested in a myriad of clinical trials as an anti-cancer therapy for lung [230], ovarian [231,232,233], prostate [234], renal cell carcinoma [235], urothelial [236], and several types of sarcomas [237,238,239,240,241,242,243,244,245,246,247,248].
It is well-known that Metformin (MET) stimulates AMP-activated protein kinase (AMPK) and is widely used for the treatment of hyperglycemia. However, recent studies have described that MET interferes with the Hippo signaling pathway. Wu et al. have reported that MET activates the AMPKα, which alters the YAP/TEAD4/CCNE1/2 axis signaling, inducing cell cycle arrest and reducing cell growth of bladder cancer cells [249]. Jin et al. showed that MET controls miR-381/YAP activity and reduces the malignant phenotype of non-small cell lung cancers (NSCLCs) cells [250]. Another mechanism has been reported by Liu et al. where MET induces activation of the Hippo pathway through Scribble (SCRIB). Upregulation of SCRIB expression recruits MST1/2 and LATS1 to the plasma membrane, leading to YAP phosphorylation and its retention within the cytoplasm and finally inhibiting cell proliferation and invasion in human breast cancer cell lines [251]. Another recent study described that MET treatment downregulated YAP/TAZ expression and enhanced YAP phosphorylation in melanoma cells [252]. Thus, recent studies have examined the potential use of MET in cancer patients to decrease tumor growth, reduce the risk of cancer and improve prognosis [253,254,255]. The anti-cancer effects of MET treatment have also been observed in several types of sarcoma cell lines, such as OS [256,257,258,259,260], EwS [259,261], RMS [259,262], and endometrial [263]. In addition, MET is currently under several clinical trials in cancer, including colorectal [264], endometrial [265], ovarian [266], esophageal [267], and CS [268].
4.3. Inhibition of TEAD-Transcription Activity
TEAD transcription factors (TEAD1-4), as the downstream effectors for YAP/TAZ activity, are very attractive therapeutic targets to disturb Hippo-induced transcriptional activity. They are composed of two well-structured and conserved domains, the YAP-binding domain (YBD) and the DNA binding domain (DBD), separated by a proline-rich region [269]. The YBD is stabilized by S-palmitoylation and is required for its function in hippo pathway signaling [270]. Thus, TEAD lipidation status is a regulator of protein homeostasis, and its modulation can be regulated by small molecules [271,272].
Remarkably, a small molecule inhibitor of TEAD, IK-930, that prevents palmitate binding has been very recently described. In preclinical models, IK-930 demonstrates antitumor activity in mouse xenograft models with Hippo pathway genetic alterations such as NF2 deficiency and gene fusion involving YAP1 and WWTR1. IK-930 is under clinical investigation, Phase 1, as an oral TEAD inhibitor agent in patients with advanced solid tumors. This study began in January 2022 and is currently recruiting [273].
5. Conclusions
The Hippo pathway signaling represents a potential opportunity for cancer treatment. As has been discussed in this review, the Hippo pathway is dysregulated in many types of sarcomas and has been associated with tumor progression, malignancy, and poor prognosis. The research efforts for unveiling the Hippo pathway implications in sarcoma development and clinical behavior will provide new therapeutic insights. The identification of new drugs targeting this signaling pathway is, to date, a challenge for pharmaceutical companies and the sarcoma community.
Author Contributions
C.S.-A., J.O.-P., A.T.A., E.d.Á. and J.D.-M. designed and wrote the manuscript; J.D.-M. and E.d.Á. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
E.d.Á.’s laboratory is supported by ISCIIIFEDER (PI20/00003) to E.d.Á., Consejería de Salud y Familias, Junta de Andalucía (PE-0186-2018) to E.d.Á. and (PI-0061-2020) to C.S.-A. and E.d.Á., GEIS (Beca José María Buesa) to A.T.A., GEIS-Fundación Mari Paz Jiménez Casado (Beca Trienal a la Investigación en Sarcomas) to J.D.-M., Fundación CRIS Contra el Cáncer, Asociación Pablo Ugarte, Fundación María García Estrada and CIBERONC. C.S.-A. is supported by the European Social Fund and the Junta de Andalucía (Talento Doctores 2020, DOC_01473), J.O.-P. is granted by a pre-doctoral fellowship from the VI Plan Propio from the Universidad de Sevilla; A.T.A. is supported Juan de la Cierva Incorporación fellowship (IJC-2018-036767-I), and J.D.-M. is supported by CIBERONC (CB16/12/00361).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pan, D. The Hippo Signaling Pathway in Development and Cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.-X.; Zhao, B.; Guan, K.-L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef] [PubMed]
- Justice, R.W.; Zilian, O.; Woods, D.F.; Noll, M.; Bryant, P.J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995, 9, 534–546. [Google Scholar] [CrossRef]
- Xu, T.; Wang, W.; Zhang, S.; Stewart, R.A.; Yu, W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development 1995, 121, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Udan, R.S.; Kango-Singh, M.; Nolo, R.; Tao, C.; Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003, 5, 914–920. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. CB 2007, 17, 2054–2060. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef]
- Cordenonsi, M.; Zanconato, F.; Azzolin, L.; Forcato, M.; Rosato, A.; Frasson, C.; Inui, M.; Montagner, M.; Parenti, A.R.; Poletti, A.; et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011, 147, 759–772. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Wang, H.; Shi, Z.; Dong, A.; Zhang, W.; Song, X.; He, F.; Wang, Y.; Zhang, Z.; Wang, W.; et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 2014, 25, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.N.; Curtis, S.J.; Fillmore, C.M.; Rowbotham, S.P.; Mohseni, M.; Wagner, D.E.; Beede, A.M.; Montoro, D.T.; Sinkevicius, K.W.; Walton, Z.E.; et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. EMBO J. 2014, 33, 468–481. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Tretiakova, M.S.; Silvis, M.R.; Lucas, J.; Klezovitch, O.; Coleman, I.; Bolouri, H.; Kutyavin, V.I.; Morrissey, C.; True, L.D.; et al. ERG Activates the YAP1 Transcriptional Program and Induces the Development of Age-Related Prostate Tumors. Cancer Cell 2015, 27, 797–808. [Google Scholar] [CrossRef]
- Yimlamai, D.; Christodoulou, C.; Galli, G.G.; Yanger, K.; Pepe-Mooney, B.; Gurung, B.; Shrestha, K.; Cahan, P.; Stanger, B.Z.; Camargo, F.D. Hippo pathway activity influences liver cell fate. Cell 2014, 157, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep. 2018, 25, 1304–1317.e1305. [Google Scholar] [CrossRef]
- Poma, A.M.; Torregrossa, L.; Bruno, R.; Basolo, F.; Fontanini, G. Hippo pathway affects survival of cancer patients: Extensive analysis of TCGA data and review of literature. Sci. Rep. 2018, 8, 10623. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.; Tapon, N. The Salvador-Warts-Hippo pathway—An emerging tumour-suppressor network. Nat. Rev. Cancer 2007, 7, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.G.; Koh, E.; Chen, X.; Gumbiner, B.M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA 2011, 108, 11930–11935. [Google Scholar] [CrossRef]
- Ma, L.; Cui, J.; Xi, H.; Bian, S.; Wei, B.; Chen, L. Fat4 suppression induces Yap translocation accounting for the promoted proliferation and migration of gastric cancer cells. Cancer Biol. Ther. 2016, 17, 36–47. [Google Scholar] [CrossRef]
- Kim, M.; Jho, E.H. Cross-talk between Wnt/β-catenin and Hippo signaling pathways: A brief review. BMB Rep. 2014, 47, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Azzolin, L.; Zanconato, F.; Bresolin, S.; Forcato, M.; Basso, G.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Role of TAZ as mediator of Wnt signaling. Cell 2012, 151, 1443–1456. [Google Scholar] [CrossRef] [PubMed]
- Azzolin, L.; Panciera, T.; Soligo, S.; Enzo, E.; Bicciato, S.; Dupont, S.; Bresolin, S.; Frasson, C.; Basso, G.; Guzzardo, V.; et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 2014, 158, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Varelas, X.; Samavarchi-Tehrani, P.; Narimatsu, M.; Weiss, A.; Cockburn, K.; Larsen, B.G.; Rossant, J.; Wrana, J.L. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev. Cell 2010, 19, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Das Thakur, M.; Feng, Y.; Jagannathan, R.; Seppa, M.J.; Skeath, J.B.; Longmore, G.D. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. CB 2010, 20, 657–662. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, H.; Ge, X.; Chen, Q.; Yuan, D.; Chen, Q.; Leng, W.; Chen, L.; Tang, Q.; Bi, F. CD44 acts through RhoA to regulate YAP signaling. Cell. Signal. 2014, 26, 2504–2513. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef]
- Yin, F.; Yu, J.; Zheng, Y.; Chen, Q.; Zhang, N.; Pan, D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 2013, 154, 1342–1355. [Google Scholar] [CrossRef]
- Wilson, K.E.; Li, Y.W.; Yang, N.; Shen, H.; Orillion, A.R.; Zhang, J. PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J. Biol. Chem. 2014, 289, 23693–23700. [Google Scholar] [CrossRef]
- Sorrentino, G.; Ruggeri, N.; Specchia, V.; Cordenonsi, M.; Mano, M.; Dupont, S.; Manfrin, A.; Ingallina, E.; Sommaggio, R.; Piazza, S.; et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 2014, 16, 357–366. [Google Scholar] [CrossRef]
- Fang, C.Y.; Lai, T.C.; Hsiao, M.; Chang, Y.C. The Diverse Roles of TAO Kinases in Health and Diseases. Int. J. Mol. Sci. 2020, 21, 7463. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Lu, Q.; Wang, L.H.; Liu, C.Y.; Lei, Q.; Guan, K.L. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011, 25, 51–63. [Google Scholar] [CrossRef]
- Boggiano, J.C.; Vanderzalm, P.J.; Fehon, R.G. Tao-1 Phosphorylates Hippo/MST Kinases to Regulate the Hippo-Salvador-Warts Tumor Suppressor Pathway. Dev. Cell 2011, 21, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Praskova, M.; Khoklatchev, A.; Ortiz-Vega, S.; Avruch, J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 2004, 381, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.S.; Park, H.W.; Guan, K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Plouffe, S.W.; Meng, Z.; Lin, K.C.; Lin, B.; Hong, A.W.; Chun, J.V.; Guan, K.L. Characterization of Hippo Pathway Components by Gene Inactivation. Mol. Cell 2016, 64, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Irvine, K.D. Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 2013, 6, ra81. [Google Scholar] [CrossRef]
- Hergovich, A. MOB control: Reviewing a conserved family of kinase regulators. Cell. Signal. 2011, 23, 1433–1440. [Google Scholar] [CrossRef]
- Morice, S.; Danieau, G.; Rédini, F.; Brounais-Le-Royer, B.; Verrecchia, F. Hippo/YAP Signaling Pathway: A Promising Therapeutic Target in Bone Paediatric Cancers? Cancers 2020, 12, 645. [Google Scholar] [CrossRef]
- Muslin, A.J.; Xing, H. 14-3-3 proteins: Regulation of subcellular localization by molecular interference. Cell. Signal. 2000, 12, 703–709. [Google Scholar] [CrossRef]
- Basu, S.; Totty, N.F.; Irwin, M.S.; Sudol, M.; Downward, J. Akt Phosphorylates the Yes-Associated Protein, YAP, to Induce Interaction with 14-3-3 and Attenuation of p73-Mediated Apoptosis. Mol. Cell 2003, 11, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Jang, J.W.; Bae, S.C. DNA binding partners of YAP/TAZ. BMB Rep. 2018, 51, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, A.; Kaneko, K.J.; Shu, H.; Zhao, Y.; DePamphilis, M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001, 15, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef]
- Ferrigno, O.; Lallemand, F.; Verrecchia, F.; L’Hoste, S.; Camonis, J.; Atfi, A.; Mauviel, A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene 2002, 21, 4879–4884. [Google Scholar] [CrossRef]
- Varelas, X.; Sakuma, R.; Samavarchi-Tehrani, P.; Peerani, R.; Rao, B.M.; Dembowy, J.; Yaffe, M.B.; Zandstra, P.W.; Wrana, J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008, 10, 837–848. [Google Scholar] [CrossRef]
- Komuro, A.; Nagai, M.; Navin, N.E.; Sudol, M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003, 278, 33334–33341. [Google Scholar] [CrossRef]
- Rosenbluh, J.; Nijhawan, D.; Cox, A.G.; Li, X.; Neal, J.T.; Schafer, E.J.; Zack, T.I.; Wang, X.; Tsherniak, A.; Schinzel, A.C.; et al. β-Catenin-Driven Cancers Require a YAP1 Transcriptional Complex for Survival and Tumorigenesis. Cell 2012, 151, 1457–1473. [Google Scholar] [CrossRef]
- Murakami, M.; Nakagawa, M.; Olson, E.N.; Nakagawa, O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl. Acad. Sci. USA 2005, 102, 18034–18039. [Google Scholar] [CrossRef]
- Qiao, Y.; Lin, S.J.; Chen, Y.; Voon, D.C.; Zhu, F.; Chuang, L.S.; Wang, T.; Tan, P.; Lee, S.C.; Yeoh, K.G.; et al. RUNX3 is a novel negative regulator of oncogenic TEAD-YAP complex in gastric cancer. Oncogene 2016, 35, 2664–2674. [Google Scholar] [CrossRef]
- Chuang, L.S.H.; Ito, Y. The Multiple Interactions of RUNX with the Hippo-YAP Pathway. Cells 2021, 10, 2925. [Google Scholar] [CrossRef] [PubMed]
- Zagurovskaya, M.; Shareef, M.M.; Das, A.; Reeves, A.; Gupta, S.; Sudol, M.; Bedford, M.T.; Prichard, J.; Mohiuddin, M.; Ahmed, M.M. EGR-1 forms a complex with YAP-1 and upregulates Bax expression in irradiated prostate carcinoma cells. Oncogene 2009, 28, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Ma, Y.; Yang, L.; Wang, T.; Meng, X.; Zong, Z.; Sun, X.; Hua, X.; Li, H. Yes-associated protein (YAP) binds to HIF-1α and sustains HIF-1α protein stability to promote hepatocellular carcinoma cell glycolysis under hypoxic stress. J. Exp. Clin. Cancer Res. CR 2018, 37, 216. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Chen, L.F.; Shigesada, K.; Murakami, Y.; Ito, Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999, 18, 2551–2562. [Google Scholar] [CrossRef] [PubMed]
- Strano, S.; Munarriz, E.; Rossi, M.; Castagnoli, L.; Shaul, Y.; Sacchi, A.; Oren, M.; Sudol, M.; Cesareni, G.; Blandino, G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001, 276, 15164–15173. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Du, S.; Lei, T.; Wang, H.; He, X.; Tong, R.; Wang, Y. Multifaceted regulation and functions of YAP/TAZ in tumors (Review). Oncol. Rep. 2018, 40, 16–28. [Google Scholar] [CrossRef]
- Zanconato, F.; Battilana, G.; Cordenonsi, M.; Piccolo, S. YAP/TAZ as therapeutic targets in cancer. Curr. Opin. Pharmacol. 2016, 29, 26–33. [Google Scholar] [CrossRef]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef]
- Lamar, J.M.; Xiao, Y.; Norton, E.; Jiang, Z.G.; Gerhard, G.M.; Kooner, S.; Warren, J.S.A.; Hynes, R.O. SRC tyrosine kinase activates the YAP/TAZ axis and thereby drives tumor growth and metastasis. J. Biol. Chem. 2019, 294, 2302–2317. [Google Scholar] [CrossRef]
- Warren, J.S.A.; Xiao, Y.; Lamar, J.M. YAP/TAZ Activation as a Target for Treating Metastatic Cancer. Cancers 2018, 10, 115. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours, 5th ed.; WHO Classification of Tumours Series; International Agency for Research on Cancer: Lyon, France, 2020; Volume 3, Available online: https://publications.iarc.fr/588 (accessed on 14 November 2022).
- Szulzewsky, F.; Holland, E.C.; Vasioukhin, V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev. Biol. 2021, 475, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Errani, C.; Zhang, L.; Sung, Y.S.; Hajdu, M.; Singer, S.; Maki, R.G.; Healey, J.H.; Antonescu, C.R. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosom. Cancer 2011, 50, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Le Loarer, F.; Mosquera, J.M.; Sboner, A.; Zhang, L.; Chen, C.L.; Chen, H.W.; Pathan, N.; Krausz, T.; Dickson, B.C.; et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosom. Cancer 2013, 52, 775–784. [Google Scholar] [CrossRef]
- Tremblay, A.M.; Missiaglia, E.; Galli, G.G.; Hettmer, S.; Urcia, R.; Carrara, M.; Judson, R.N.; Thway, K.; Nadal, G.; Selfe, J.L.; et al. The Hippo Transducer YAP1 Transforms Activated Satellite Cells and Is a Potent Effector of Embryonal Rhabdomyosarcoma Formation. Cancer Cell 2014, 26, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.; Schagdarsurengin, U.; Blümke, K.; Würl, P.; Pfeifer, G.P.; Hauptmann, S.; Taubert, H.; Dammann, R. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol. Carcinog. 2007, 46, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Fullenkamp, C.A.; Hall, S.L.; Jaber, O.I.; Pakalniskis, B.L.; Savage, E.C.; Savage, J.M.; Ofori-Amanfo, G.K.; Lambertz, A.M.; Ivins, S.D.; Stipp, C.S.; et al. TAZ and YAP are frequently activated oncoproteins in sarcomas. Oncotarget 2016, 7, 30094–30108. [Google Scholar] [CrossRef] [PubMed]
- Isfort, I.; Elges, S.; Cyra, M.; Berthold, R.; Renner, M.; Mechtersheimer, G.; Aman, P.; Larsson, O.; Ratner, N.; Hafner, S.; et al. Prevalence of the Hippo Effectors YAP1/TAZ in Tumors of Soft Tissue and Bone. Sci. Rep. 2019, 9, 19704. [Google Scholar] [CrossRef]
- Rodríguez-Núñez, P.; Romero-Pérez, L.; Amaral, A.T.; Puerto-Camacho, P.; Jordán, C.; Marcilla, D.; Grünewald, T.G.; Alonso, J.; de Alava, E.; Díaz-Martín, J. Hippo pathway effectors YAP1/TAZ induce an EWS-FLI1-opposing gene signature and associate with disease progression in Ewing sarcoma. J. Pathol. 2020, 250, 374–386. [Google Scholar] [CrossRef]
- Bouvier, C.; Macagno, N.; Nguyen, Q.; Loundou, A.; Jiguet-Jiglaire, C.; Gentet, J.C.; Jouve, J.L.; Rochwerger, A.; Mattei, J.C.; Bouvard, D.; et al. Prognostic value of the Hippo pathway transcriptional coactivators YAP/TAZ and β1-integrin in conventional osteosarcoma. Oncotarget 2016, 7, 64702–64710. [Google Scholar] [CrossRef]
- Desai, C.; Thomason, J.; Kohlmeyer, J.L.; Reisetter, A.C.; Ahirwar, P.; Jahanseir, K.; Leidinger, M.; Ofori-Amanfo, G.; Fritchie, K.; Velu, S.E.; et al. Prognostic and therapeutic value of the Hippo pathway, RABL6A, and p53-MDM2 axes in sarcomas. Oncotarget 2021, 12, 740–755. [Google Scholar] [CrossRef]
- Chan, L.H.; Wang, W.; Yeung, W.; Deng, Y.; Yuan, P.; Mak, K.K. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene 2014, 33, 4857–4866. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.K.; Gadi, A.; Maurizi, G.; Roy, U.B.; Mansukhani, A.; Basilico, C. Myeloid Zinc Finger 1 and GA Binding Protein Co-Operate with Sox2 in Regulating the Expression of Yes-Associated Protein 1 in Cancer Cells. Stem Cells 2017, 35, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chen, C.; Xu, R.; Li, Y.; Hu, R.; Li, Z.; Zhu, X. Knockdown of HuR represses osteosarcoma cells migration, invasion and stemness through inhibition of YAP activation and increases susceptibility to chemotherapeutic agents. Biomed. Pharmacother. 2018, 102, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Hu, R.; Xu, R.; Xu, W. LncRNA B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and migration via enhancing YAP transcriptional activity. Cell Prolif. 2018, 51, e12504. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Huang, K.; Jie, Z.; Wu, Y.; Chen, J.; Chen, Z.; Fang, X.; Shen, S. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol. Cancer 2018, 17, 170. [Google Scholar] [CrossRef]
- Luo, Z.; Wu, G.; Zhang, D.; Liu, J.; Ran, R. microRNA-625 targets Yes-associated protein 1 to suppress cell proliferation and invasion of osteosarcoma. Mol. Med. Rep. 2018, 17, 2005–2011. [Google Scholar] [CrossRef]
- Cheng, L.; Wu, B.; Zhang, L.; Bian, E.; An, R.; Yu, S.; Liu, W.; Xiong, Z. Gankyrin promotes osteosarcoma tumorigenesis by forming a positive feedback loop with YAP. Cell. Signal. 2020, 65, 109460. [Google Scholar] [CrossRef]
- Yi, X.; Deng, X.; Zhao, Y.; Deng, B.; Deng, J.; Fan, H.; Du, Y.; Hao, L. Ubiquitin-like protein FAT10 promotes osteosarcoma growth by modifying the ubiquitination and degradation of YAP1. Exp. Cell Res. 2020, 387, 111804. [Google Scholar] [CrossRef]
- Zucchini, C.; Manara, M.C.; Cristalli, C.; Carrabotta, M.; Greco, S.; Pinca, R.S.; Ferrari, C.; Landuzzi, L.; Pasello, M.; Lollini, P.L.; et al. ROCK2 deprivation leads to the inhibition of tumor growth and metastatic potential in osteosarcoma cells through the modulation of YAP activity. J. Exp. Clin. Cancer Res. CR 2019, 38, 503. [Google Scholar] [CrossRef]
- Deng, X.; Yi, X.; Deng, J.; Zou, Y.; Wang, S.; Shan, W.; Liu, P.; Zhang, Z.; Chen, L.; Hao, L. ROCK2 promotes osteosarcoma growth and metastasis by modifying PFKFB3 ubiquitination and degradation. Exp. Cell Res. 2019, 385, 111689. [Google Scholar] [CrossRef]
- Deng, X.; Yi, X.; Huang, D.; Liu, P.; Chen, L.; Du, Y.; Hao, L. ROCK2 mediates osteosarcoma progression and TRAIL resistance by modulating O-GlcNAc transferase degradation. Am. J. Cancer Res. 2020, 10, 781–798. [Google Scholar] [PubMed]
- McClatchey, A.I.; Saotome, I.; Mercer, K.; Crowley, D.; Gusella, J.F.; Bronson, R.T.; Jacks, T. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes Dev. 1998, 12, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, M.; Robanus-Maandag, E.; van der Valk, M.; Niwa-Kawakita, M.; Abramowski, V.; Goutebroze, L.; Woodruff, J.M.; Berns, A.; Thomas, G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000, 14, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Morrison, H.; Sherman, L.S.; Legg, J.; Banine, F.; Isacke, C.; Haipek, C.A.; Gutmann, D.H.; Ponta, H.; Herrlich, P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001, 15, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cai, X.; Wu, C.; Wu, L.; Wang, Y.; Liu, Y.; Yu, Z.; Qin, S.; Ma, F.; Thiery, J.P.; et al. Adhesion glycoprotein CD44 functions as an upstream regulator of a network connecting ERK, AKT and Hippo-YAP pathways in cancer progression. Oncotarget 2015, 6, 2951–2965. [Google Scholar] [CrossRef]
- Gvozdenovic, A.; Arlt, M.J.; Campanile, C.; Brennecke, P.; Husmann, K.; Born, W.; Muff, R.; Fuchs, B. Silencing of CD44 gene expression in human 143-B osteosarcoma cells promotes metastasis of intratibial tumors in SCID mice. PLoS ONE 2013, 8, e60329. [Google Scholar] [CrossRef]
- Ma, J.; Klemm, J.; Gerardo-Ramírez, M.; Frappart, L.; Castven, D.; Becker, D.; Zoch, A.; Parent, R.; Bartosch, B.; Minnich, K.; et al. Cluster of differentiation 44 promotes osteosarcoma progression in mice lacking the tumor suppressor Merlin. Int. J. Cancer 2020, 147, 2564–2577. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef]
- Maurizi, G.; Verma, N.; Gadi, A.; Mansukhani, A.; Basilico, C. Sox2 is required for tumor development and cancer cell proliferation in osteosarcoma. Oncogene 2018, 37, 4626–4632. [Google Scholar] [CrossRef]
- Zhou, Y.; Jin, Q.; Xiao, W.; Sun, C. Tankyrase1 antisense oligodeoxynucleotides suppress the proliferation, migration and invasion through Hippo/YAP pathway in human osteosarcoma cells. Pathol. Res. Pract. 2019, 215, 152381. [Google Scholar] [CrossRef]
- Su, X.; Teng, J.; Jin, G.; Li, J.; Zhao, Z.; Cao, X.; Guo, Y.; Guo, M.; Li, X.; Wu, J.; et al. ELK1-induced upregulation of long non-coding RNA MIR100HG predicts poor prognosis and promotes the progression of osteosarcoma by epigenetically silencing LATS1 and LATS2. Biomed. Pharmacother. 2019, 109, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Liu, H.; Sun, N.; Xie, Y.; Yan, F.; Cai, L. Polyethylenimine-dextran-coated magnetic nanoparticles loaded with miR-302b suppress osteosarcoma in vitro and in vivo. Nanomedicine 2020, 15, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xiang, H.; Cong, W.; Yang, H.; Zhang, G.; Wang, Y.; Guo, Z.; Shen, Y.; Chen, B. PLOD1, a target of miR-34c, contributes to cell growth and metastasis via repressing LATS1 phosphorylation and inactivating Hippo pathway in osteosarcoma. Biochem. Biophys. Res. Commun. 2020, 527, 29–36. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Zhu, T.; Yin, R. RASSF4 Overexpression Inhibits the Proliferation, Invasion, EMT, and Wnt Signaling Pathway in Osteosarcoma Cells. Oncol. Res. 2017, 25, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.H.; Yang, C.Q.; Zhang, C.L.; Gao, Y.; Yuan, H.B.; Wang, C. RASSF5 inhibits growth and invasion and induces apoptosis in osteosarcoma cells through activation of MST1/LATS1 signaling. Oncol. Rep. 2014, 32, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Katschnig, A.M.; Kauer, M.O.; Schwentner, R.; Tomazou, E.M.; Mutz, C.N.; Linder, M.; Sibilia, M.; Alonso, J.; Aryee, D.N.T.; Kovar, H. EWS-FLI1 perturbs MRTFB/YAP-1/TEAD target gene regulation inhibiting cytoskeletal autoregulatory feedback in Ewing sarcoma. Oncogene 2017, 36, 5995–6005. [Google Scholar] [CrossRef]
- Bierbaumer, L.; Katschnig, A.M.; Radic-Sarikas, B.; Kauer, M.O.; Petro, J.A.; Högler, S.; Gurnhofer, E.; Pedot, G.; Schäfer, B.W.; Schwentner, R.; et al. YAP/TAZ inhibition reduces metastatic potential of Ewing sarcoma cells. Oncogenesis 2021, 10, 2. [Google Scholar] [CrossRef]
- Gharanei, S.; Brini, A.T.; Vaiyapuri, S.; Alholle, A.; Dallol, A.; Arrigoni, E.; Kishida, T.; Hiruma, T.; Avigad, S.; Grimer, R.; et al. RASSF2 methylation is a strong prognostic marker in younger age patients with Ewing sarcoma. Epigenetics 2013, 8, 893–898. [Google Scholar] [CrossRef]
- Avigad, S.; Shukla, S.; Naumov, I.; Cohen, I.J.; Ash, S.; Meller, I.; Kollender, Y.; Issakov, J.; Yaniv, I. Aberrant methylation and reduced expression of RASSF1A in Ewing sarcoma. Pediatr. Blood Cancer 2009, 53, 1023–1028. [Google Scholar] [CrossRef]
- Lamar, J.M.; Motilal Nehru, V.; Weinberg, G. Epithelioid Hemangioendothelioma as a Model of YAP/TAZ-Driven Cancer: Insights from a Rare Fusion Sarcoma. Cancers 2018, 10, 229. [Google Scholar] [CrossRef]
- Rosenbaum, E.; Jadeja, B.; Xu, B.; Zhang, L.; Agaram, N.P.; Travis, W.; Singer, S.; Tap, W.D.; Antonescu, C.R. Prognostic stratification of clinical and molecular epithelioid hemangioendothelioma subsets. Mod. Pathol. 2020, 33, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, M.; Cheng, Y.Y.; Jensen, P.; Azoitei, N.; Brunner, I.; Hullein, J.; Slabicki, M.; Isfort, I.; Cyra, M.; Berthold, R.; et al. Requirement for YAP1 signaling in myxoid liposarcoma. EMBO Mol. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Berthold, R.; Isfort, I.; Erkut, C.; Heinst, L.; Grünewald, I.; Wardelmann, E.; Kindler, T.; Åman, P.; Grünewald, T.G.P.; Cidre-Aranaz, F.; et al. Fusion protein-driven IGF-IR/PI3K/AKT signals deregulate Hippo pathway promoting oncogenic cooperation of YAP1 and FUS-DDIT3 in myxoid liposarcoma. Oncogenesis 2022, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Arai, Y.; Tanzawa, Y.; Wakai, S.; Hama, N.; Kawai, A.; Shibata, T. KMT2A (MLL) fusions in aggressive sarcomas in young adults. Histopathology 2019, 75, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Puls, F.; Agaimy, A.; Flucke, U.; Mentzel, T.; Sumathi, V.P.; Ploegmakers, M.; Stoehr, R.; Kindblom, L.G.; Hansson, M.; Sydow, S.; et al. Recurrent Fusions Between YAP1 and KMT2A in Morphologically Distinct Neoplasms Within the Spectrum of Low-grade Fibromyxoid Sarcoma and Sclerosing Epithelioid Fibrosarcoma. Am. J. Surg. Pathol. 2020, 44, 594–606. [Google Scholar] [CrossRef]
- Kao, Y.C.; Lee, J.C.; Zhang, L.; Sung, Y.S.; Swanson, D.; Hsieh, T.H.; Liu, Y.R.; Agaram, N.P.; Huang, H.Y.; Dickson, B.C.; et al. Recurrent YAP1 and KMT2A Gene Rearrangements in a Subset of MUC4-negative Sclerosing Epithelioid Fibrosarcoma. Am. J. Surg. Pathol. 2020, 44, 368–377. [Google Scholar] [CrossRef]
- Massoth, L.R.; Hung, Y.P.; Nardi, V.; Nielsen, G.P.; Hasserjian, R.P.; Louissaint, A., Jr.; Fisch, A.S.; Deshpande, V.; Zukerberg, L.R.; Lennerz, J.K.; et al. Pan-sarcoma genomic analysis of KMT2A rearrangements reveals distinct subtypes defined by YAP1-KMT2A-YAP1 and VIM-KMT2A fusions. Mod. Pathol. 2020, 33, 2307–2317. [Google Scholar] [CrossRef]
- Watson, S.; Perrin, V.; Guillemot, D.; Reynaud, S.; Coindre, J.M.; Karanian, M.; Guinebretiere, J.M.; Freneaux, P.; Le Loarer, F.; Bouvet, M.; et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J. Pathol. 2018, 245, 29–40. [Google Scholar] [CrossRef]
- Muntean, A.G.; Tan, J.; Sitwala, K.; Huang, Y.; Bronstein, J.; Connelly, J.A.; Basrur, V.; Elenitoba-Johnson, K.S.; Hess, J.L. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 2010, 17, 609–621. [Google Scholar] [CrossRef]
- Harada, K.; Toyooka, S.; Maitra, A.; Maruyama, R.; Toyooka, K.O.; Timmons, C.F.; Tomlinson, G.E.; Mastrangelo, D.; Hay, R.J.; Minna, J.D.; et al. Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene 2002, 21, 4345–4349. [Google Scholar] [CrossRef]
- Slemmons, K.K.; Yeung, C.; Baumgart, J.T.; Juarez, J.O.M.; McCalla, A.; Helman, L.J. Targeting Hippo-Dependent and Hippo-Independent YAP1 Signaling for the Treatment of Childhood Rhabdomyosarcoma. Cancer Res. 2020, 80, 3046–3056. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Zhang, L.; Sung, Y.S.; Huang, S.C.; Chen, C.L.; Bisogno, G.; Zin, A.; Agaram, N.P.; LaQuaglia, M.P.; Wexler, L.H.; et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am. J. Surg. Pathol. 2016, 40, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, J.M.; Sboner, A.; Zhang, L.; Kitabayashi, N.; Chen, C.L.; Sung, Y.S.; Wexler, L.H.; LaQuaglia, M.P.; Edelman, M.; Sreekantaiah, C.; et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosom. Cancer 2013, 52, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Whittle, S.; Venkatramani, R.; Schönstein, A.; Pack, S.D.; Alaggio, R.; Vokuhl, C.; Rudzinski, E.R.; Wulf, A.L.; Zin, A.; Gruver, J.R.; et al. Congenital spindle cell rhabdomyosarcoma: An international cooperative analysis. Eur. J. Cancer 2022, 168, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.Z.L.; Saminathan, S.N.; Chang, K.T.E.; Odoño, E.G.; Kuick, C.H.; Chen, H.; Lee, V.K.M. A rare case of congenital spindle cell rhabdomyosarcoma with TEAD1-NCOA2 fusion: A subset of spindle cell rhabdomyosarcoma with indolent behavior. Pathol. Int. 2020, 70, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.C.; Park, H.W.; Guan, K.L. Regulation of the Hippo Pathway Transcription Factor TEAD. Trends Biochem. Sci. 2017, 42, 862–872. [Google Scholar] [CrossRef]
- Mesrouze, Y.; Aguilar, G.; Bokhovchuk, F.; Martin, T.; Delaunay, C.; Villard, F.; Meyerhofer, M.; Zimmermann, C.; Fontana, P.; Wille, R.; et al. A new perspective on the interaction between the Vg/VGLL1-3 proteins and the TEAD transcription factors. Sci. Rep. 2020, 10, 17442. [Google Scholar] [CrossRef]
- Yamaguchi, N. Multiple Roles of Vestigial-Like Family Members in Tumor Development. Front. Oncol. 2020, 10, 1266. [Google Scholar] [CrossRef]
- Isfort, I.; Cyra, M.; Elges, S.; Kailayangiri, S.; Altvater, B.; Rossig, C.; Steinestel, K.; Grünewald, I.; Huss, S.; Eßeling, E.; et al. SS18-SSX-Dependent YAP/TAZ Signaling in Synovial Sarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3718–3731. [Google Scholar] [CrossRef]
- Saba, K.H.; Cornmark, L.; Hofvander, J.; Magnusson, L.; Nilsson, J.; van den Bos, H.; Spierings, D.C.; Foijer, F.; Staaf, J.; Brosjo, O.; et al. Loss of NF2 defines a genetic subgroup of non-FOS-rearranged osteoblastoma. J. Pathol. Clin. Res. 2020, 6, 231–237. [Google Scholar] [CrossRef]
- Merritt, N.M.; Fullenkamp, C.A.; Hall, S.L.; Qian, Q.; Desai, C.; Thomason, J.; Lambertz, A.M.; Dupuy, A.J.; Darbro, B.; Tanas, M.R. A comprehensive evaluation of Hippo pathway silencing in sarcomas. Oncotarget 2018, 9, 31620–31636. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Liu, Y.; Fuller, A.M.; Katti, R.; Ciotti, G.E.; Chor, S.; Alam, M.Z.; Devalaraja, S.; Lorent, K.; Weber, K.; et al. TGFβ and Hippo Pathways Cooperate to Enhance Sarcomagenesis and Metastasis through the Hyaluronan-Mediated Motility Receptor (HMMR). Mol. Cancer Res. MCR 2020, 18, 560–573. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Lawlor, M.A.; Rivera-Reyes, A.; Egolf, S.; Chor, S.; Pak, K.; Ciotti, G.E.; Lee, A.C.; Marino, G.E.; Shah, J.; et al. YAP1-Mediated Suppression of USP31 Enhances NFκB Activity to Promote Sarcomagenesis. Cancer Res. 2018, 78, 2705–2720. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, H.; Zhang, X.; Liu, Z.; Ma, X. PRMT1 potentiates chondrosarcoma development through activation of YAP activity. Mol. Carcinog. 2019, 58, 2193–2206. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.C.; Sung, Y.S.; Zhang, L.; Chen, C.L.; Huang, S.C.; Antonescu, C.R. Expanding the molecular signature of ossifying fibromyxoid tumors with two novel gene fusions: CREBBP-BCORL1 and KDM2A-WWTR1. Genes Chromosom. Cancer 2017, 56, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Lamplot, J.D.; Denduluri, S.; Qin, J.; Li, R.; Liu, X.; Zhang, H.; Chen, X.; Wang, N.; Pratt, A.; Shui, W.; et al. The Current and Future Therapies for Human Osteosarcoma. Curr. Cancer Ther. Rev. 2013, 9, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Mortus, J.R.; Zhang, Y.; Hughes, D.P.M. Developmental Pathways Hijacked by Osteosarcoma. In Current Advances in Osteosarcoma; Kleinerman, M.D.E.S., Ed.; Springer International Publishing: Cham, Switzerland, 2014; pp. 93–118. [Google Scholar]
- Martin, J.W.; Squire, J.A.; Zielenska, M. The genetics of osteosarcoma. Sarcoma 2012, 2012, 627254. [Google Scholar] [CrossRef]
- Sadikovic, B.; Yoshimoto, M.; Chilton-MacNeill, S.; Thorner, P.; Squire, J.A.; Zielenska, M. Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Hum. Mol. Genet. 2009, 18, 1962–1975. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, B.; Shen, L.; Shen, Y.; Chen, X.D. The role and clinical significance of YES-associated protein 1 in human osteosarcoma. Int. J. Immunopathol. Pharmacol. 2013, 26, 157–167. [Google Scholar] [CrossRef]
- Chai, J.; Xu, S.; Guo, F. TEAD1 mediates the oncogenic activities of Hippo-YAP1 signaling in osteosarcoma. Biochem. Biophys. Res. Commun. 2017, 488, 297–302. [Google Scholar] [CrossRef]
- Luu, A.K.; Schott, C.R.; Jones, R.; Poon, A.C.; Golding, B.; Hamed, R.; Deheshi, B.; Mutsaers, A.; Wood, G.A.; Viloria-Petit, A.M. An evaluation of TAZ and YAP crosstalk with TGFβ signalling in canine osteosarcoma suggests involvement of hippo signalling in disease progression. BMC Vet. Res. 2018, 14, 365. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, W.; Tang, P.; Jiang, D.; Gu, C.; Huang, Y.; Gong, F.; Rong, Y.; Qian, D.; Chen, J.; et al. miR-624-5p promoted tumorigenesis and metastasis by suppressing hippo signaling through targeting PTPRB in osteosarcoma cells. J. Exp. Clin. Cancer Res. CR 2019, 38, 488. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Han, E.; Rattanakorn, K.; Gadi, A.; Verma, N.; Maurizi, G.; Gunaratne, P.H.; Coarfa, C.; Kennedy, O.D.; Garabedian, M.J.; et al. PPARγ agonists promote differentiation of cancer stem cells by restraining YAP transcriptional activity. Oncotarget 2016, 7, 60954–60970. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, K.; Ma, Y.; Zhou, M.; Fan, S. The TAZ-miR-224-SMAD4 axis promotes tumorigenesis in osteosarcoma. Cell Death Dis. 2017, 8, e2539. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Huang, K.; Wu, Y.; Ma, Y.; Wang, J.; Qin, F.; Ma, J. A miR-135b-TAZ positive feedback loop promotes epithelial-mesenchymal transition (EMT) and tumorigenesis in osteosarcoma. Cancer Lett. 2017, 407, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Alava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Prim. 2018, 4, 5. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Abedalthagafi, M.; Anwar, A.E.; Bui, M.M. Akt and Hippo Pathways in Ewing’s Sarcoma Tumors and Their Prognostic Significance. J. Cancer 2015, 6, 1005–1010. [Google Scholar] [CrossRef]
- Hsu, J.H.; Lawlor, E.R. BMI-1 suppresses contact inhibition and stabilizes YAP in Ewing sarcoma. Oncogene 2011, 30, 2077–2085. [Google Scholar] [CrossRef]
- Franzetti, G.A.; Laud-Duval, K.; van der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; de Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef]
- Marques Howarth, M.; Simpson, D.; Ngok, S.P.; Nieves, B.; Chen, R.; Siprashvili, Z.; Vaka, D.; Breese, M.R.; Crompton, B.D.; Alexe, G.; et al. Long noncoding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis. J. Clin. Investig. 2014, 124, 5275–5290. [Google Scholar] [CrossRef]
- Vlahov, N.; Scrace, S.; Soto, M.S.; Grawenda, A.M.; Bradley, L.; Pankova, D.; Papaspyropoulos, A.; Yee, K.S.; Buffa, F.; Goding, C.R.; et al. Alternate RASSF1 Transcripts Control SRC Activity, E-Cadherin Contacts, and YAP-Mediated Invasion. Curr. Biol. 2015, 25, 3019–3034. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Huang, Q.; Hu, J.; Li, L.; Xiao, Y.; Yu, H.; Han, Z.; Wang, T.; Zhou, W.; Wei, H.; et al. EWS-FLI1-mediated tenascin-C expression promotes tumour progression by targeting MALAT1 through integrin alpha5beta1-mediated YAP activation in Ewing sarcoma. Br. J. Cancer 2019, 121, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, A.G.; Julian, C.M.; Konzen, S.; Treichel, S.; Lawlor, E.R.; Bailey, K.M. Microenvironmental Factors Drive Tenascin C and Src Cooperation to Promote Invadopodia Formation in Ewing Sarcoma. Neoplasia 2019, 21, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, A.; Agulnik, M. Epithelioid Hemangioendothelioma: Update on Diagnosis and Treatment. Curr. Treat. Options Oncol. 2018, 19, 19. [Google Scholar] [CrossRef]
- Mendlick, M.R.; Nelson, M.; Pickering, D.; Johansson, S.L.; Seemayer, T.A.; Neff, J.R.; Vergara, G.; Rosenthal, H.; Bridge, J.A. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am. J. Surg. Pathol. 2001, 25, 684–687. [Google Scholar] [CrossRef]
- Tanas, M.R.; Sboner, A.; Oliveira, A.M.; Erickson-Johnson, M.R.; Hespelt, J.; Hanwright, P.J.; Flanagan, J.; Luo, Y.; Fenwick, K.; Natrajan, R.; et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci. Transl. Med. 2011, 3, 98ra82. [Google Scholar] [CrossRef]
- Patel, N.R.; Salim, A.A.; Sayeed, H.; Sarabia, S.F.; Hollingsworth, F.; Warren, M.; Jakacky, J.; Tanas, M.; Oliveira, A.M.; Rubin, B.P.; et al. Molecular characterization of epithelioid haemangioendotheliomas identifies novel WWTR1-CAMTA1 fusion variants. Histopathology 2015, 67, 699–708. [Google Scholar] [CrossRef]
- Anderson, T.; Zhang, L.; Hameed, M.; Rusch, V.; Travis, W.D.; Antonescu, C.R. Thoracic epithelioid malignant vascular tumors: A clinicopathologic study of 52 cases with emphasis on pathologic grading and molecular studies of WWTR1-CAMTA1 fusions. Am. J. Surg. Pathol. 2015, 39, 132–139. [Google Scholar] [CrossRef]
- Tanas, M.R.; Ma, S.; Jadaan, F.O.; Ng, C.K.; Weigelt, B.; Reis-Filho, J.S.; Rubin, B.P. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 2016, 35, 929–938. [Google Scholar] [CrossRef]
- Dei Tos, A.P. Liposarcomas: Diagnostic pitfalls and new insights. Histopathology 2014, 64, 38–52. [Google Scholar] [CrossRef]
- Perez-Losada, J.; Pintado, B.; Gutierrez-Adan, A.; Flores, T.; Banares-Gonzalez, B.; del Campo, J.C.; Martin-Martin, J.F.; Battaner, E.; Sanchez-Garcia, I. The chimeric FUS/TLS-CHOP fusion protein specifically induces liposarcomas in transgenic mice. Oncogene 2000, 19, 2413–2422. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Cironi, L.; Provero, P.; Suva, M.L.; Stehle, J.C.; Baumer, K.; Guillou, L.; Stamenkovic, I. Expression of the FUS-CHOP fusion protein in primary mesenchymal progenitor cells gives rise to a model of myxoid liposarcoma. Cancer Res. 2006, 66, 7016–7023. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Rosenblum, M.K.; Pereira, P.; Nascimento, A.G.; Woodruff, J.M. Sclerosing Epithelioid Fibrosarcoma: A Study of 16 Cases and Confirmation of a Clinicopathologically Distinct Tumor. Am. J. Surg. Pathol. 2001, 25, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.; Fisher, C.; Thway, K. Low-grade fibromyxoid sarcoma: Clinical, morphologic and genetic features. Ann. Diagn. Pathol. 2017, 28, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Arbajian, E.; Puls, F.; Magnusson, L.; Thway, K.; Fisher, C.; Sumathi, V.P.; Tayebwa, J.; Nord, K.H.; Kindblom, L.G.; Mertens, F. Recurrent EWSR1-CREB3L1 gene fusions in sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2014, 38, 801–808. [Google Scholar] [CrossRef]
- Prieto-Granada, C.; Zhang, L.; Chen, H.W.; Sung, Y.S.; Agaram, N.P.; Jungbluth, A.A.; Antonescu, C.R. A genetic dichotomy between pure sclerosing epithelioid fibrosarcoma (SEF) and hybrid SEF/low-grade fibromyxoid sarcoma: A pathologic and molecular study of 18 cases. Genes Chromosom. Cancer 2015, 54, 28–38. [Google Scholar] [CrossRef]
- Crose, L.E.; Galindo, K.A.; Kephart, J.G.; Chen, C.; Fitamant, J.; Bardeesy, N.; Bentley, R.C.; Galindo, R.L.; Chi, J.T.; Linardic, C.M. Alveolar rhabdomyosarcoma-associated PAX3-FOXO1 promotes tumorigenesis via Hippo pathway suppression. J. Clin. Investig. 2014, 124, 285–296. [Google Scholar] [CrossRef]
- Pipes, G.C.; Creemers, E.E.; Olson, E.N. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006, 20, 1545–1556. [Google Scholar] [CrossRef]
- Honda, M.; Hidaka, K.; Fukada, S.I.; Sugawa, R.; Shirai, M.; Ikawa, M.; Morisaki, T. Vestigial-like 2 contributes to normal muscle fiber type distribution in mice. Sci. Rep. 2017, 7, 7168. [Google Scholar] [CrossRef]
- Stacchiotti, S.; Van Tine, B.A. Synovial Sarcoma: Current Concepts and Future Perspectives. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, M. Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene 2001, 20, 5755–5762. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.W.; Cleven, A.H.G.; Kroon, H.M.; Briaire-de Bruijn, I.H.; Szuhai, K.; Bovee, J. Utility of FOS as diagnostic marker for osteoid osteoma and osteoblastoma. Virchows Arch. 2020, 476, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.C.; Perez, E.A.; Franceschi, D.; Moffat, F.L., Jr.; Livingstone, A.S.; Koniaris, L.G. Outcomes for soft-tissue sarcoma in 8249 cases from a large state cancer registry. J. Surg Res. 2007, 141, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Thway, K. Sarcomas. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 327–331. [Google Scholar]
- Ballinger, M.L.; Goode, D.L.; Ray-Coquard, I.; James, P.A.; Mitchell, G.; Niedermayr, E.; Puri, A.; Schiffman, J.D.; Dite, G.S.; Cipponi, A.; et al. Monogenic and polygenic determinants of sarcoma risk: An international genetic study. Lancet Oncol. 2016, 17, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Eisinger-Mathason, T.S.K.; Mucaj, V.; Biju, K.M.; Nakazawa, M.S.; Gohil, M.; Cash, T.P.; Yoon, S.S.; Skuli, N.; Park, K.M.; Gerecht, S.; et al. Deregulation of the Hippo pathway in soft-tissue sarcoma promotes FOXM1 expression and tumorigenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E3402–E3411. [Google Scholar] [CrossRef] [PubMed]
- Mito, J.K.; Riedel, R.F.; Dodd, L.; Lahat, G.; Lazar, A.J.; Dodd, R.D.; Stangenberg, L.; Eward, W.C.; Hornicek, F.J.; Yoon, S.S.; et al. Cross Species Genomic Analysis Identifies a Mouse Model as Undifferentiated Pleomorphic Sarcoma/Malignant Fibrous Histiocytoma. PLoS ONE 2009, 4, e8075. [Google Scholar] [CrossRef]
- Kelleher, F.C.; O’Sullivan, H. FOXM1 in sarcoma: Role in cell cycle, pluripotency genes and stem cell pathways. Oncotarget 2016, 7, 42792–42804. [Google Scholar] [CrossRef]
- Bakkar, N.; Wang, J.; Ladner, K.J.; Wang, H.; Dahlman, J.M.; Carathers, M.; Acharyya, S.; Rudnicki, M.A.; Hollenbach, A.D.; Guttridge, D.C. IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 2008, 180, 787–802. [Google Scholar] [CrossRef]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef]
- Rivera-Reyes, A.; Ye, S.; Marino, G.E.; Egolf, S.; Ciotti, G.E.; Chor, S.; Liu, Y.; Posimo, J.M.; Park, P.M.C.; Pak, K.; et al. YAP1 enhances NF-κB-dependent and independent effects on clock-mediated unfolded protein responses and autophagy in sarcoma. Cell Death Dis. 2018, 9, 1108. [Google Scholar] [CrossRef]
- Nazeri, E.; Gouran Savadkoohi, M.; Majidzadeh, A.K.; Esmaeili, R. Chondrosarcoma: An overview of clinical behavior, molecular mechanisms mediated drug resistance and potential therapeutic targets. Crit. Rev. Oncol. Hematol. 2018, 131, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.C.; Lee, J.C.; Huang, H.Y. What is new about the molecular genetics in matrix-producing soft tissue tumors? -The contributions to pathogenetic understanding and diagnostic classification. Virchows. Arch. 2020, 476, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Schneider, N.; Fisher, C.; Thway, K. Ossifying fibromyxoid tumor: Morphology, genetics, and differential diagnosis. Ann. Diagn. Pathol. 2016, 20, 52–58. [Google Scholar] [CrossRef]
- Lin, L.; Sabnis, A.J.; Chan, E.; Olivas, V.; Cade, L.; Pazarentzos, E.; Asthana, S.; Neel, D.; Yan, J.J.; Lu, X.; et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 2015, 47, 250–256. [Google Scholar] [CrossRef]
- Li, Z.; Razavi, P.; Li, Q.; Toy, W.; Liu, B.; Ping, C.; Hsieh, W.; Sanchez-Vega, F.; Brown, D.N.; Da Cruz Paula, A.F.; et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 2018, 34, 893–905.e898. [Google Scholar] [CrossRef]
- Guenther, L.M.; Dharia, N.V.; Ross, L.; Conway, A.; Robichaud, A.L.; Catlett, J.L., 2nd; Wechsler, C.S.; Frank, E.S.; Goodale, A.; Church, A.J.; et al. A Combination CDK4/6 and IGF1R Inhibitor Strategy for Ewing Sarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 1343–1357. [Google Scholar] [CrossRef]
- Bressler, N.M.; Bressler, S.B. Photodynamic therapy with verteporfin (Visudyne): Impact on ophthalmology and visual sciences. Investig. Ophthalmol. Vis. Sci. 2000, 41, 624–628. [Google Scholar]
- Liu-Chittenden, Y.; Huang, B.; Shim, J.S.; Chen, Q.; Lee, S.-J.; Anders, R.A.; Liu, J.O.; Pan, D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012, 26, 1300–1305. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, X.; Feng, W.; Yu, Y.; Jeong, K.; Guo, W.; Lu, Y.; Mills, G.B. Verteporfin inhibits YAP function through up-regulating 14-3-3σ sequestering YAP in the cytoplasm. Am. J. Cancer Res. 2016, 6, 27–37. [Google Scholar]
- Wang, B.; Shao, W.; Shi, Y.; Liao, J.; Chen, X.; Wang, C. Verteporfin induced SUMOylation of YAP1 in endometrial cancer. Am. J. Cancer Res. 2020, 10, 1207–1217. [Google Scholar] [PubMed]
- Wei, H.; Wang, F.; Wang, Y.; Li, T.; Xiu, P.; Zhong, J.; Sun, X.; Li, J. Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma via disrupting the YAP-TEAD complex. Cancer Sci. 2017, 108, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Gou, J.; Jia, J.; Yi, T.; Cui, T.; Li, Z. Verteporfin, a suppressor of YAP-TEAD complex, presents promising antitumor properties on ovarian cancer. OncoTargets Ther. 2016, 9, 5371–5381. [Google Scholar] [CrossRef]
- Giraud, J.; Molina-Castro, S.; Seeneevassen, L.; Sifré, E.; Izotte, J.; Tiffon, C.; Staedel, C.; Boeuf, H.; Fernandez, S.; Barthelemy, P.; et al. Verteporfin targeting YAP1/TAZ-TEAD transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int. J. Cancer 2020, 146, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Lin, F.; Wu, W.; Liu, Y.; Huang, W. Verteporfin inhibits YAP-induced bladder cancer cell growth and invasion via Hippo signaling pathway. Int. J. Med. Sci. 2018, 15, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Li, X. Verteporfin inhibits cell proliferation and induces apoptosis in different subtypes of breast cancer cell lines without light activation. BMC Cancer 2020, 20, 1042. [Google Scholar] [CrossRef]
- Lui, J.W.; Xiao, S.; Ogomori, K.; Hammarstedt, J.E.; Little, E.C.; Lang, D. The Efficiency of Verteporfin as a Therapeutic Option in Pre-Clinical Models of Melanoma. J. Cancer 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Isakoff, S.J.; Rogers, G.S.; Hill, S.; McMullan, P.; Habin, K.R.; Park, H.; Bartenstein, D.W.; Chen, S.T.; Barry, W.T.; Overmoyer, B. An open label, phase II trial of continuous low-irradiance photodynamic therapy (CLIPT) using verteporfin for the treatment of cutaneous breast cancer metastases. J. Clin. Oncol. 2017, 35, TPS1121. [Google Scholar] [CrossRef]
- Oku, Y.; Nishiya, N.; Shito, T.; Yamamoto, R.; Yamamoto, Y.; Oyama, C.; Uehara, Y. Small molecules inhibiting the nuclear localization of YAP/TAZ for chemotherapeutics and chemosensitizers against breast cancers. FEBS Open Bio 2015, 5, 542–549. [Google Scholar] [CrossRef]
- Shor, A.C.; Keschman, E.A.; Lee, F.Y.; Muro-Cacho, C.; Letson, G.D.; Trent, J.C.; Pledger, W.J.; Jove, R. Dasatinib inhibits migration and invasion in diverse human sarcoma cell lines and induces apoptosis in bone sarcoma cells dependent on SRC kinase for survival. Cancer Res. 2007, 67, 2800–2808. [Google Scholar] [CrossRef]
- Mukaihara, K.; Tanabe, Y.; Kubota, D.; Akaike, K.; Hayashi, T.; Mogushi, K.; Hosoya, M.; Sato, S.; Kobayashi, E.; Okubo, T.; et al. Cabozantinib and dastinib exert anti-tumor activity in alveolar soft part sarcoma. PLoS ONE 2017, 12, e0185321. [Google Scholar] [CrossRef]
- Lopez-Acevedo, M.; Grace, L.; Teoh, D.; Whitaker, R.; Adams, D.J.; Jia, J.; Nixon, A.B.; Secord, A.A. Dasatinib (BMS-35482) potentiates the activity of gemcitabine and docetaxel in uterine leiomyosarcoma cell lines. Gynecol. Oncol. Res. Pract. 2014, 1, 2. [Google Scholar] [CrossRef][Green Version]
- Timeus, F.; Crescenzio, N.; Fandi, A.; Doria, A.; Foglia, L.; Cordero di Montezemolo, L. In vitro antiproliferative and antimigratory activity of dasatinib in neuroblastoma and Ewing sarcoma cell lines. Oncol. Rep. 2008, 19, 353–359. [Google Scholar] [CrossRef][Green Version]
- Kawakita, T.; Masato, N.; Takiguchi, E.; Abe, A.; Irahara, M. Cytotoxic effects of 15-deoxy-Δ12,14-prostaglandin J2 alone and in combination with dasatinib against uterine sarcoma in vitro. Exp. Ther. Med. 2017, 13, 2939–2945. [Google Scholar] [CrossRef]
- Abaza, Y.; Kantarjian, H.; Alwash, Y.; Borthakur, G.; Champlin, R.; Kadia, T.; Garcia-Manero, G.; Daver, N.; Ravandi, F.; Verstovsek, S.; et al. Phase I/II study of dasatinib in combination with decitabine in patients with accelerated or blast phase chronic myeloid leukemia. Am. J. Hematol. 2020, 95, 1288–1295. [Google Scholar] [CrossRef]
- Kimura, S.; Imagawa, J.; Murai, K.; Hino, M.; Kitawaki, T.; Okada, M.; Tanaka, H.; Shindo, M.; Kumagai, T.; Ikezoe, T.; et al. Treatment-free remission after first-line dasatinib discontinuation in patients with chronic myeloid leukaemia (first-line DADI trial): A single-arm, multicentre, phase 2 trial. Lancet. Haematol. 2020, 7, e218–e225. [Google Scholar] [CrossRef]
- Cortes, J.E.; Jiang, Q.; Wang, J.; Weng, J.; Zhu, H.; Liu, X.; Hochhaus, A.; Kim, D.W.; Radich, J.; Savona, M.; et al. Dasatinib vs. imatinib in patients with chronic myeloid leukemia in chronic phase (CML-CP) who have not achieved an optimal response to 3 months of imatinib therapy: The DASCERN randomized study. Leukemia 2020, 34, 2064–2073. [Google Scholar] [CrossRef]
- Foà, R.; Bassan, R.; Vitale, A.; Elia, L.; Piciocchi, A.; Puzzolo, M.C.; Canichella, M.; Viero, P.; Ferrara, F.; Lunghi, M.; et al. Dasatinib-Blinatumomab for Ph-Positive Acute Lymphoblastic Leukemia in Adults. N. Engl. J. Med. 2020, 383, 1613–1623. [Google Scholar] [CrossRef]
- Morris, P.G.; Rota, S.; Cadoo, K.; Zamora, S.; Patil, S.; D’Andrea, G.; Gilewski, T.; Bromberg, J.; Dang, C.; Dickler, M.; et al. Phase II Study of Paclitaxel and Dasatinib in Metastatic Breast Cancer. Clin. Breast Cancer 2018, 18, 387–394. [Google Scholar] [CrossRef]
- Creelan, B.C.; Gray, J.E.; Tanvetyanon, T.; Chiappori, A.A.; Yoshida, T.; Schell, M.J.; Antonia, S.J.; Haura, E.B. Phase 1 trial of dasatinib combined with afatinib for epidermal growth factor receptor- (EGFR-) mutated lung cancer with acquired tyrosine kinase inhibitor (TKI) resistance. Br. J. Cancer 2019, 120, 791–796. [Google Scholar] [CrossRef]
- Kelley, M.J.; Jha, G.; Shoemaker, D.; Herndon, J.E., 2nd; Gu, L.; Barry, W.T.; Crawford, J.; Ready, N. Phase II Study of Dasatinib in Previously Treated Patients with Advanced Non-Small Cell Lung Cancer. Cancer Investig. 2017, 35, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Schuetze, S.M.; Wathen, J.K.; Lucas, D.R.; Choy, E.; Samuels, B.L.; Staddon, A.P.; Ganjoo, K.N.; von Mehren, M.; Chow, W.A.; Loeb, D.M.; et al. SARC009: Phase 2 study of dasatinib in patients with previously treated, high-grade, advanced sarcoma. Cancer 2016, 122, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Schuetze, S.M.; Bolejack, V.; Choy, E.; Ganjoo, K.N.; Staddon, A.P.; Chow, W.A.; Tawbi, H.A.; Samuels, B.L.; Patel, S.R.; von Mehren, M.; et al. Phase 2 study of dasatinib in patients with alveolar soft part sarcoma, chondrosarcoma, chordoma, epithelioid sarcoma, or solitary fibrous tumor. Cancer 2017, 123, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, M.; Cioffi, A.; Dômont, J.; Rutkowski, P.; Roth, A.D.; von Moos, R.; Inauen, R.; Toulmonde, M.; Burkhard, R.O.; Knuesli, C.; et al. Long-term outcome of dasatinib first-line treatment in gastrointestinal stromal tumor: A multicenter, 2-stage phase 2 trial (Swiss Group for Clinical Cancer Research 56/07). Cancer 2018, 124, 1449–1454. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Shoushtari, A.N.; Keohan, M.L.; Dickson, M.A.; Gounder, M.M.; Chi, P.; Loo, J.K.; Gaffney, L.; Schneider, L.; Patel, Z.; et al. Combined KIT and CTLA-4 Blockade in Patients with Refractory GIST and Other Advanced Sarcomas: A Phase Ib Study of Dasatinib plus Ipilimumab. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 2972–2980. [Google Scholar] [CrossRef]
- Kato, S.; Jardim, D.L.; Johnson, F.M.; Subbiah, V.; Piha-Paul, S.; Tsimberidou, A.M.; Falchook, G.S.; Karp, D.; Zinner, R.; Wheler, J.; et al. Phase I study of the combination of crizotinib (as a MET inhibitor) and dasatinib (as a c-SRC inhibitor) in patients with advanced cancer. Investig. New Drugs 2018, 36, 416–423. [Google Scholar] [CrossRef]
- Demierre, M.F.; Higgins, P.D.; Gruber, S.B.; Hawk, E.; Lippman, S.M. Statins and cancer prevention. Nat. Rev. Cancer 2005, 5, 930–942. [Google Scholar] [CrossRef]
- Matusewicz, L.; Czogalla, A.; Sikorski, A.F. Attempts to use statins in cancer therapy: An update. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2020, 42, 1010428320941760. [Google Scholar] [CrossRef]
- Yang, J.; Li, C.; Shen, Y.; Zhou, H.; Shao, Y.; Zhu, W.; Chen, Y. Impact of statin use on cancer-specific mortality and recurrence: A meta-analysis of 60 observational studies. Medicine 2020, 99, e19596. [Google Scholar] [CrossRef]
- Gachpazan, M.; Kashani, H.; Khazaei, M.; Hassanian, S.M.; Rezayi, M.; Asgharzadeh, F.; Ghayour-Mobarhan, M.; Ferns, G.A.; Avan, A. The Impact of Statin Therapy on the Survival of Patients with Gastrointestinal Cancer. Curr. Drug Targets 2019, 20, 738–747. [Google Scholar] [CrossRef]
- Borgquist, S.; Broberg, P.; Tojjar, J.; Olsson, H. Statin use and breast cancer survival—A Swedish nationwide study. BMC Cancer 2019, 19, 54. [Google Scholar] [CrossRef]
- Nilsson, S.; Huelsenbeck, J.; Fritz, G. Mevalonate pathway inhibitors affect anticancer drug-induced cell death and DNA damage response of human sarcoma cells. Cancer Lett. 2011, 304, 60–69. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, W. New molecular insights into osteosarcoma targeted therapy. Curr. Opin. Oncol. 2013, 25, 398–406. [Google Scholar] [CrossRef]
- Majidi, A.; Na, R.; Jordan, S.J.; De Fazio, A.; Webb, P.M. Statin use and survival following a diagnosis of ovarian cancer: A prospective observational study. Int. J. Cancer 2020, 148, 1608–1615. [Google Scholar] [CrossRef]
- Brånvall, E.; Ekberg, S.; Eloranta, S.; Wästerlid, T.; Birmann, B.M.; Smedby, K.E. Statin use is associated with improved survival in multiple myeloma: A Swedish population-based study of 4315 patients. Am. J. Hematol. 2020, 95, 652–661. [Google Scholar] [CrossRef]
- Allott, E.H.; Ebot, E.M.; Stopsack, K.H.; Gonzalez-Feliciano, A.G.; Markt, S.C.; Wilson, K.M.; Ahearn, T.U.; Gerke, T.A.; Downer, M.K.; Rider, J.R.; et al. Statin Use Is Associated with Lower Risk of PTEN-Null and Lethal Prostate Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 1086–1093. [Google Scholar] [CrossRef]
- Pourlotfi, A.; Ahl, R.; Sjolin, G.; Forssten, M.P.; Bass, G.A.; Cao, Y.; Matthiessen, P.; Mohseni, S. Statin therapy and postoperative short-term mortality after rectal cancer surgery. Color. Dis. Off. J. Assoc. Coloproctology Great Br. Irel. 2020, 23, 875–881. [Google Scholar] [CrossRef]
- Raymakers, A.; Sin, D.D.; Sadatsafavi, M.; FitzGerald, J.M.; Marra, C.A.; Lynd, L.D. Statin use and lung cancer risk in chronic obstructive pulmonary disease patients: A population-based cohort study. Respir. Res. 2020, 21, 118. [Google Scholar] [CrossRef]
- Alexandre, L.; Clark, A.B.; Walton, S.; Lewis, M.P.; Kumar, B.; Cheong, E.C.; Warren, H.; Kadirkamanathan, S.S.; Parsons, S.L.; Dresner, S.M.; et al. Adjuvant statin therapy for oesophageal adenocarcinoma: The STAT-ROC feasibility study. BJS Open 2020, 4, 59–70. [Google Scholar] [CrossRef]
- Jameson, M.B.; Gormly, K.; Espinoza, D.; Hague, W.; Asghari, G.; Jeffery, G.M.; Price, T.J.; Karapetis, C.S.; Arendse, M.; Armstrong, J.; et al. SPAR—A randomised, placebo-controlled phase II trial of simvastatin in addition to standard chemotherapy and radiation in preoperative treatment for rectal cancer: An AGITG clinical trial. BMC Cancer 2019, 19, 1229. [Google Scholar] [CrossRef]
- Davidson, B.A.; Secord, A.A. Profile of pazopanib and its potential in the treatment of epithelial ovarian cancer. Int. J. Women’s Health 2014, 6, 289–300. [Google Scholar] [CrossRef]
- Bao, Y.; Nakagawa, K.; Yang, Z.; Ikeda, M.; Withanage, K.; Ishigami-Yuasa, M.; Okuno, Y.; Hata, S.; Nishina, H.; Hata, Y. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J. Biochem. 2011, 150, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, S.; Horiuchi, K.; Yoda, M.; Nakayama, R.; Tohmonda, T.; Susa, M.; Nakamura, M.; Chiba, K.; Toyama, Y.; Morioka, H. A novel multi-kinase inhibitor pazopanib suppresses growth of synovial sarcoma cells through inhibition of the PI3K-AKT pathway. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2012, 30, 1493–1498. [Google Scholar] [CrossRef]
- Kim, S.T.; Jang, H.L.; Lee, S.J.; Lee, J.; Choi, Y.L.; Kim, K.M.; Cho, J.; Park, S.H.; Park, Y.S.; Lim, H.Y.; et al. Pazopanib, a novel multitargeted kinase inhibitor, shows potent in vitro antitumor activity in gastric cancer cell lines with FGFR2 amplification. Mol. Cancer Ther. 2014, 13, 2527–2536. [Google Scholar] [CrossRef] [PubMed]
- Sloan, B.; Scheinfeld, N.S. Pazopanib, a VEGF receptor tyrosine kinase inhibitor for cancer therapy. Curr. Opin. Investig. Drugs 2008, 9, 1324–1335. [Google Scholar] [PubMed]
- Zhu, G.; Zhao, M.; Han, Q.; Tan, Y.; Sun, Y.U.; Bouvet, M.; Singh, S.R.; Ye, J.; Hoffman, R.M. Pazopanib Inhibits Tumor Growth, Lymph-node Metastasis and Lymphangiogenesis of an Orthotopic Mouse of Colorectal Cancer. Cancer Genom. Proteom. 2020, 17, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.M.; Lee, K.H.; Kim, B.S.; Kim, H.G.; Min, Y.J.; Yi, S.Y.; Yun, H.J.; Jung, S.H.; Lee, S.H.; Ahn, J.S.; et al. Pazopanib maintenance after first-line etoposide and platinum chemotherapy in patients with extensive disease small-cell lung cancer: A multicentre, randomised, placebo-controlled Phase II study (KCSG-LU12-07). Br. J. Cancer 2018, 118, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Dinkic, C.; Eichbaum, M.; Schmidt, M.; Grischke, E.M.; Gebauer, G.; Fricke, H.C.; Lenz, F.; Wallwiener, M.; Marme, F.; Schneeweiss, A.; et al. Pazopanib (GW786034) and cyclophosphamide in patients with platinum-resistant, recurrent, pre-treated ovarian cancer—Results of the PACOVAR-trial. Gynecol. Oncol. 2017, 146, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.D.; Banerjee, S.; Hall, M.; Clamp, A.R.; Zhou, C.; Hasan, J.; Orbegoso, C.; Taylor, S.; Tugwood, J.; Lyon, A.R.; et al. Pazopanib and Fosbretabulin in recurrent ovarian cancer (PAZOFOS): A multi-centre, phase 1b and open-label, randomised phase 2 trial. Gynecol. Oncol. 2020, 156, 545–551. [Google Scholar] [CrossRef]
- Richardson, D.L.; Sill, M.W.; Coleman, R.L.; Sood, A.K.; Pearl, M.L.; Kehoe, S.M.; Carney, M.E.; Hanjani, P.; Van Le, L.; Zhou, X.C.; et al. Paclitaxel with and Without Pazopanib for Persistent or Recurrent Ovarian Cancer: A Randomized Clinical Trial. JAMA Oncol. 2018, 4, 196–202. [Google Scholar] [CrossRef]
- Maughan, B.L.; Pal, S.K.; Gill, D.; Boucher, K.; Martin, C.; Salgia, M.; Nussenzveig, R.; Liu, T.; Hawks, J.L.; Batten, J.; et al. Modulation of Premetastatic Niche by the Vascular Endothelial Growth Factor Receptor Tyrosine Kinase Inhibitor Pazopanib in Localized High-Risk Prostate Cancer Followed by Radical Prostatectomy: A Phase II Randomized Trial. Oncology 2018, 23, 1413-e1151. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Haas, N.B.; Donskov, F.; Gross-Goupil, M.; Varlamov, S.; Kopyltsov, E.; Lee, J.L.; Melichar, B.; Rini, B.I.; Choueiri, T.K.; et al. Randomized Phase III Trial of Adjuvant Pazopanib Versus Placebo After Nephrectomy in Patients with Localized or Locally Advanced Renal Cell Carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 3916–3923. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.J.; Hussain, S.A.; Protheroe, A.S.; Birtle, A.; Chakraborti, P.; Huddart, R.A.; Jagdev, S.; Bahl, A.; Stockdale, A.; Sundar, S.; et al. Randomized Phase II Study Investigating Pazopanib Versus Weekly Paclitaxel in Relapsed or Progressive Urothelial Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 1770–1777. [Google Scholar] [CrossRef]
- Chow, W.; Frankel, P.; Ruel, C.; Araujo, D.M.; Milhem, M.; Okuno, S.; Hartner, L.; Undevia, S.; Staddon, A. Results of a prospective phase 2 study of pazopanib in patients with surgically unresectable or metastatic chondrosarcoma. Cancer 2020, 126, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Grünwald, V.; Karch, A.; Schuler, M.; Schöffski, P.; Kopp, H.G.; Bauer, S.; Kasper, B.; Lindner, L.H.; Chemnitz, J.M.; Crysandt, M.; et al. Randomized Comparison of Pazopanib and Doxorubicin as First-Line Treatment in Patients with Metastatic Soft Tissue Sarcoma Age 60 Years or Older: Results of a German Intergroup Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 3555–3564. [Google Scholar] [CrossRef]
- Hirbe, A.C.; Eulo, V.; Moon, C.I.; Luo, J.; Myles, S.; Seetharam, M.; Toeniskoetter, J.; Kershner, T.; Haarberg, S.; Agulnik, M.; et al. A phase II study of pazopanib as front-line therapy in patients with non-resectable or metastatic soft-tissue sarcomas who are not candidates for chemotherapy. Eur. J. Cancer 2020, 137, 1–9. [Google Scholar] [CrossRef]
- Kim, M.; Kim, T.M.; Keam, B.; Kim, Y.J.; Paeng, J.C.; Moon, K.C.; Kim, D.W.; Heo, D.S. A Phase II Trial of Pazopanib in Patients with Metastatic Alveolar Soft Part Sarcoma. Oncology 2019, 24, 20-e29. [Google Scholar] [CrossRef]
- Martin-Broto, J.; Cruz, J.; Penel, N.; Le Cesne, A.; Hindi, N.; Luna, P.; Moura, D.S.; Bernabeu, D.; de Alava, E.; Lopez-Guerrero, J.A.; et al. Pazopanib for treatment of typical solitary fibrous tumours: A multicentre, single-arm, phase 2 trial. Lancet. Oncol. 2020, 21, 456–466. [Google Scholar] [CrossRef]
- Martin-Broto, J.; Stacchiotti, S.; Lopez-Pousa, A.; Redondo, A.; Bernabeu, D.; de Alava, E.; Casali, P.G.; Italiano, A.; Gutierrez, A.; Moura, D.S.; et al. Pazopanib for treatment of advanced malignant and dedifferentiated solitary fibrous tumour: A multicentre, single-arm, phase 2 trial. Lancet Oncol 2019, 20, 134–144. [Google Scholar] [CrossRef]
- Mehta, C.R.; Liu, L.; Theuer, C. An adaptive population enrichment phase III trial of TRC105 and pazopanib versus pazopanib alone in patients with advanced angiosarcoma (TAPPAS trial). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 103–108. [Google Scholar] [CrossRef]
- Pautier, P.; Penel, N.; Ray-Coquard, I.; Italiano, A.; Bompas, E.; Delcambre, C.; Bay, J.O.; Bertucci, F.; Delaye, J.; Chevreau, C.; et al. A phase II of gemcitabine combined with pazopanib followed by pazopanib maintenance, as second-line treatment in patients with advanced leiomyosarcomas: A unicancer French Sarcoma Group study (LMS03 study). Eur. J. Cancer 2020, 125, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Samuels, B.L.; Chawla, S.P.; Somaiah, N.; Staddon, A.P.; Skubitz, K.M.; Milhem, M.M.; Kaiser, P.E.; Portnoy, D.C.; Priebat, D.A.; Walker, M.S.; et al. Results of a prospective phase 2 study of pazopanib in patients with advanced intermediate-grade or high-grade liposarcoma. Cancer 2017, 123, 4640–4647. [Google Scholar] [CrossRef]
- Stacchiotti, S.; Ferrari, S.; Redondo, A.; Hindi, N.; Palmerini, E.; Vaz Salgado, M.A.; Frezza, A.M.; Casali, P.G.; Gutierrez, A.; Lopez-Pousa, A.; et al. Pazopanib for treatment of advanced extraskeletal myxoid chondrosarcoma: A multicentre, single-arm, phase 2 trial. Lancet. Oncol. 2019, 20, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Urakawa, H.; Kawai, A.; Goto, T.; Hiraga, H.; Ozaki, T.; Tsuchiya, H.; Nakayama, R.; Naka, N.; Matsumoto, Y.; Kobayashi, E.; et al. Phase II trial of pazopanib in patients with metastatic or unresectable chemoresistant sarcomas: A Japanese Musculoskeletal Oncology Group study. Cancer Sci. 2020, 111, 3303–3312. [Google Scholar] [CrossRef]
- Weiss, A.R.; Chen, Y.L.; Scharschmidt, T.J.; Chi, Y.Y.; Tian, J.; Black, J.O.; Davis, J.L.; Fanburg-Smith, J.C.; Zambrano, E.; Anderson, J.; et al. Pathological response in children and adults with large unresected intermediate-grade or high-grade soft tissue sarcoma receiving preoperative chemoradiotherapy with or without pazopanib (ARST1321): A multicentre, randomised, open-label, phase 2 trial. Lancet. Oncol. 2020, 21, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zheng, Q.; Li, Y.; Wang, G.; Gao, S.; Zhang, X.; Yan, X.; Zhang, X.; Xie, J.; Wang, Y.; et al. Metformin targets a YAP1-TEAD4 complex via AMPKα to regulate CCNE1/2 in bladder cancer cells. J. Exp. Clin. Cancer Res. CR 2019, 38, 376. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Guo, J.; Wu, Y.; Chen, W.; Du, J.; Yang, L.; Wang, X.; Gong, K.; Dai, J.; Miao, S.; et al. Metformin-repressed miR-381-YAP-snail axis activity disrupts NSCLC growth and metastasis. J. Exp. Clin. Cancer Res. CR 2020, 39, 6. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Chen, H.; Wang, R.; Li, P.; Miao, Y.; Liu, P. Metformin suppresses proliferation and invasion of drug-resistant breast cancer cells by activation of the Hippo pathway. J. Cell. Mol. Med. 2020, 24, 5786–5796. [Google Scholar] [CrossRef]
- Hajimoradi Javarsiani, M.; Sajedianfard, J.; Haghjooy Javanmard, S. The effects of metformin on the hippo pathway in the proliferation of melanoma cancer cells: A preclinical study. Arch. Physiol. Biochem. 2020, 128, 1150–1155. [Google Scholar] [CrossRef]
- Gandini, S.; Guerrieri-Gonzaga, A.; Puntoni, M.; Decensi, A. Metformin and breast cancer risk. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 973–974. [Google Scholar] [CrossRef]
- De, A.; Kuppusamy, G. Metformin in breast cancer: Preclinical and clinical evidence. Curr. Probl. Cancer 2020, 44, 100488. [Google Scholar] [CrossRef] [PubMed]
- Mu, N.; Xu, T.; Gao, M.; Dong, M.; Tang, Q.; Hao, L.; Wang, G.; Li, Z.; Wang, W.; Yang, Y.; et al. Therapeutic effect of metformin in the treatment of endometrial cancer. Oncol. Lett. 2020, 20, 156. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, J.; Wang, Y.; Zhou, L.; Che, J.; Wang, F.; Peng, S.; Zhang, G.; Shang, P. Metformin Suppresses Self-Renewal Ability and Tumorigenicity of Osteosarcoma Stem Cells via Reactive Oxygen Species-Mediated Apoptosis and Autophagy. Oxidative Med. Cell. Longev. 2019, 2019, 9290728. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhou, P.; Xu, K.; Chen, T.; Jiao, J.; Wei, H.; Yang, X.; Xu, W.; Wan, W.; Xiao, J. Metformin induces cell cycle arrest, apoptosis and autophagy through ROS/JNK signaling pathway in human osteosarcoma. Int. J. Biol. Sci. 2020, 16, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Eikawa, S.; Nishida, M.; Kunisada, Y.; Yoshida, A.; Fujiwara, T.; Kunisada, T.; Ozaki, T.; Udono, H. Metformin induces CD11b+-cell-mediated growth inhibition of an osteosarcoma: Implications for metabolic reprogramming of myeloid cells and anti-tumor effects. Int. Immunol. 2019, 31, 187–198. [Google Scholar] [CrossRef]
- Garofalo, C.; Capristo, M.; Manara, M.C.; Mancarella, C.; Landuzzi, L.; Belfiore, A.; Lollini, P.L.; Picci, P.; Scotlandi, K. Metformin as an adjuvant drug against pediatric sarcomas: Hypoxia limits therapeutic effects of the drug. PLoS ONE 2013, 8, e83832. [Google Scholar] [CrossRef]
- Chen, X.; Hu, C.; Zhang, W.; Shen, Y.; Wang, J.; Hu, F.; Yu, P. Metformin inhibits the proliferation, metastasis, and cancer stem-like sphere formation in osteosarcoma MG63 cells in vitro. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 9873–9883. [Google Scholar] [CrossRef]
- Nan, X.; Wang, J.; Cheng, H.; Yin, Z.; Sheng, J.; Qiu, B.; Lau, C.C.; Yustein, J.T.; Zhao, H.; Wong, S.T.C. Imatinib revives the therapeutic potential of metformin on ewing sarcoma by attenuating tumor hypoxic response and inhibiting convergent signaling pathways. Cancer Lett. 2020, 469, 195–206. [Google Scholar] [CrossRef]
- Duan, C.; Evison, A.; Taylor, L.; Onur, S.; Morten, K.; Townley, H. The common diabetes drug metformin can diminish the action of citral against Rhabdomyosarcoma cells in vitro. Phytother. Res. PTR 2020, 35, 1378–1388. [Google Scholar] [CrossRef]
- Ezewuiro, O.; Grushko, T.A.; Kocherginsky, M.; Habis, M.; Hurteau, J.A.; Mills, K.A.; Hunn, J.; Olopade, O.I.; Fleming, G.F.; Romero, I.L. Association of Metformin Use with Outcomes in Advanced Endometrial Cancer Treated with Chemotherapy. PLoS ONE 2016, 11, e0147145. [Google Scholar] [CrossRef]
- Bragagnoli, A.C.; Araujo, R.L.C.; Ferraz, M.W.; Dos Santos, L.V.; Abdalla, K.C.; Comar, F.; Santos, F.A.; Oliveira, M.A.; Carvalheira, J.B.C.; Cárcano, F.M.; et al. Metformin plus lrinotecan in patients with refractory colorectal cancer: A phase 2 clinical trial. Br. J. Cancer 2021, 124, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Petchsila, K.; Prueksaritanond, N.; Insin, P.; Yanaranop, M.; Chotikawichean, N. Effect of Metformin for Decreasing Proliferative Marker in Women with Endometrial Cancer: A Randomized Double-blind Placebo-Controlled Trial. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Chan, D.K.; Shank, J.J.; Griffith, K.A.; Fan, H.; Szulawski, R.; Yang, K.; Reynolds, R.K.; Johnston, C.; McLean, K.; et al. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight 2020, 5, e133247. [Google Scholar] [CrossRef]
- Wang, S.; Lin, Y.; Xiong, X.; Wang, L.; Guo, Y.; Chen, Y.; Chen, S.; Wang, G.; Lin, P.; Chen, H.; et al. Low-Dose Metformin Reprograms the Tumor Immune Microenvironment in Human Esophageal Cancer: Results of a Phase II Clinical Trial. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 4921–4932. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Coelen, R.J.S.; Khurshed, M.; Roos, E.; Caan, M.W.A.; van Linde, M.E.; Kouwenhoven, M.; Bramer, J.A.M.; Bovée, J.; Mathôt, R.A.; et al. Study protocol of a phase IB/II clinical trial of metformin and chloroquine in patients with IDH1-mutated or IDH2-mutated solid tumours. BMJ Open 2017, 7, e014961. [Google Scholar] [CrossRef]
- Holden, J.K.; Cunningham, C.N. Targeting the Hippo Pathway and Cancer through the TEAD Family of Transcription Factors. Cancers 2018, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Noland, C.L.; Gierke, S.; Schnier, P.D.; Murray, J.; Sandoval, W.N.; Sagolla, M.; Dey, A.; Hannoush, R.N.; Fairbrother, W.J.; Cunningham, C.N. Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling. Structure 2016, 24, 179–186. [Google Scholar] [CrossRef]
- Holden, J.K.; Crawford, J.J.; Noland, C.L.; Schmidt, S.; Zbieg, J.R.; Lacap, J.A.; Zang, R.; Miller, G.M.; Zhang, Y.; Beroza, P.; et al. Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling. Cell Rep. 2020, 31, 107809. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, L.; Tao, Z.; Jarugumilli, G.K.; Erb, H.; Singh, A.; Li, Q.; Cotton, J.L.; Greninger, P.; Egan, R.K.; et al. Pharmacological blockade of TEAD–YAP reveals its therapeutic limitation in cancer cells. Nat. Commun. 2022, 13, 6744. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Lakhani, N.J.; McKean, M.; Lingaraj, T.; Victor, L.; Sanchez-Martin, M.; Kacena, K.; Malek, K.S.; Santillana, S. A phase 1, first-in-human study of IK-930, an oral TEAD inhibitor targeting the Hippo pathway in subjects with advanced solid tumors. J. Clin. Oncol. 2022, 40, TPS3168. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).