Diverse Roles and Targets of miRNA in the Pathogenesis of Testicular Germ Cell Tumour
Abstract
:Simple Summary
Abstract
1. Introduction
2. Male Germ Cell Development
3. TGCT Development
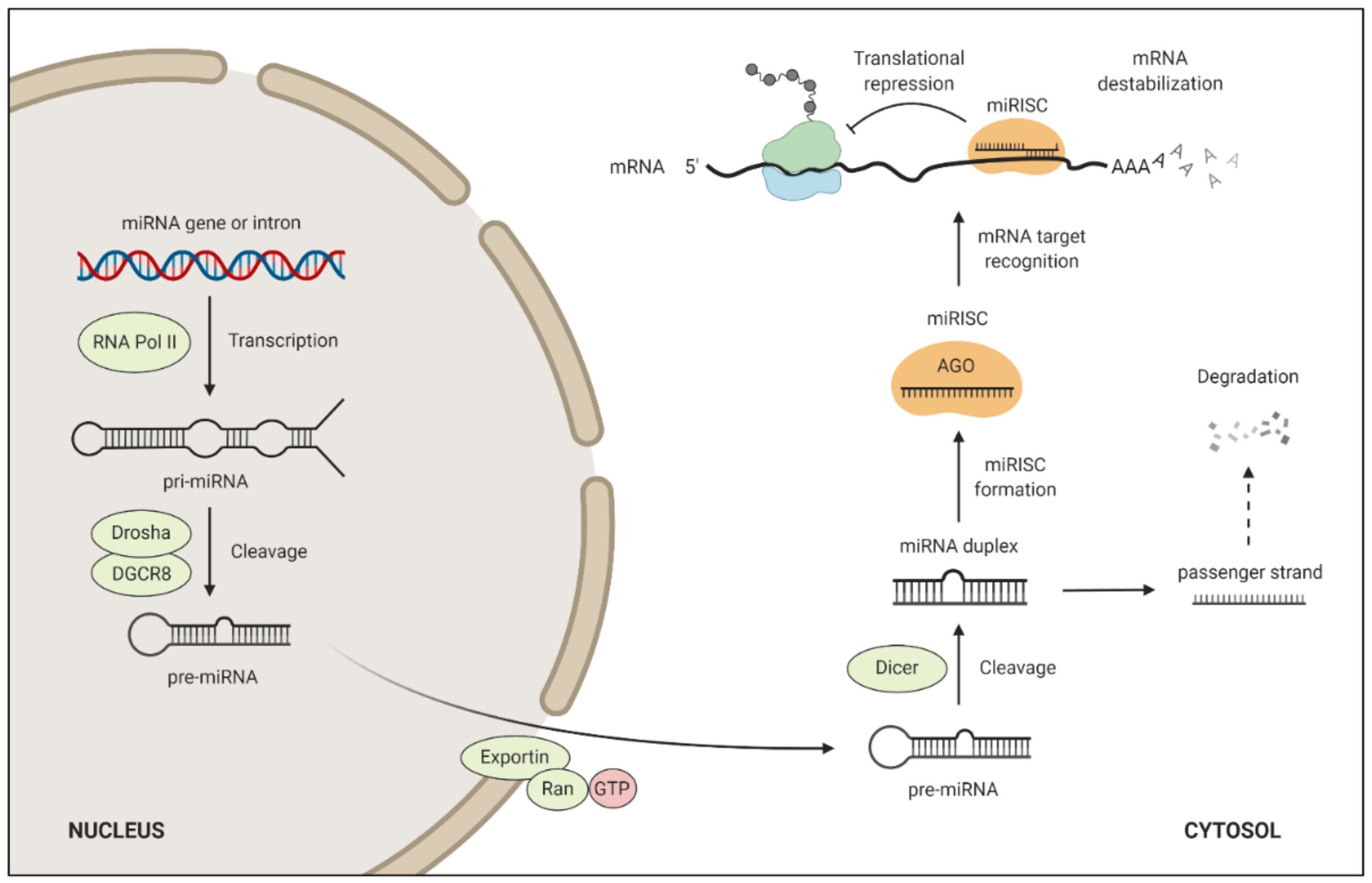
4. miRNA Target Regulation and Function
5. miRNAs in TGCTs
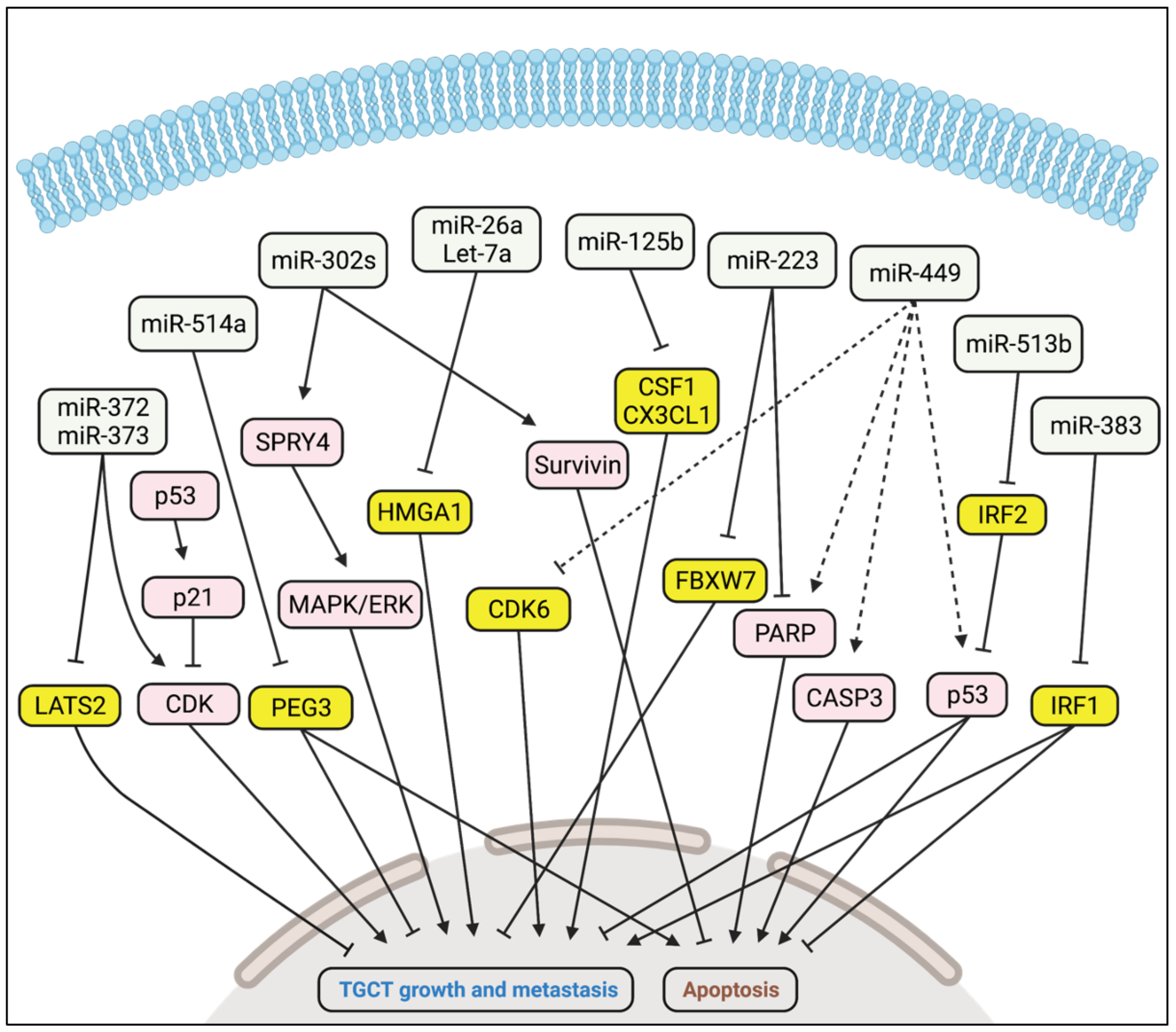
5.1. Oncogenic Activity
5.2. Tumour Suppressor Activity
| miRNA | Target | Function in TGCT | Reference |
|---|---|---|---|
| miR-372 miR-373 | LATS2 | Promote TGCT growth and metastasis | [62] |
| miR-302a miR-302b miR-302c | Unknown | Promote TGCT growth and metastasis via SPRY4 and MAPK/ERK | [63] |
| miR-223 | FBXW7 | Promotes TGCT growth | [67] |
| miR-449 | CDK6 | Inhibits TGCT growth | [69] |
| miR-26a Let-7a | HMGA1 | Inhibit TGCT growth | [70] |
| miR-125b | CSF1, CX3CL1 | Inhibits TGCT growth | [71] |
| miR-514a | PEG3 | Inhibits TGCT growth | [72] |
| miR-383 | IRF1 | Inhibits TGCT growth | [73] |
| miR-513b | IRF2 | Inhibits TGCT growth | [75] |
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| EC | Embryonal carcinoma |
| GCNIS | Germ cell neoplasia in situ |
| miRNA | MicroRNA |
| miRISC | miRNA-induced silencing complex |
| PGC | Primordial germ cell |
| pri-miRNA | Primary miRNA transcript |
| TGCT | Testicular germ cell tumour |
References
- Siegel, R.; Ward, E.; Brawley, O.; Jemal, A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011, 61, 212–236. [Google Scholar] [CrossRef] [PubMed]
- Shanmugalingam, T.; Soultati, A.; Chowdhury, S.; Rudman, S.; Van Hemelrijck, M. Global incidence and outcome of testicular cancer. Clin. Epidemiol. 2013, 5, 417–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Leeuwen, M.; Kieffer, J.M.; Efficace, F.; Fossa, S.D.; Bolla, M.; Collette, L.; Colombel, M.; De Giorgi, U.; Holzner, B.; van de Poll-Franse, L.V.; et al. International evaluation of the psychometrics of health-related quality of life questionnaires for use among long-term survivors of testicular and prostate cancer. Health Qual. Life Outcomes 2017, 15, 97. [Google Scholar] [CrossRef] [Green Version]
- Rajpert-De Meyts, E. Developmental model for the pathogenesis of testicular carcinoma in situ: Genetic and environmental aspects. Hum. Reprod. Update 2006, 12, 303–323. [Google Scholar] [CrossRef] [Green Version]
- Hemminki, K.; Rawal, R.; Chen, B.; Bermejo, J.L. Genetic epidemiology of cancer: From families to heritable genes. Int. J. Cancer 2004, 111, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Holroyd, A.; Lloyd, A.; Broderick, P.; Nsengimana, J.; Eeles, R.; Easton, D.F.; Dudakia, D.; Bishop, D.T.; Reid, A.; et al. Identification of four new susceptibility loci for testicular germ cell tumour. Nat. Commun. 2015, 6, 8690. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Loveday, C.; Levy, M.; Dudakia, D.; Rapley, E.; Nsengimana, J.; Bishop, D.T.; Reid, A.; Huddart, R.; Broderick, P.; et al. Large-scale Sequencing of Testicular Germ Cell Tumour (TGCT) Cases Excludes Major TGCT Predisposition Gene. Eur. Urol. 2018, 73, 828–831. [Google Scholar] [CrossRef]
- Pluta, J.; Pyle, L.C.; Nead, K.T.; Wilf, R.; Li, M.; Mitra, N.; Weathers, B.; D’Andrea, K.; Almstrup, K.; Anson-Cartwright, L.; et al. Identification of 22 susceptibility loci associated with testicular germ cell tumors. Nat. Commun. 2021, 12, 4487. [Google Scholar] [CrossRef] [PubMed]
- Das, M.K.; Kleppa, L.; Haugen, T.B. Functions of genes related to testicular germ cell tumour development. Andrology 2019, 7, 527–535. [Google Scholar] [CrossRef]
- Litchfield, K.; Levy, M.; Huddart, R.A.; Shipley, J.; Turnbull, C. The genomic landscape of testicular germ cell tumours: From susceptibility to treatment. Nat. Rev. Urol. 2016, 13, 409–419. [Google Scholar] [CrossRef]
- Litchfield, K.; Levy, M.; Orlando, G.; Loveday, C.; Law, P.J.; Migliorini, G.; Holroyd, A.; Broderick, P.; Karlsson, R.; Haugen, T.B.; et al. Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor. Nat. Genet. 2017, 49, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; McGlynn, K.A.; Rajpert-De Meyts, E.; Bishop, D.T.; Chung, C.C.; Dalgaard, M.D.; Greene, M.H.; Gupta, R.; Grotmol, T.; Haugen, T.B.; et al. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat. Genet. 2017, 49, 1141–1147. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Sanchez, A.; Amatruda, J.F. Zebrafish Germ Cell Tumors. Adv. Exp. Med. Biol. 2016, 916, 479–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, L.C.; Little, C.C. Spontaneous Testicular Teratomas in an Inbred Strain of Mice. Proc. Natl. Acad. Sci. USA 1954, 40, 1080–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lennartsson, J.; Ronnstrand, L. Stem cell factor receptor/c-Kit: From basic science to clinical implications. Physiol. Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef] [Green Version]
- Gondos, B.; Hobel, C.J. Ultrastructure of germ cell development in the human fetal testis. Z. Zellforsch. Mikrosk. Anat. 1971, 119, 1–20. [Google Scholar] [CrossRef]
- Stukenborg, J.B.; Colon, E.; Soder, O. Ontogenesis of testis development and function in humans. Sex. Dev. 2010, 4, 199–212. [Google Scholar] [CrossRef]
- Ehmcke, J.; Wistuba, J.; Schlatt, S. Spermatogonial stem cells: Questions, models and perspectives. Hum. Reprod. Update 2006, 12, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Clermont, Y. Kinetics of spermatogenesis in mammals: Seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev. 1972, 52, 198–236. [Google Scholar] [CrossRef]
- Amann, R.P. The cycle of the seminiferous epithelium in humans: A need to revisit? J. Androl. 2008, 29, 469–487. [Google Scholar] [CrossRef] [PubMed]
- Stukenborg, J.B.; Kjartansdottir, K.R.; Reda, A.; Colon, E.; Albersmeier, J.P.; Soder, O. Male germ cell development in humans. Horm. Res. Paediatr. 2014, 81, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Xiaoxia, L.; Jiaozi, F.; Shi, Y.; Yuming, Z.; Lihong, G. Clinical applications of stem cells from human exfoliated deciduous teeth in stem cell therapy. Hua Xi Kou Qiang Yi Xue Za Zhi 2017, 35, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Poon, J.; Wessel, G.M.; Yajima, M. An unregulated regulator: Vasa expression in the development of somatic cells and in tumorigenesis. Dev. Biol. 2016, 415, 24–32. [Google Scholar] [CrossRef]
- Kvist, K.; Clasen-Linde, E.; Langballe, O.; Hansen, S.H.; Cortes, D.; Thorup, J. The Expression of Markers for Intratubular Germ Cell Neoplasia in Normal Infantile Testes. Front. Endocrinol. 2018, 9, 286. [Google Scholar] [CrossRef]
- Baroni, T.; Arato, I.; Mancuso, F.; Calafiore, R.; Luca, G. On the Origin of Testicular Germ Cell Tumors: From Gonocytes to Testicular Cancer. Front. Endocrinol. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef] [Green Version]
- Fendler, A.; Stephan, C.; Yousef, G.M.; Kristiansen, G.; Jung, K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat. Rev. Urol. 2016, 13, 734–752. [Google Scholar] [CrossRef]
- Tang, F.; Kaneda, M.; O’Carroll, D.; Hajkova, P.; Barton, S.C.; Sun, Y.A.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007, 21, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.; Chuva de Sousa Lopes, S.M.; Kaneda, M.; Tang, F.; Hajkova, P.; Lao, K.; O’Carroll, D.; Das, P.P.; Tarakhovsky, A.; Miska, E.A.; et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 2008, 3, e1738. [Google Scholar] [CrossRef] [Green Version]
- Bouhallier, F.; Allioli, N.; Lavial, F.; Chalmel, F.; Perrard, M.H.; Durand, P.; Samarut, J.; Pain, B.; Rouault, J.P. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 2010, 16, 720–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Xu, C. Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction 2015, 149, R127–R137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, Y.; Meikar, O.; Papaioannou, M.D.; Conne, B.; Grey, C.; Weier, M.; Pralong, F.; De Massy, B.; Kaessmann, H.; Vassalli, J.D.; et al. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS ONE 2011, 6, e25241. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Song, R.; Ortogero, N.; Zheng, H.; Evanoff, R.; Small, C.L.; Griswold, M.D.; Namekawa, S.H.; Royo, H.; Turner, J.M.; et al. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J. Biol. Chem. 2012, 287, 25173–25190; discussion 172–163. [Google Scholar] [CrossRef] [Green Version]
- Looijenga, L.H.; Zafarana, G.; Grygalewicz, B.; Summersgill, B.; Debiec-Rychter, M.; Veltman, J.; Schoenmakers, E.F.; Rodriguez, S.; Jafer, O.; Clark, J.; et al. Role of gain of 12p in germ cell tumour development. APMIS 2003, 111, 161–171; discussion 172–163. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Summersgill, B.; Osin, P.; Lu, Y.J.; Huddart, R.; Shipley, J. Chromosomal imbalances associated with carcinoma in situ and associated testicular germ cell tumours of adolescents and adults. Br. J. Cancer 2001, 85, 213–220. [Google Scholar] [CrossRef]
- Atkin, N.B.; Baker, M.C. Specific chromosome change, i(12p), in testicular tumours? Lancet 1982, 2, 1349. [Google Scholar] [CrossRef]
- Skakkebaek, N.E.; Berthelsen, J.G. Carcinoma-in-situ of the testis and invasive growth of different types of germ cell tumours. A revised germ cell theory. Int. J. Androl. 1981, 4 (Suppl. S4), 26–33. [Google Scholar] [CrossRef]
- Rajpert-De Meyts, E.; Kvist, M.; Skakkebaek, N.E. Heterogeneity of expression of immunohistochemical tumour markers in testicular carcinoma in situ: Pathogenetic relevance. Virchows Arch. 1996, 428, 133–139. [Google Scholar] [CrossRef]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef] [PubMed]
- De Graaff, W.E.; Oosterhuis, J.W.; de Jong, B.; Dam, A.; van Putten, W.L.; Castedo, S.M.; Sleijfer, D.T.; Schraffordt Koops, H. Ploidy of testicular carcinoma in situ. Lab. Investig. 1992, 66, 166–168. [Google Scholar] [PubMed]
- De Jong, B.; Oosterhuis, J.W.; Castedo, S.M.; Vos, A.; te Meerman, G.J. Pathogenesis of adult testicular germ cell tumors. A cytogenetic model. Cancer Genet. Cytogenet. 1990, 48, 143–167. [Google Scholar] [CrossRef]
- Srigley, J.R.; Mackay, B.; Toth, P.; Ayala, A. The ultrastructure and histogenesis of male germ neoplasia with emphasis on seminoma with early carcinomatous features. Ultrastruct. Pathol. 1988, 12, 67–86. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Fu, G.; Brkic, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Chong, M.M.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, S.; Mannsperger, H.; Zhang, J.D.; Horvat, E.A.; Schmidt, C.; Kublbeck, M.; Henjes, F.; Ward, A.; Tschulena, U.; Zweig, K.; et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012, 8, 570. [Google Scholar] [CrossRef]
- Mestdagh, P.; Bostrom, A.K.; Impens, F.; Fredlund, E.; Van Peer, G.; De Antonellis, P.; von Stedingk, K.; Ghesquiere, B.; Schulte, S.; Dews, M.; et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol. Cell 2010, 40, 762–773. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Gillis, A.J.; Stoop, H.J.; Hersmus, R.; Oosterhuis, J.W.; Sun, Y.; Chen, C.; Guenther, S.; Sherlock, J.; Veltman, I.; Baeten, J.; et al. High-throughput microRNAome analysis in human germ cell tumours. J. Pathol. 2007, 213, 319–328. [Google Scholar] [CrossRef]
- Murray, M.J.; Saini, H.K.; van Dongen, S.; Palmer, R.D.; Muralidhar, B.; Pett, M.R.; Piipari, M.; Thornton, C.M.; Nicholson, J.C.; Enright, A.J.; et al. The two most common histological subtypes of malignant germ cell tumour are distinguished by global microRNA profiles, associated with differential transcription factor expression. Mol. Cancer 2010, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Palmer, R.D.; Murray, M.J.; Saini, H.K.; van Dongen, S.; Abreu-Goodger, C.; Muralidhar, B.; Pett, M.R.; Thornton, C.M.; Nicholson, J.C.; Enright, A.J.; et al. Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010, 70, 2911–2923. [Google Scholar] [CrossRef] [Green Version]
- Rounge, T.B.; Furu, K.; Skotheim, R.I.; Haugen, T.B.; Grotmol, T.; Enerly, E. Profiling of the small RNA populations in human testicular germ cell tumors shows global loss of piRNAs. Mol. Cancer 2015, 14, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, M.K.; Evensen, H.S.F.; Furu, K.; Haugen, T.B. miRNA-302s may act as oncogenes in human testicular germ cell tumours. Sci. Rep. 2019, 9, 9189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, M.K.; Furu, K.; Evensen, H.F.; Haugen, O.P.; Haugen, T.B. Knockdown of SPRY4 and SPRY4-IT1 inhibits cell growth and phosphorylation of Akt in human testicular germ cell tumours. Sci. Rep. 2018, 8, 2462. [Google Scholar] [CrossRef] [Green Version]
- Altieri, D.C. Targeting survivin in cancer. Cancer Lett. 2013, 332, 225–228. [Google Scholar] [CrossRef] [Green Version]
- Weikert, S.; Schrader, M.; Krause, H.; Schulze, W.; Muller, M.; Miller, K. The inhibitor of apoptosis (IAP) survivin is expressed in human testicular germ cell tumors and normal testes. Cancer Lett. 2005, 223, 331–337. [Google Scholar] [CrossRef]
- Liu, J.; Shi, H.; Li, X.; Chen, G.; Larsson, C.; Lui, W.O. miR2233p regulates cell growth and apoptosis via FBXW7 suggesting an oncogenic role in human testicular germ cell tumors. Int. J. Oncol. 2017, 50, 356–364. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, K.; Nihira, N.T.; Inuzuka, H.; Wei, W. Physiological functions of FBW7 in cancer and metabolism. Cell Signal. 2018, 46, 15–22. [Google Scholar] [CrossRef]
- Lize, M.; Pilarski, S.; Dobbelstein, M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010, 17, 452–458. [Google Scholar] [CrossRef] [Green Version]
- De Martino, M.; Esposito, F.; Pellecchia, S.; Cortez Cardoso Penha, R.; Botti, G.; Fusco, A.; Chieffi, P. HMGA1-Regulating microRNAs Let-7a and miR-26a are Downregulated in Human Seminomas. Int. J. Mol. Sci. 2020, 21, 3014. [Google Scholar] [CrossRef]
- Batool, A.; Wang, Y.Q.; Hao, X.X.; Chen, S.R.; Liu, Y.X. A miR-125b/CSF1-CX3CL1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 2018, 9, 962. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Li, X.; Lee, L.; Liu, J.; Warsito, D.; Hajeri, P.; Hultman, I.; Fotouhi, O.; Marklund, S.; Ahrlund-Richter, L.; et al. Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death Dis. 2017, 8, e2759. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Tian, H.; Liu, L.; Zhang, X.S.; Li, W.Q.; Deng, Y.M.; Yao, G.D.; Yin, M.M.; Sun, F. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis. 2010, 1, e94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Tian, H.; Cao, Y.X.; He, X.; Chen, L.; Song, X.; Ping, P.; Huang, H.; Sun, F. Downregulation of miR-320a/383-sponge-like long non-coding RNA NLC1-C (narcolepsy candidate-region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death Dis. 2015, 6, e1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, X.; Wang, G.; Wang, L.; Lin, Y.; Sun, F. Hsa-miR-513b-5p suppresses cell proliferation and promotes P53 expression by targeting IRF2 in testicular embryonal carcinoma cells. Gene 2017, 626, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Flor, I.; Spiekermann, M.; Loning, T.; Dieckmann, K.P.; Belge, G.; Bullerdiek, J. Expression of microRNAs of C19MC in Different Histological Types of Testicular Germ Cell Tumour. Cancer Genomics Proteomics 2016, 13, 281–289. [Google Scholar]
- Port, M.; Glaesener, S.; Ruf, C.; Riecke, A.; Bokemeyer, C.; Meineke, V.; Honecker, F.; Abend, M. Micro-RNA expression in cisplatin resistant germ cell tumor cell lines. Mol. Cancer 2011, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Barchi, M.; Bielli, P.; Dolci, S.; Rossi, P.; Grimaldi, P. Non-Coding RNAs and Splicing Activity in Testicular Germ Cell Tumors. Life 2021, 11, 736. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, M.K.; Haugen, Ø.P.; Haugen, T.B. Diverse Roles and Targets of miRNA in the Pathogenesis of Testicular Germ Cell Tumour. Cancers 2022, 14, 1190. https://doi.org/10.3390/cancers14051190
Das MK, Haugen ØP, Haugen TB. Diverse Roles and Targets of miRNA in the Pathogenesis of Testicular Germ Cell Tumour. Cancers. 2022; 14(5):1190. https://doi.org/10.3390/cancers14051190
Chicago/Turabian StyleDas, Mrinal K., Øyvind P. Haugen, and Trine B. Haugen. 2022. "Diverse Roles and Targets of miRNA in the Pathogenesis of Testicular Germ Cell Tumour" Cancers 14, no. 5: 1190. https://doi.org/10.3390/cancers14051190
APA StyleDas, M. K., Haugen, Ø. P., & Haugen, T. B. (2022). Diverse Roles and Targets of miRNA in the Pathogenesis of Testicular Germ Cell Tumour. Cancers, 14(5), 1190. https://doi.org/10.3390/cancers14051190






