Characterization of the pVHL Interactome in Human Testis Using High-Throughput Library Screening
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Two-Hybrid (Y2H) Assay and Bait Vector Construction
2.2. Library Screening by Yeast Mating
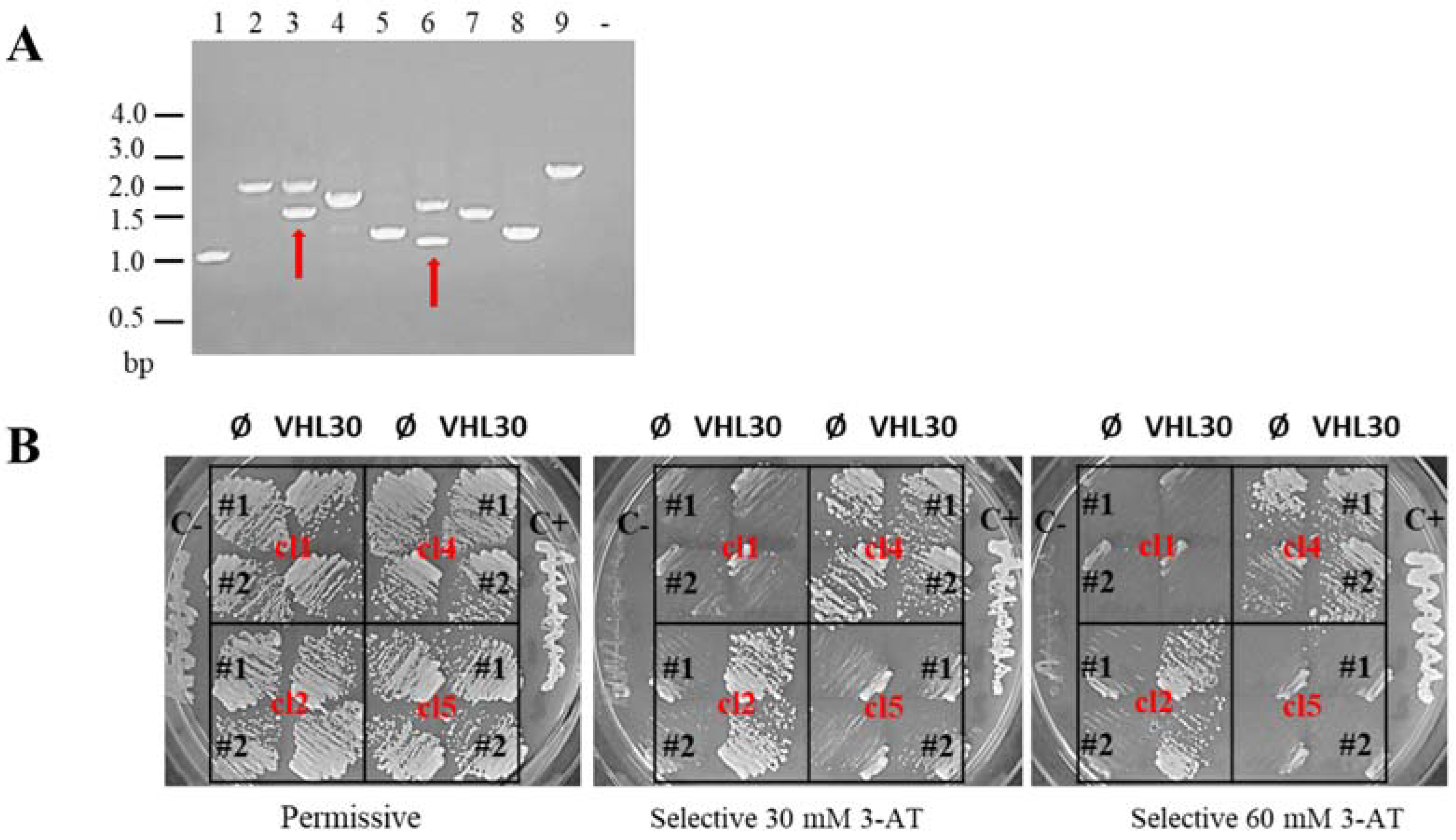
2.3. Positive Clone Analysis
2.4. Positive Clone Identification and Bioinformatic Analysis
3. Results
3.1. Yeast Two-Hybrid (Y2H) Library Screening in Testis Tissue
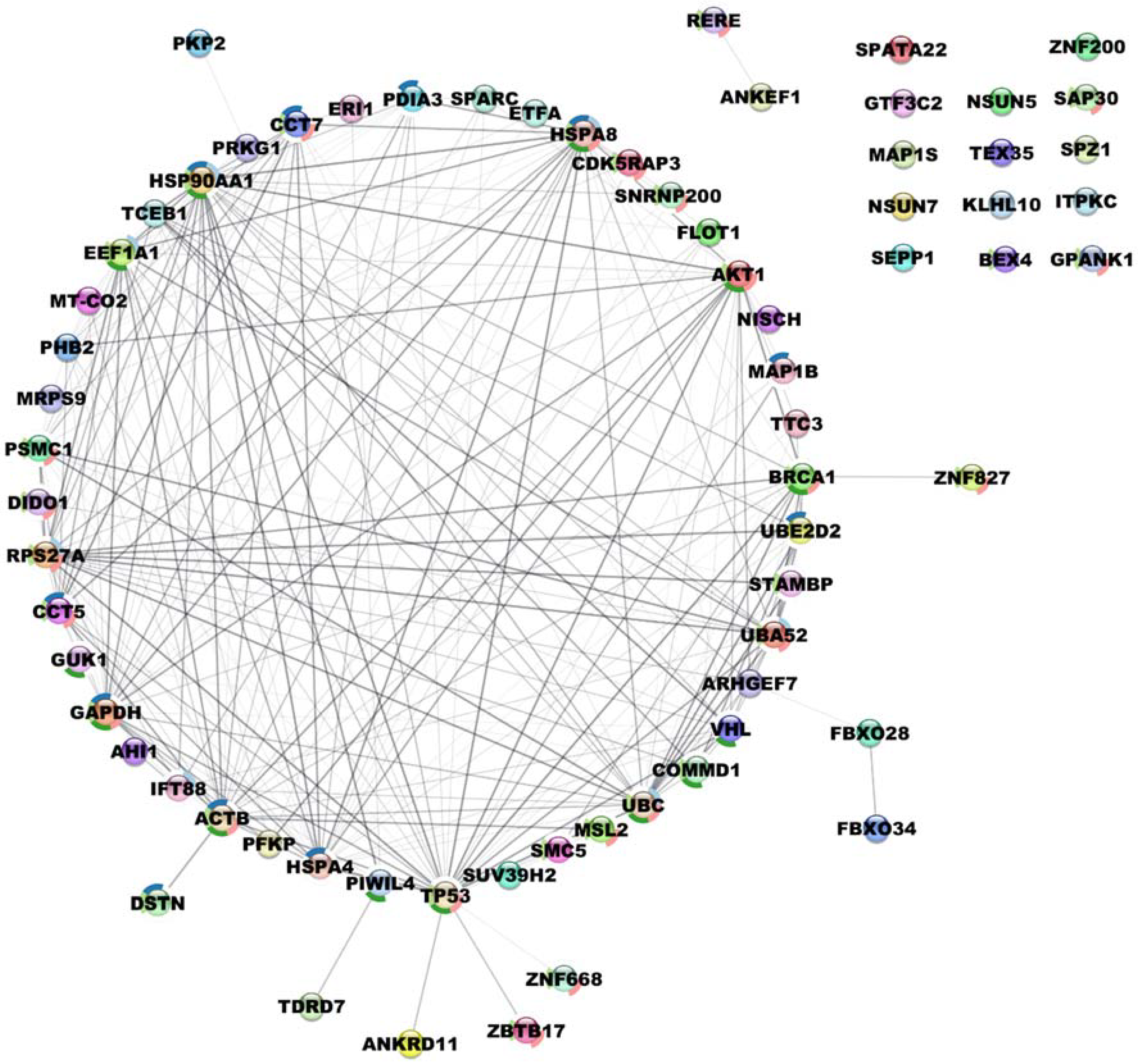
3.2. Identification and Characterization of pVHL30 Interactors
3.3. Predicted Sub-Cellular Distribution of the New Identified pVHL30 Interactors
3.4. Prediction of pVHL30 Binding Motifs
3.5. Mutations Found in Cancer Affect the pVHL30 Binding Motifs
3.6. Pathway Analysis of pVHL30 Binding Interactors
3.7. pVHL30 Interaction with 3’- or 5’-Untraslated Regions (UTR)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shuin, T.; Yamasaki, I.; Tamura, K.; Okuda, H.; Furihata, M.; Ashida, S. Von Hippel-Lindau Disease: Molecular Pathological Basis, Clinical Criteria, Genetic Testing, Clinical Features of Tumors and Treatment. Jpn. J. Clin. Oncol. 2006, 36, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Nordstrom-O’Brien, M.; van der Luijt, R.B.; van Rooijen, E.; van den Ouweland, A.M.; Majoor-Krakauer, D.F.; Lolkema, M.P.; van Brussel, A.; Voest, E.E.; Giles, R.H. Genetic Analysis of von Hippel-Lindau Disease. Hum. Mutat. 2010, 31, 521–537. [Google Scholar] [CrossRef]
- Crespigio, J.; Berbel, L.C.L.; Dias, M.A.; Berbel, R.F.; Pereira, S.S.; Pignatelli, D.; Mazzuco, T.L. Von Hippel–Lindau Disease: A Single Gene, Several Hereditary Tumors. J. Endocrinol. Investig. 2017, 41, 21–31. [Google Scholar] [CrossRef]
- Latif, F.; Tory, K.; Gnarra, J.; Yao, M.; Duh, F.M.; Orcutt, M.L.; Stackhouse, T.; Kuzmin, I.; Modi, W.; Geil, L. Identification of the von Hippel-Lindau Disease Tumor Suppressor Gene. Science 1993, 260, 1317–1320. [Google Scholar] [CrossRef]
- Iliopoulos, O.; Ohh, M.; Kaelin, W.G. PVHL19 Is a Biologically Active Product of the von Hippel-Lindau Gene Arising from Internal Translation Initiation. Proc. Natl. Acad. Sci. USA 1998, 95, 11661–11666. [Google Scholar] [CrossRef] [Green Version]
- Chesnel, F.; Hascoet, P.; Gagné, J.P.; Couturier, A.; Jouan, F.; Poirier, G.G.; Le Goff, C.; Vigneau, C.; Danger, Y.; Verite, F.; et al. The von Hippel–Lindau Tumour Suppressor Gene: Uncovering the Expression of the PVHL172 Isoform. Br. J. Cancer 2015, 113, 336–344. [Google Scholar] [CrossRef]
- Min, J.-H.; Yang, H.; Ivan, M.; Gertler, F.; Kaelin, W.G.; Pavletich, N.P. Structure of an HIF-1alpha-PVHL Complex: Hydroxyproline Recognition in Signaling. Science 2002, 296, 1886–1889. [Google Scholar] [CrossRef]
- Minervini, G.; Mazzotta, G.M.; Masiero, A.; Sartori, E.; Corrà, S.; Potenza, E.; Costa, R.; Tosatto, S.C.E. Isoform-Specific Interactions of the von Hippel-Lindau Tumor Suppressor Protein. Sci. Rep. 2015, 5, 12605. [Google Scholar] [CrossRef]
- Kamura, T.; Koepp, D.M.; Conrad, M.N.; Skowyra, D.; Moreland, R.J.; Iliopoulos, O.; Lane, W.S.; Kaelin, W.G.; Elledge, S.J.; Conaway, R.C.; et al. Rbx1, a Component of the VHL Tumor Suppressor Complex and SCF Ubiquitin Ligase. Science 1999, 284, 657–661. [Google Scholar] [CrossRef]
- Stebbins, C.E.; Kaelin, W.G.; Pavletich, N.P. Structure of the VHL-ElonginC-ElonginB Complex: Implications for VHL Tumor Suppressor Function. Science 1999, 284, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Epstein, A.C.R.; Gleadle, J.M.; McNeill, L.A.; Hewitson, K.S.; O’Rourke, J.; Mole, D.R.; Mukherji, M.; Metzen, E.; Wilson, M.I.; Dhanda, A.; et al. C. Elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases That Regulate HIF by Prolyl Hydroxylation. Cell 2001, 107, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Ohh, M.; Yauch, R.L.; Lonergan, K.M.; Whaley, J.M.; Stemmer-Rachamimov, A.O.; Louis, D.N.; Gavin, B.J.; Kley, N.; Kaelin, W.G., Jr.; Iliopoulos, O. The von Hippel-Lindau Tumor Suppressor Protein Is Required for Proper Assembly of an Extracellular Fibronectin Matrix. Mol. Cell 1998, 1, 959–968. [Google Scholar] [CrossRef]
- Young, A.P.; Schlisio, S.; Minamishima, Y.A.; Zhang, Q.; Li, L.; Grisanzio, C.; Signoretti, S.; Kaelin, W.G. VHL Loss Actuates a HIF-Independent Senescence Programme Mediated by Rb and P400. Nat. Cell Biol. 2008, 10, 361–369. [Google Scholar] [CrossRef]
- Guo, J.; Chakraborty, A.A.; Liu, P.; Gan, W.; Zheng, X.; Inuzuka, H.; Wang, B.; Zhang, J.; Zhang, L.; Yuan, M.; et al. PVHL Suppresses Kinase Activity of Akt in a Proline-Hydroxylation-Dependent Manner. Science 2016, 353, 929–932. [Google Scholar] [CrossRef] [Green Version]
- Minervini, G.; Pennuto, M.; Tosatto, S.C.E. The PVHL Neglected Functions, a Tale of Hypoxia-Dependent and -Independent Regulations in Cancer. Open Biol. 2020, 10, 200109. [Google Scholar] [CrossRef]
- Roe, J.-S.; Youn, H.-D. The Positive Regulation of P53 by the Tumor Suppressor VHL. Cell Cycle 2006, 5, 2054–2056. [Google Scholar] [CrossRef] [Green Version]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct Molecular Interaction Database in 2012. Nucleic Acids Res. 2012, 40, D841–D846. [Google Scholar] [CrossRef]
- Tabaro, F.; Minervini, G.; Sundus, F.; Quaglia, F.; Leonardi, E.; Piovesan, D.; Tosatto, S.C.E. VHLdb: A Database of von Hippel-Lindau Protein Interactors and Mutations. Sci. Rep. 2016, 6, 31128. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Tessarollo, L.; Hong, S.-B.; Baba, M.; Southon, E.; Back, T.C.; Spence, S.; Lobe, C.G.; Sharma, N.; Maher, G.W.; et al. Hepatic Vascular Tumors, Angiectasis in Multiple Organs, and Impaired Spermatogenesis in Mice with Conditional Inactivation of the VHL Gene. Cancer Res. 2003, 63, 5320–5328. [Google Scholar]
- Choyke, P.L.; Glenn, G.M.; Wagner, J.P.; Lubensky, I.A.; Thakore, K.; Zbar, B.; Linehan, W.M.; Walther, M.M. Epididymal Cystadenomas in von Hippel-Lindau Disease. Urology 1997, 49, 926–931. [Google Scholar] [CrossRef]
- Papadakis, G.Z.; Millo, C.; Sadowski, S.M.; Bagci, U.; Patronas, N.J. Epididymal Cystadenomas in von Hippel-Lindau Disease Showing Increased Activity on 68Ga-DOTA-TATE PET/CT. Clin. Nucl. Med. 2016, 41, 781–782. [Google Scholar] [CrossRef] [Green Version]
- Cox, R.; Vang, R.; Epstein, J.I. Papillary Cystadenoma of the Epididymis and Broad Ligament: Morphologic and Immunohistochemical Overlap with Clear Cell Papillary Renal Cell Carcinoma. Am. J. Surg. Pathol. 2014, 38, 713–718. [Google Scholar] [CrossRef]
- Schermer, B.; Ghenoiu, C.; Bartram, M.; Müller, R.U.; Kotsis, F.; Höhne, M.; Kühn, W.; Rapka, M.; Nitschke, R.; Zentgraf, H.; et al. The von Hippel-Lindau Tumor Suppressor Protein Controls Ciliogenesis by Orienting Microtubule Growth. J. Cell Biol. 2006, 175, 547–554. [Google Scholar] [CrossRef] [Green Version]
- Girardet, L.; Augière, C.; Asselin, M.-P.; Belleannée, C. Primary Cilia: Biosensors of the Male Reproductive Tract. Andrology 2019, 7, 588–602. [Google Scholar] [CrossRef] [Green Version]
- Higgins, M.; Obaidi, I.; McMorrow, T. Primary Cilia and Their Role in Cancer (Review). Oncol. Lett. 2019, 17, 3041–3047. [Google Scholar] [CrossRef]
- Adamiok-Ostrowska, A.; Piekiełko-Witkowska, A. Ciliary Genes in Renal Cystic Diseases. Cells 2020, 9, 907. [Google Scholar] [CrossRef] [Green Version]
- States, D.J.; Gish, W. Combined Use of Sequence Similarity and Codon Bias for Coding Region Identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Chatr-aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K.; O’Donnell, L.; Oster, S.; Theesfeld, C.; Sellam, A.; et al. The BioGRID Interaction Database: 2017 Update. Nucleic Acids Res. 2016, 45, D369–D379. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Greene, C.S.; Krishnan, A.; Wong, A.K.; Ricciotti, E.; Zelaya, R.A.; Himmelstein, D.S.; Zhang, R.; Hartmann, B.M.; Zaslavsky, E.; Sealfon, S.C.; et al. Understanding Multicellular Function and Disease with Human Tissue-Specific Networks. Nat. Genet. 2015, 47, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—the Eukaryotic Linear Motif Resource in 2020. Nucleic Acids Res. 2020, 48, D296–D306. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Chen, D.; Shiloh, A.; Luo, J.; Nikolaev, A.Y.; Qin, J.; Gu, W. Deubiquitination of P53 by HAUSP Is an Important Pathway for P53 Stabilization. Nature 2002, 416, 648–653. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 Promotes the Rapid Degradation of P53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef]
- Falconieri, A.; Minervini, G.; Bortolotto, R.; Piovesan, D.; Lopreiato, R.; Sartori, G.; Pennuto, M.; Tosatto, S.C.E. The E3 Ubiquitin-Protein Ligase MDM2 Is a Novel Interactor of the von Hippel–Lindau Tumor Suppressor. Sci. Rep. 2020, 10, 15850. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, D.; Messing, E.M.; Wu, G. VHL Protein-Interacting Deubiquitinating Enzyme 2 Deubiquitinates and Stabilizes HIF-1alpha. EMBO Rep. 2005, 6, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, D.; Na, X.; Schoen, S.R.; Messing, E.M.; Wu, G. Identification of a Deubiquitinating Enzyme Subfamily as Substrates of the von Hippel-Lindau Tumor Suppressor. Biochem. Biophys. Res. Commun. 2002, 294, 700–709. [Google Scholar] [CrossRef]
- Ye, Y.; Vasavada, S.; Kuzmin, I.; Stackhouse, T.; Zbar, B.; Williams, B.R. Subcellular Localization of the von Hippel-Lindau Disease Gene Product Is Cell Cycle-Dependent. Int. J. Cancer 1998, 78, 62–69. [Google Scholar] [CrossRef]
- Bouhamdani, N.; Comeau, D.; Coholan, A.; Cormier, K.; Turcotte, S. Targeting Lysosome Function Causes Selective Cytotoxicity in VHL-Inactivated Renal Cell Carcinomas. Carcinogenesis 2020, 41, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Neduva, V.; Russell, R.B. DILIMOT: Discovery of Linear Motifs in Proteins. Nucleic Acids Res. 2006, 34, W350–W355. [Google Scholar] [CrossRef] [Green Version]
- Khacho, M.; Mekhail, K.; Pilon-Larose, K.; Pause, A.; Côté, J.; Lee, S. EEF1A Is a Novel Component of the Mammalian Nuclear Protein Export Machinery. Mol. Biol. Cell 2008, 19, 5296–5308. [Google Scholar] [CrossRef] [Green Version]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic Cancer Genetics at High-Resolution. Nucleic Acids Res. 2016, 45, D777–D783. [Google Scholar] [CrossRef]
- Rousseau, A.; Bertolotti, A. Regulation of Proteasome Assembly and Activity in Health and Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef] [Green Version]
- Hergovich, A.; Lisztwan, J.; Barry, R.; Ballschmieter, P.; Krek, W. Regulation of Microtubule Stability by the von Hippel-Lindau Tumour Suppressor Protein PVHL. Nat. Cell Biol. 2003, 5, 64–70. [Google Scholar] [CrossRef]
- Grote, M.; Wolf, E.; Will, C.L.; Lemm, I.; Agafonov, D.E.; Schomburg, A.; Fischle, W.; Urlaub, H.; Lührmann, R. Molecular Architecture of the Human Prp19/CDC5L Complex. Mol. Cell. Biol. 2010, 30, 2105–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Woods, N.T.; Kim, D.; Sweet, M.; Monteiro, A.N.A.; Karchin, R. Yeast Two-Hybrid Junk Sequences Contain Selected Linear Motifs. Nucleic Acids Res. 2011, 39, e128. [Google Scholar] [CrossRef] [PubMed]
- Roberts, G.G.; Parrish, J.R.; Mangiola, B.A.; Finley, R.L. High-Throughput Yeast Two-Hybrid Screening. Methods Mol. Biol. 2012, 812, 39–61. [Google Scholar] [CrossRef]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.G.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivagurunathan, S.; Palanisamy, K.; Arunachalam, J.P.; Chidambaram, S. Possible Role of HIWI2 in Modulating Tight Junction Proteins in Retinal Pigment Epithelial Cells through Akt Signaling Pathway. Mol. Cell. Biochem. 2017, 427, 145–156. [Google Scholar] [CrossRef]
- Henaoui, I.S.; Jacovetti, C.; Guerra Mollet, I.; Guay, C.; Sobel, J.; Eliasson, L.; Regazzi, R. PIWI-Interacting RNAs as Novel Regulators of Pancreatic Beta Cell Function. Diabetologia 2017, 60, 1977–1986. [Google Scholar] [CrossRef]
- Strub, G.M.; Paillard, M.; Liang, J.; Gomez, L.; Allegood, J.C.; Hait, N.C.; Maceyka, M.; Price, M.M.; Chen, Q.; Simpson, D.C.; et al. Sphingosine-1-Phosphate Produced by Sphingosine Kinase 2 in Mitochondria Interacts with Prohibitin 2 to Regulate Complex IV Assembly and Respiration. FASEB J. 2011, 25, 600–612. [Google Scholar] [CrossRef] [Green Version]
- Peña, F.J.; Ortiz-Rodríguez, J.M.; Gaitskell-Phillips, G.L.; Gil, M.C.; Ortega-Ferrusola, C.; Martín-Cano, F.E. An Integrated Overview on the Regulation of Sperm Metabolism (Glycolysis-Krebs Cycle-Oxidative Phosphorylation). Anim. Reprod. Sci. 2021, 106805. [Google Scholar] [CrossRef]
- Vasudev, N.S.; Sim, S.; Cairns, D.A.; Ferguson, R.E.; Craven, R.A.; Stanley, A.; Cartledge, J.; Thompson, D.; Selby, P.J.; Banks, R.E. Pre-Operative Urinary Cathepsin D Is Associated with Survival in Patients with Renal Cell Carcinoma. Br. J. Cancer 2009, 101, 1175–1182. [Google Scholar] [CrossRef] [Green Version]
- Young, A.C.; Craven, R.A.; Cohen, D.; Taylor, C.; Booth, C.; Harnden, P.; Cairns, D.A.; Astuti, D.; Gregory, W.; Maher, E.R.; et al. Analysis of VHL Gene Alterations and Their Relationship to Clinical Parameters in Sporadic Conventional Renal Cell Carcinoma. Clin. Cancer Res. 2009, 15, 7582–7592. [Google Scholar] [CrossRef] [Green Version]
- Gossage, L.; Eisen, T. Alterations in VHL as Potential Biomarkers in Renal-Cell Carcinoma. Nat. Rev. Clin. Oncol. 2010, 7, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.L.; Hsieh, A.C. The Untranslated Regions of MRNAs in Cancer. Trends Cancer 2019, 5, 245–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Protein Name | UniProt ID | Function | n° Hits |
|---|---|---|---|
| Elongation factor 1-alpha 1 (EEF1A1) | P68104 | protein biosynthesis | 17 |
| Elongin C (ELOC) | Q15369 | protein degradation | 6 |
| T-complex protein 1 subunit eta (CCT7) | Q99832 | actin/tubulin folding | 4 |
| Ubiquitin-coniugating enzyme E2 D2 (UBE2D2) | P62837 | protein ubiquitination | 2 |
| T-complex protein 1 subunit epsilon (CCT5) | P48643 | actin/tubulin folding | 1 |
| U5 small nuclear ribonucleoprotein 200 kDa helicase (SNRNP200) | O75643 | RNA splicing | 1 |
| Putative methyltransferase (NSU7) | Q8NE18 | methylation | 12 |
| Spermatogenesis-associated protein 22 (SPATA22) | Q8NHS9 | germ cell division | 10 |
| Zinc finger and BTB domain-containing protein 17 (ZBTB17) | Q13105 | cell cycle regulator | 9 |
| Microtubule-associated protein 1S (MAP1S) | Q66K74 | apoptosis | 4 |
| Protein disulfide-isomerase A3 (PDIA3) | P30101 | protein folding | 4 |
| Death-inducer obliterator 1 (DIDO1) | Q9BTC0 | tumor suppressor | 3 |
| Electron transfer flavoprotein subunit alpha, mitochondrial (ETFA) | P13804 | electron transport | 3 |
| 26S proteasome regulatory subunit 4 (PSMC1) | P62191 | protein degradation | 3 |
| Histone deacetylase complex subunit (SAP30) | O75446 | deacetylation | 3 |
| E3 ubiquitin-protein ligase (TTC3) | P53804 | ubiquitination/protein degradation | 3 |
| Ankyrin repeat and EF-hand domain-containing protein 1 (ANKEF1) | Q9NU02 | n.d. | 2 |
| Ankyrin repeat domain-containing protein 11 (ANKRD11) | Q6UB99 | chromatin regulator | 2 |
| Guanylate kinase (GUK1) | B1ANH3 | phosphorylation | 2 |
| Protein BEX4 (BEX4) | Q9NWD9 | microtubule deacetylation | 2 |
| Cytochrome c oxidase subunit 2 (MT-CO2) | P00403 | oxygen reduction | 2 |
| Prohibitin-2 (PHB2) | Q99623 | transcription inhibitor | 2 |
| Piwi-like protein 4 (PIWIL4) | Q7Z3Z4 | tumor enhancer | 2 |
| cGMP-dependent protein kinase 1 (PRKG1) | Q13976 | protein phosphorylation | 2 |
| Arginine-glutamic acid dipeptide repeats protein (RERE) | Q9P2R6 | cell survival control | 2 |
| Structural maintenance of chromosomes protein 5 (SMC5) | Q8IY18 | DNA repair | 2 |
| STAM-binding protein (STAMBP) | O95630 | protein degradation | 2 |
| Testis-expressed protein 35 (TEX35) | Q5T0J7 | n.d. | 2 |
| Jouberin (AHI1) | Q8N157 | ciliogenesis | 1 |
| Rho guanine nucleotide exchange factor 7 (ARHGEF7) | Q14155 | apoptosis | 1 |
| Protein BEX2 (BEX2) | Q9BXY8-2 | cell cycle regulator | 1 |
| Breast cancer type 1 susceptibility protein (BRCA1) | P38398 | E3-ub lig/DNA repair | 1 |
| CDK5 regulatory subunit-associated protein 3 (CDK5RAP3) | J3QRX0 | n.d. | 1 |
| COMM domain-containing protein 1 (COMMD1) | Q8N668 | protein ubiquitination regulator | 1 |
| Copine-5 (CPNE5) | A0A0J9YWA1 | dendrite formation | 1 |
| Destrin (DSTN) | P60981 | actin depolymerization | 1 |
| 3’-5’ exoribonuclease 1 (ERI1) | Q8IV48 | RNA exonuclease | 1 |
| F-box only protein 28 (FBXO28) | Q9NVF7 | ubiquitination/protein degradation | 1 |
| F-box only protein 34 (FBXO34) | Q9NWN3 | SRP of E3-ub complex | 1 |
| Flotillin-1 (FLOT1) | O75955 | caveolae formation | 1 |
| G patch domain and ankyrin repeat-containing protein 1 (GPANK1) | O95872 | n.d. | 1 |
| General transcription factor 3C polypeptide 2 (GTF3C2) | Q8WUA4 | DNA transcription | 1 |
| Intraflagellar transport protein 88 homolog (IFT88) | Q13099 | ciliogenesis | 1 |
| Inositol-trisphosphate 3-kinase (ITPKC) | Q96DU7 | phosphorylation | 1 |
| Kelch-like protein 10 (KLHL10) | Q6JEL2 | ubiquitination/protein degradation | 1 |
| Microtubule-associated protein 1B (MAP1B) | P46821 | microtubule stabilization | 1 |
| 28S ribosomal protein S9, mitochondrial (MRPS9) | P82933 | n.d. | 1 |
| E3 ubiquitin-protein ligase MSL2 (MSL2) | Q9HCI7 | ubiquitination/protein degradation | 1 |
| Nischarin (NISCH) | Q9Y2I1 | cell survival/migration | 1 |
| Probable 28S rRNA (cytosine-C(5))-methyltransferase (NSUN5) | Q96P11 | methylation | 1 |
| ATP-dependent 6-phosphofructokinase, platelet type (PFKP) | Q01813 | glycolysis | 1 |
| Plakophilin-2 (PKP2) | Q99959 | cell-cell adhesion | 1 |
| Selenoprotein P (SELENOP) | P49908 | selenium transport | 1 |
| SPARC | P09486 | cell growth | 1 |
| Spermatogenic leucine zipper protein 1 (SPZ1) | Q9BXG8 | germ cell proliferation and differentiation | 1 |
| Histone-lysine N-methyltransferase (SUV39H2) | Q9H5I1 | chromatin regulator | 1 |
| Tudor domain-containing protein 7 (TDRD7) | Q8NHU6 | post-transcription regulator | 1 |
| Zinc finger protein 200 (ZNF200) | P98182 | spermatogenesis | 1 |
| Zinc finger protein 668 (ZNF668) | Q96K58 | transcription regulator | 1 |
| Zinc finger protein 827 (ZNF827) | Q17R98 | transcription regulator | 1 |
| Motif | Scons | N° Protein | p Value |
|---|---|---|---|
| VGxxxK | 2.22 × 10−29 | 4 | 2.84 × 10−5 |
| PxxxVxxN | 3.12 × 10−24 | 4 | 2.20 × 10−5 |
| GxKxxK | 1.42 × 10−22 | 4 | 2.48 × 10−4 |
| KKKxK | 1.13 × 10−20 | 4 | 9.00 × 10−6 |
| KxKxKxK | 1.39 × 10−18 | 4 | 3.81 × 10−6 |
| KxxxPK | 3.10 ×10−18 | 5 | 1.29 × 10−5 |
| KxxKxxxP | 1.20 ×10−17 | 4 | 2.37 × 10−4 |
| KNxxxK | 1.63 × 10−16 | 4 | 3.15 × 10−4 |
| AxxVP | 3.45 × 10−16 | 4 | 2.18 × 10−4 |
| KKK | 4.05 × 10−16 | 5 | 2.76 × 10−4 |
| Protein | Variants | Pathologic Condition |
|---|---|---|
| SPATA 22 | p.Arg89Ile | glioma and adenocarcinoma (large intestine) |
| ZBTB17 | p.Arg562Cys | carcinoma (endometrium, thyroid) |
| p.Arg625Trp | carcinoma (large cell) | |
| p.Ser497 = | adenoma (large intestine) and carcinoma (upper aerodigestive tract) | |
| CCT7 | p.Pro235Ser | malignant melanoma |
| p.Glu316Lys | adenocarcinoma (lung, urinary tract) | |
| MAP1S | p.Arg863Asn | adenocarcinoma (large intestine) |
| p.Gly891Ser | carcinoma (urinary tract) | |
| PDIA3 | p.Leu361 = | ccRCC (Kidney) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falconieri, A.; Minervini, G.; Quaglia, F.; Sartori, G.; Tosatto, S.C.E. Characterization of the pVHL Interactome in Human Testis Using High-Throughput Library Screening. Cancers 2022, 14, 1009. https://doi.org/10.3390/cancers14041009
Falconieri A, Minervini G, Quaglia F, Sartori G, Tosatto SCE. Characterization of the pVHL Interactome in Human Testis Using High-Throughput Library Screening. Cancers. 2022; 14(4):1009. https://doi.org/10.3390/cancers14041009
Chicago/Turabian StyleFalconieri, Antonella, Giovanni Minervini, Federica Quaglia, Geppo Sartori, and Silvio C. E. Tosatto. 2022. "Characterization of the pVHL Interactome in Human Testis Using High-Throughput Library Screening" Cancers 14, no. 4: 1009. https://doi.org/10.3390/cancers14041009
APA StyleFalconieri, A., Minervini, G., Quaglia, F., Sartori, G., & Tosatto, S. C. E. (2022). Characterization of the pVHL Interactome in Human Testis Using High-Throughput Library Screening. Cancers, 14(4), 1009. https://doi.org/10.3390/cancers14041009







