The Antianginal Drug Perhexiline Displays Cytotoxicity against Colorectal Cancer Cells In Vitro: A Potential for Drug Repurposing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Lines and Culture
2.3. Patient-Derived Organoids
2.4. Crystal Violet Viability Assay
2.5. Apoptosis Assay by Annexin V/Propidium Iodide Staining
2.6. Caspase 3/7 Activation Assay
2.7. AlamarBlue Viability Assay for Patient-Derived Organoids
2.8. Statistical Analysis
3. Results
3.1. Effect of Perhexiline and Its Enatiomers on CRC Monolayer Growth
3.2. Effect of Perhexiline and Its Enantiomers on Annexin V Induction
3.3. Kinetics of Apoptosis Induction by Perhexiline and Its Enantiomers
3.4. Effect of Perhexiline and Its Enantiomers on CRC Spheroids
3.5. Effect of Perhexiline and Its Enantiomers on Patient-Derived CRC Organoids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Patterns and Trends in Colorectal Cancer Incidence and Mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Ashrafian, H.; Horowitz, J.D.; Frenneaux, M.P. Perhexiline. Cardiovasc. Drug Rev. 2007, 25, 76–97. [Google Scholar] [CrossRef]
- Unger, S.A.; Robinson, M.A.; Horowitz, J.D. Perhexiline Improves Symptomatic Status in Elderly Patients with Severe Aortic Stenosis. Aust. N. Z. J. Med. 1997, 27, 24–28. [Google Scholar] [CrossRef]
- Lee, L.; Campbell, R.; Scheuermann-Freestone, M.; Taylor, R.; Gunaruwan, P.; Williams, L.; Ashrafian, H.; Horowitz, J.; Fraser, A.G.; Clarke, K. Metabolic Modulation with Perhexiline in Chronic Heart Failure: A Randomized, Controlled Trial of Short-Term Use of a Novel Treatment. Circulation 2005, 112, 3280–3288. [Google Scholar] [CrossRef]
- Gould, B.J.; Amoah, A.G.B.; Parke, D.V. Stereoselective Pharmacokinetics of Perhexiline. Xenobiotica 1986, 16, 491–502. [Google Scholar] [CrossRef]
- Davies, B.J.; Herbert, M.K.; Culbert, J.A.; Pyke, S.M.; Coller, J.K.; Somogyi, A.A.; Milne, R.W.; Sallustio, B.C. Enantioselective Assay for the Determination of Perhexiline Enantiomers in Human Plasma by Liquid Chromatography. J. Chromatogr. B 2006, 832, 114–120. [Google Scholar] [CrossRef]
- Narasimhan, V.; Wright, J.A.; Churchill, M.; Wang, T.; Rosati, R.; Lannagan, T.R.M.; Vrbanac, L.; Richardson, A.B.; Kobayashi, H.; Price, T.; et al. Medium-Throughput Drug Screening of Patient-Derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy. Clin. Cancer Res. 2020, 26, 3662–3670. [Google Scholar] [CrossRef]
- Palethorpe, H.M.; Drew, P.A.; Smith, E. Androgen Signaling in Esophageal Adenocarcinoma Cell Lines in Vitro. Dig. Dis. Sci. 2017, 62, 3402–3414. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.-H.; Wang, F.; Wang, Y.-N.; He, M.-M.; Wu, Q.-N.; Lu, Y.-X.; Yu, H.-E.; Chen, Z.-H.; Zhao, Q. Inhibition of Fatty Acid Catabolism Augments the Efficacy of Oxaliplatin-Based Chemotherapy in Gastrointestinal Cancers. Cancer Lett. 2020, 473, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The Third Dimension Bridges the Gap between Cell Culture and Live Tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Weaver, V.M.; Lelièvre, S.; Lakins, J.N.; Chrenek, M.A.; Jones, J.C.R.; Giancotti, F.; Werb, Z.; Bissell, M.J. B4 Integrin-Dependent Formation of Polarized Three-Dimensional Architecture Confers Resistance to Apoptosis in Normal and Malignant Mammary Epithelium. Cancer Cell 2002, 2, 205–216. [Google Scholar] [CrossRef]
- Osswald, A.; Sun, Z.; Grimm, V.; Ampem, G.; Riegel, K.; Westendorf, A.M.; Sommergruber, W.; Otte, K.; Dürre, P.; Riedel, C.U. Three-Dimensional Tumor Spheroids for in Vitro Analysis of Bacteria as Gene Delivery Vectors in Tumor Therapy. Microb. Cell Factories 2015, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-Derived Organoids Can Predict Response to Chemotherapy in Metastatic Colorectal Cancer Patients. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Mozolewska, P.; Duzowska, K.; Pakiet, A.; Mika, A.; Śledziński, T. Inhibitors of Fatty Acid Synthesis and Oxidation as Potential Anticancer Agents in Colorectal Cancer Treatment. Anticancer Res. 2020, 40, 4843–4856. [Google Scholar] [CrossRef]
- Ceccarelli, S.M.; Chomienne, O.; Gubler, M.; Arduini, A. Carnitine Palmitoyltransferase (CPT) Modulators: A Medicinal Chemistry Perspective on 35 Years of Research. J. Med. Chem. 2011, 54, 3109–3152. [Google Scholar] [CrossRef]
- Kant, S.; Kesarwani, P.; Guastella, A.R.; Kumar, P.; Graham, S.F.; Buelow, K.L.; Nakano, I.; Chinnaiyan, P. Perhexiline Demonstrates FYN-Mediated Anti-Tumor Activity in Glioblastoma. Mol. Cancer Ther. 2020, 19, 7. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, W.; Devarakonda, T.; Zhou, H.; Wang, X.-Y.; Salloum, F.N.; Spiegel, S.; Fang, X. Functional Analysis of Molecular and Pharmacological Modulators of Mitochondrial Fatty Acid Oxidation. Sci. Rep. 2020, 10, 1450. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Liu, J.; Jiang, W.Q.; Carew, J.S.; Ogasawara, M.A.; Pelicano, H.; Croce, C.M.; Estrov, Z.; Xu, R.H.; Keating, M.J. Elimination of Chronic Lymphocytic Leukemia Cells in Stromal Microenvironment by Targeting CPT with an Antiangina Drug Perhexiline. Oncogene 2016, 35, 5663–5673. [Google Scholar] [CrossRef] [PubMed]
- Schnell, S.A.; Ambesi-Impiombato, A.; Sanchez-Martin, M.; Belver, L.; Xu, L.; Qin, Y.; Kageyama, R.; Ferrando, A.A. Therapeutic Targeting of HES1 Transcriptional Programs in T-ALL. Blood 2015, 125, 2806–2814. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wu, G.; Song, L.; Cao, L.; Tan, Z.; Tang, M.; Li, Z.; Shi, D.; Zhang, S.; Li, J. NKX2-8 Deletion-Induced Reprogramming of Fatty Acid Metabolism Confers Chemoresistance in Epithelial Ovarian Cancer. EBioMedicine 2019, 43, 238–252. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional Culture System of Cancer Cells Combined with Biomaterials for Drug Screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef]
- Pennarossa, G.; Arcuri, S.; De Iorio, T.; Gandolfi, F.; Brevini, T.A.L. Current Advances in 3D Tissue and Organ Reconstruction. Int. J. Mol. Sci. 2021, 22, 830. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; den Braver, M.W.; Herpers, B.; Terpstra, V.; Commandeur, J.N.M.; van de Water, B.; Price, L.S. A 3D in Vitro Model of Differentiated HepG2 Cell Spheroids with Improved Liver-like Properties for Repeated Dose High-Throughput Toxicity Studies. Arch. Toxicol. 2014, 88, 1083–1095. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. A Cancer Invasion Model Combined with Cancer-Associated Fibroblasts Aggregates Incorporating Gelatin Hydrogel Microspheres Containing a P53 Inhibitor. Tissue. Eng. Part C Methods 2019, 25, 711–720. [Google Scholar] [CrossRef]
- Duarte, A.A.; Gogola, E.; Sachs, N.; Barazas, M.; Annunziato, S.; de Ruiter, J.R.; Velds, A.; Blatter, S.; Houthuijzen, J.M.; van de Ven, M.; et al. BRCA-Deficient Mouse Mammary Tumor Organoids to Study Cancer-Drug Resistance. Nat. Methods 2018, 15, 134–140. [Google Scholar] [CrossRef]
- Hu, H.; Gehart, H.; Artegiani, B.; LÖpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606.e19. [Google Scholar] [CrossRef]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Killalea, S.M.; Krum, H. Systematic Review of the Efficacy and Safety of Perhexiline in the Treatment of Ischemic Heart Disease. Am. J. Cardiovasc. Drugs 2001, 1, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Phuong, H.; Choi, B.Y.; Chong, C.-R.; Raman, B.; Horowitz, J.D. Can Perhexiline Be Utilized Without Long-Term Toxicity? A Clinical Practice Audit. Ther. Drug Monit. 2016, 38, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Drury, N.E.; Licari, G.; Chong, C.-R.; Howell, N.J.; Frenneaux, M.P.; Horowitz, J.D.; Pagano, D.; Sallustio, B.C. Relationship between Plasma, Atrial and Ventricular Perhexiline Concentrations in Humans: Insights into Factors Affecting Myocardial Uptake. Br. J. Clin. Pharmacol. 2014, 77, 789–795. [Google Scholar] [CrossRef][Green Version]
- Licari, G.; Milne, R.W.; Somogyi, A.A.; Sallustio, B.C. Enantioselectivity in the Tissue Distribution of Perhexiline Contributes to Different Effects on Hepatic Histology and Peripheral Neural Function in Rats. Pharmacol. Res. Perspect. 2018, 6, e00406. [Google Scholar] [CrossRef]
- Chong, C.-R.; Drury, N.E.; Licari, G.; Frenneaux, M.P.; Horowitz, J.D.; Pagano, D.; Sallustio, B.C. Stereoselective Handling of Perhexiline: Implications Regarding Accumulation within the Human Myocardium. Eur. J. Clin. Pharmacol. 2015, 71, 1485–1491. [Google Scholar] [CrossRef]
- Ren, X.-R.; Wang, J.; Osada, T.; Mook, R.A.; Morse, M.A.; Barak, L.S.; Lyerly, H.K.; Chen, W. Perhexiline Promotes HER3 Ablation through Receptor Internalization and Inhibits Tumor Growth. Breast Cancer Res. 2015, 17, 20. [Google Scholar] [CrossRef]
- Davies, B.J.; Coller, J.K.; Somogyi, A.A.; Milne, R.W.; Sallustio, B.C. CYP2B6, CYP2D6, and CYP3A4 Catalyze the Primary Oxidative Metabolism of Perhexiline Enantiomers by Human Liver Microsomes. Drug Metab. Dispos. 2007, 35, 128–138. [Google Scholar] [CrossRef]
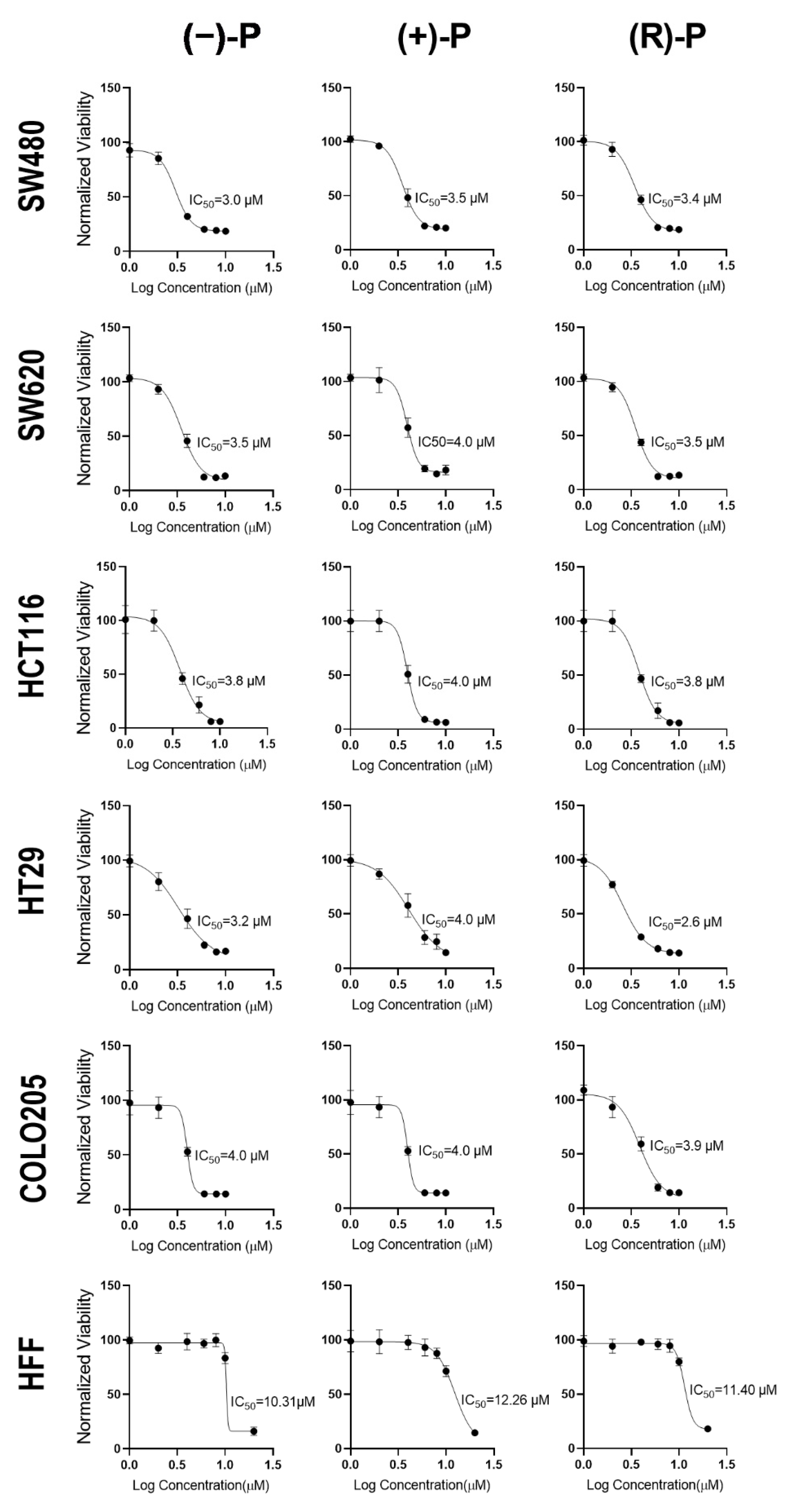
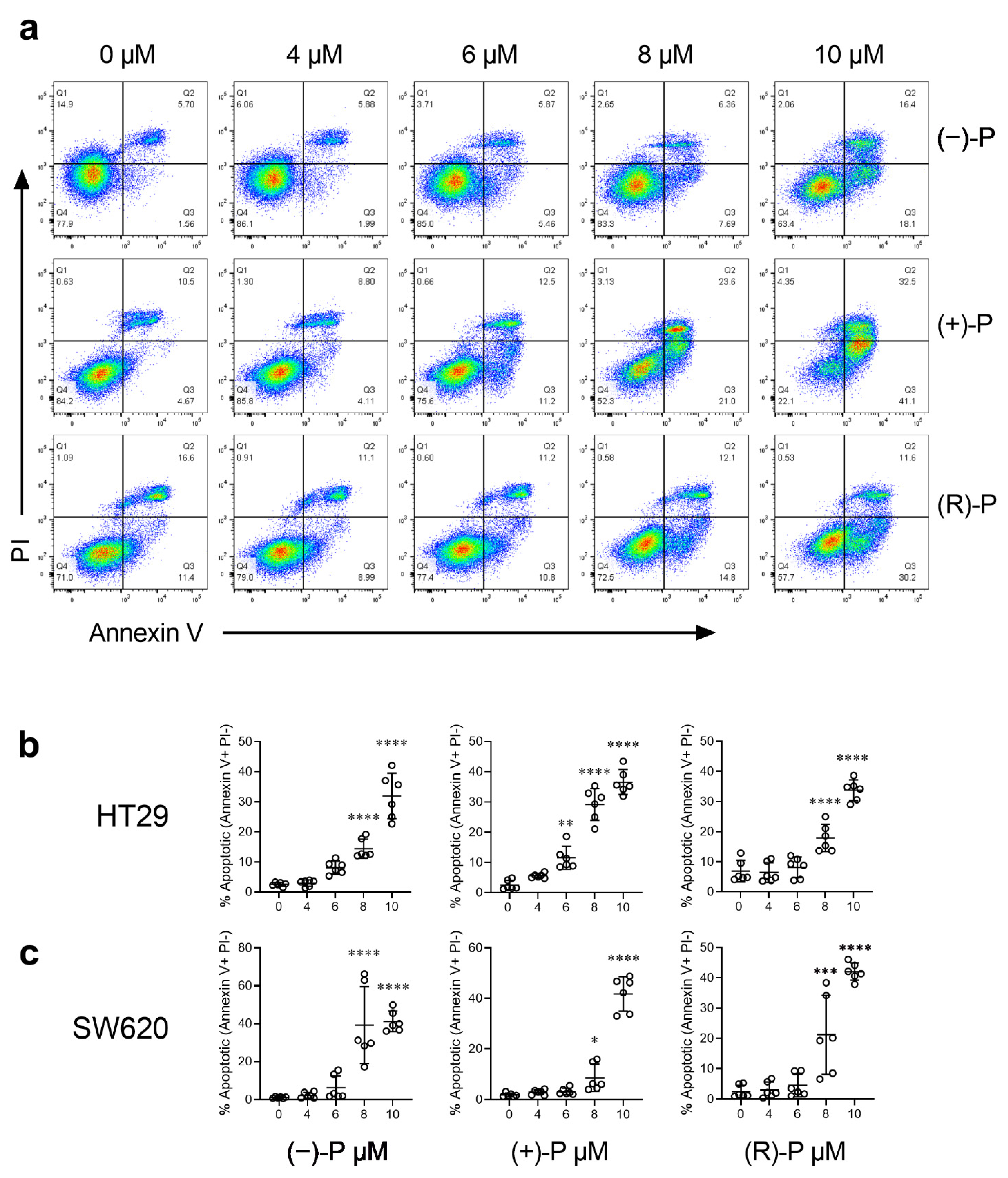

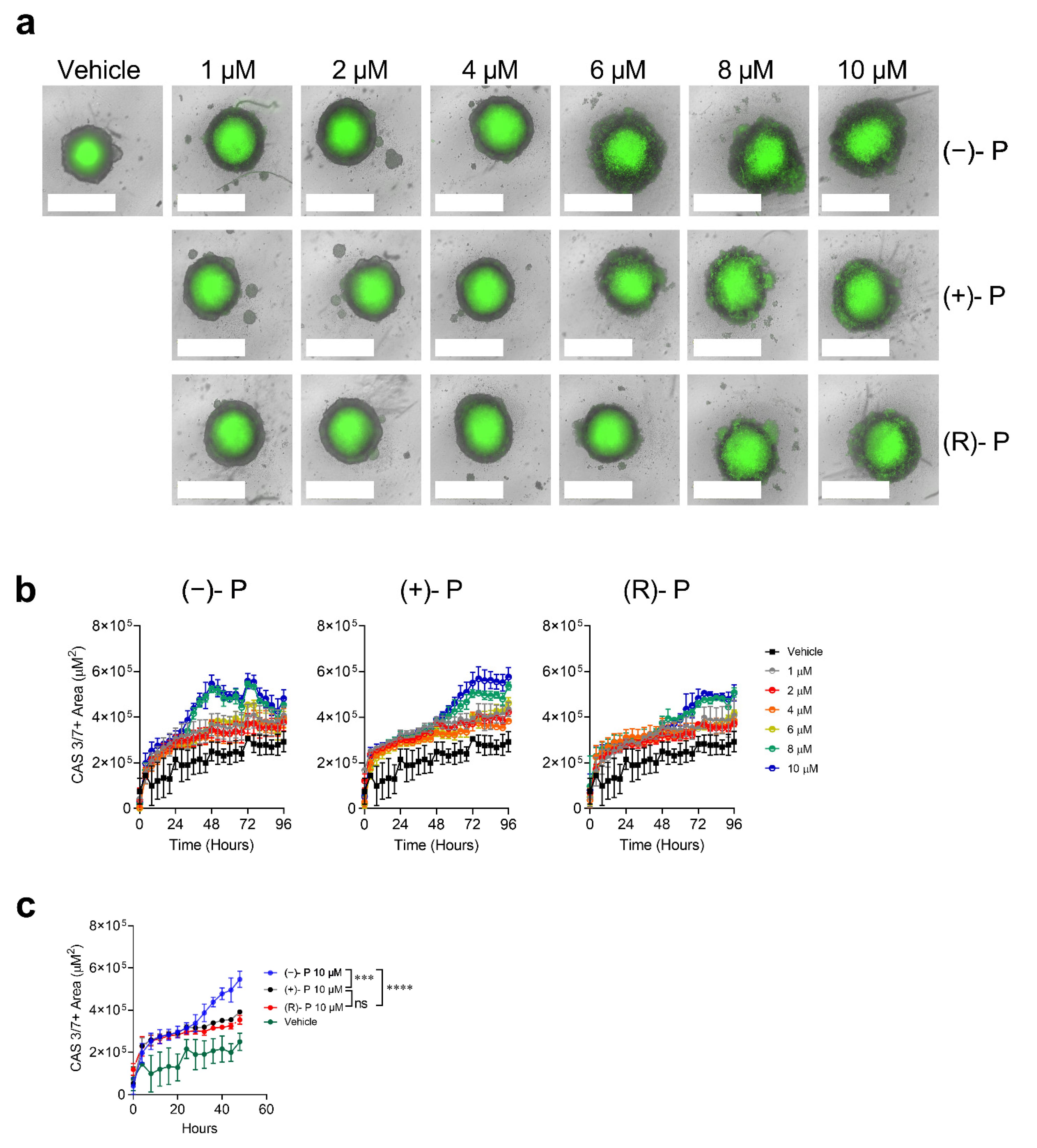
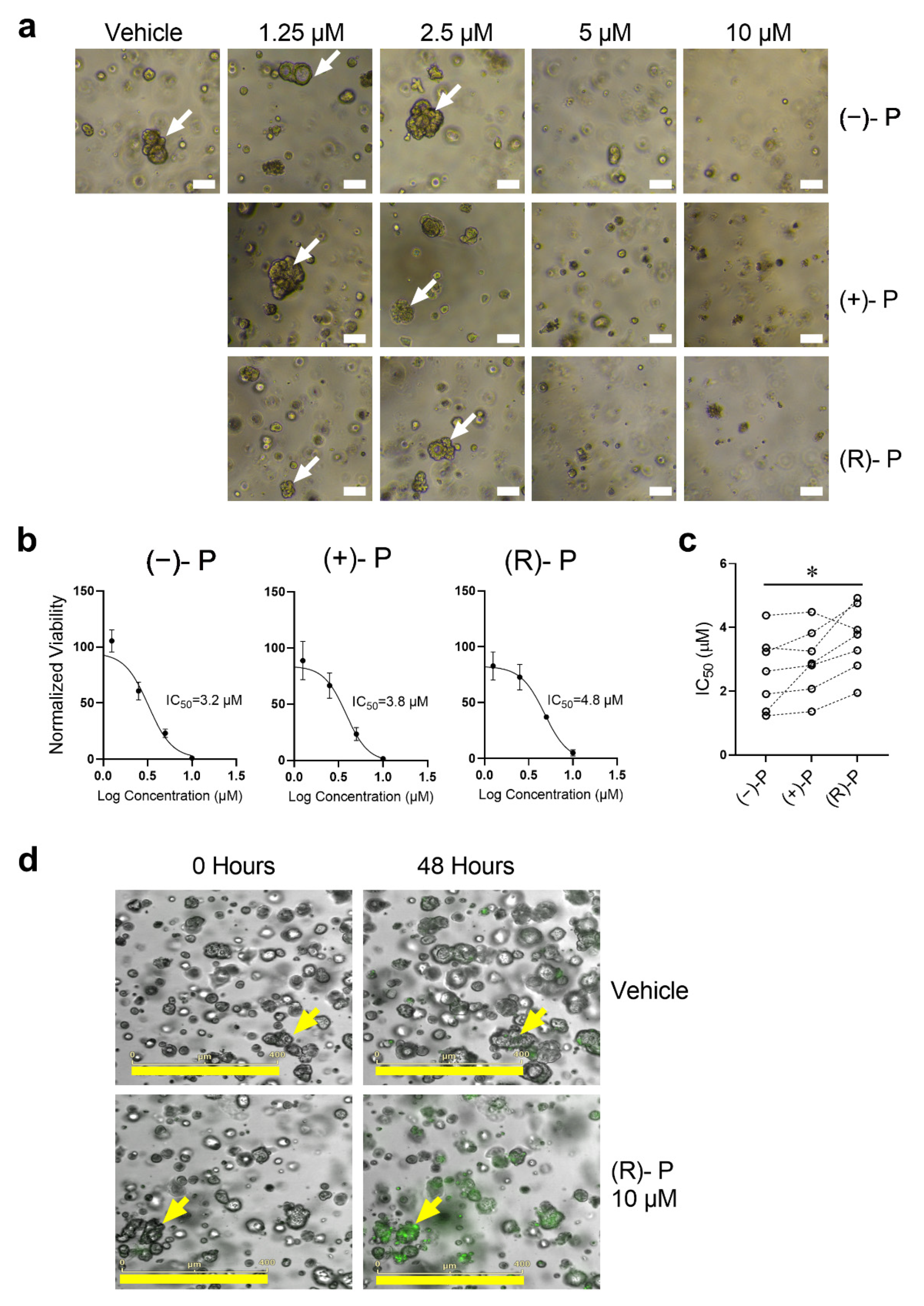
| Cell Line | Cell Type | (−)-P | (+)-P | (R)-P |
|---|---|---|---|---|
| COLO205 | CRC | 4.27 ± 0.51 | 4.39 ± 0.47 | 3.29 ± 0.90 |
| HCT116 | CRC | 3.37 ± 0.82 | 5.01 ± 1.91 | 3.45 ± 0.74 |
| HT29 | CRC | 3.81 ± 1.00 | 4.25 ± 0.32 | 3.08 ± 0.41 |
| SW480 | CRC | 3.70 ± 1.30 | 3.92 ± 0.74 | 3.92 ± 0.99 |
| SW620 | CRC | 3.88 ± 0.48 | 4.30 ± 0.70 | 3.62 ± 0.91 |
| HFF | Fibroblast | 11.06 ± 1.04 | 12.04 ± 0.53 | 11.34 ± 0.85 |
| Cell Line | (−)-P | (+)-P | (R)-P |
|---|---|---|---|
| COLO205 | 2.59 | 2.74 | 3.45 |
| HCT116 | 3.28 | 2.40 | 3.29 |
| HT29 | 2.90 | 2.83 | 3.68 |
| SW480 | 2.99 | 3.07 | 2.89 |
| SW620 | 2.85 | 2.80 | 3.13 |
| Sample | Sex | Age | Location | Stage | Primary/Metastasis | MMR * | IC50 (µM) | ||
|---|---|---|---|---|---|---|---|---|---|
| (−)-P | (+)-P | (R)-P | |||||||
| TQEH 196 | M | 59 | Liver | T4 | Metastasis | MSS | 3.23 | 3.82 | 4.76 |
| TQEH 198 | M | 68 | Liver | T4 | Metastasis | MSS | 1.36 | 2.87 | 3.78 |
| SAH01 | F | 50 | Lung | T4 | Metastasis | MSS | 1.23 | 1.36 | 1.95 |
| RAH038 | F | 87 | Colon (Ascending) | T3 | Primary | MSI (Absent MLH1 and PMS2) | 4.38 | 4.48 | 3.93 |
| RAH057 | F | 65 | Colon (Sigmoid) | T3 | Primary | MSS | 2.62 | 2.81 | 3.28 |
| RAP05 | M | 71 | Colon (Caecum) | T3 | Primary | MSS | 3.36 | 3.25 | 4.93 |
| RAH51 | F | 82 | Colon (Hepatic Flexure) | T4 | Primary | MSI (Absent MLH1 and PMS2) | 1.91 | 2.07 | 2.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhakal, B.; Li, C.M.Y.; Li, R.; Yeo, K.; Wright, J.A.; Gieniec, K.A.; Vrbanac, L.; Sammour, T.; Lawrence, M.; Thomas, M.; et al. The Antianginal Drug Perhexiline Displays Cytotoxicity against Colorectal Cancer Cells In Vitro: A Potential for Drug Repurposing. Cancers 2022, 14, 1043. https://doi.org/10.3390/cancers14041043
Dhakal B, Li CMY, Li R, Yeo K, Wright JA, Gieniec KA, Vrbanac L, Sammour T, Lawrence M, Thomas M, et al. The Antianginal Drug Perhexiline Displays Cytotoxicity against Colorectal Cancer Cells In Vitro: A Potential for Drug Repurposing. Cancers. 2022; 14(4):1043. https://doi.org/10.3390/cancers14041043
Chicago/Turabian StyleDhakal, Bimala, Celine Man Ying Li, Runhao Li, Kenny Yeo, Josephine A. Wright, Krystyna A. Gieniec, Laura Vrbanac, Tarik Sammour, Matthew Lawrence, Michelle Thomas, and et al. 2022. "The Antianginal Drug Perhexiline Displays Cytotoxicity against Colorectal Cancer Cells In Vitro: A Potential for Drug Repurposing" Cancers 14, no. 4: 1043. https://doi.org/10.3390/cancers14041043
APA StyleDhakal, B., Li, C. M. Y., Li, R., Yeo, K., Wright, J. A., Gieniec, K. A., Vrbanac, L., Sammour, T., Lawrence, M., Thomas, M., Lewis, M., Perry, J., Worthley, D. L., Woods, S. L., Drew, P., Sallustio, B. C., Smith, E., Horowitz, J. D., Maddern, G. J., ... Fenix, K. (2022). The Antianginal Drug Perhexiline Displays Cytotoxicity against Colorectal Cancer Cells In Vitro: A Potential for Drug Repurposing. Cancers, 14(4), 1043. https://doi.org/10.3390/cancers14041043







