Overview on Molecular Biomarkers for Laryngeal Cancer: Looking for New Answers to an Old Problem
Abstract
Simple Summary
Abstract
1. Introduction
2. Histopathological Features of LSCC
3. Molecular Markers for Clinical Management of LSCC
3.1. Mutated Genes and Abnormal Protein Expression in LSCC
3.2. miRNA and lncRNA Signatures as Predictors of LSCC Spreading
3.2.1. miRNAs as Tissue Biomarkers
3.2.2. miRNAs as Circulating Biomarkers
3.2.3. lncRNAs as Tissue Biomarkers
3.2.4. lncRNAs as Circulating Biomarkers
3.3. Circulating Tumor DNA and Circulating Tumor Cells: Prospective Search to Improve the Clinical Management of LSCC Patients
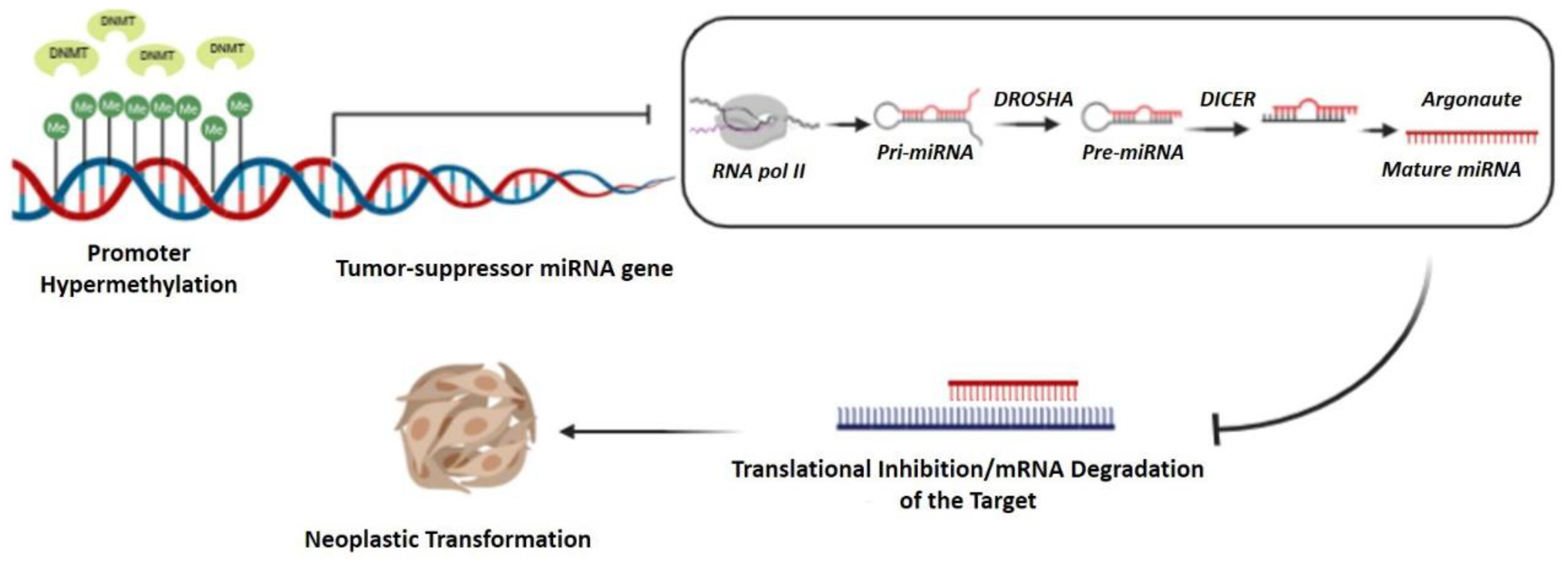
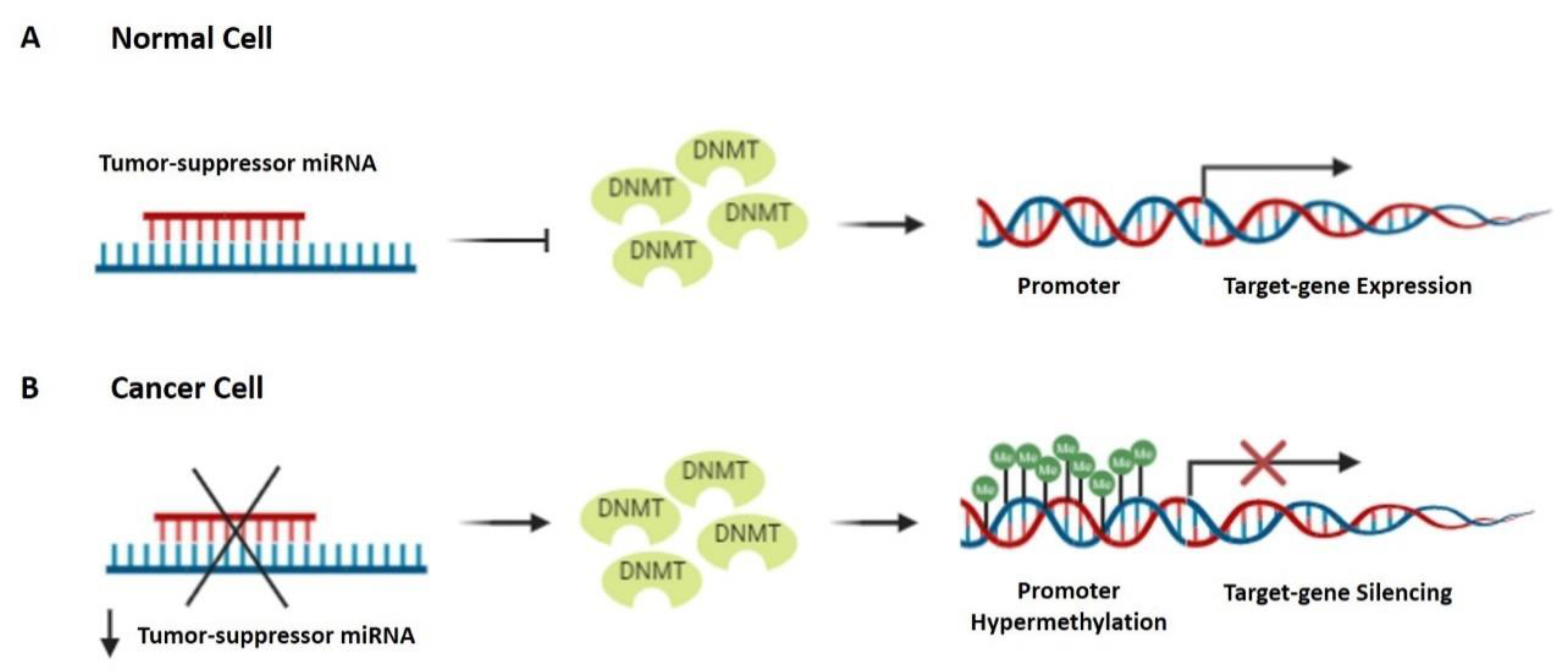
3.4. Major Epigenetic Changes as a Molecular Signature of LSCC
4. Molecular Markers of Drug Resistance in LSCC
5. Microenvironment-Derived Inflammatory Markers and Immune-Related Factors in LSCC
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Piotrowski, I.; Zhu, X.; Saccon, T.D.; Ashiqueali, S.; Schneider, A.; de Carvalho Nunes, A.D.; Noureddine, S.; Sobecka, A.; Barczak, W.; Szewczyk, M.; et al. MiRNAs as Biomarkers for Diagnosing and Predicting Survival of Head and Neck Squamous Cell Carcinoma Patients. Cancers 2021, 13, 3980. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cancer of the Larynx—Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/laryn.html (accessed on 19 January 2022).
- Kreimer, A.R.; Clifford, G.M.; Boyle, P.; Franceschi, S. Human Papillomavirus Types in Head and Neck Squamous Cell Carcinomas Worldwide: A Systematic Review. Cancer Epidemiol. Prev. Biomark. 2005, 14, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Orell-Kotikangas, H.; Österlund, P.; Mäkitie, O.; Saarilahti, K.; Ravasco, P.; Schwab, U.; Mäkitie, A.A. Cachexia at Diagnosis Is Associated with Poor Survival in Head and Neck Cancer Patients. Acta OtoLaryngol. 2017, 137, 778–785. [Google Scholar] [CrossRef]
- O’Neill, J.P.; Shaha, A.R. Nutrition Management of Patients with Malignancies of the Head and Neck. Surg. Clin. N. Am. 2011, 91, 631–639. [Google Scholar] [CrossRef]
- Santos, M.; Monteiro, E. Time between Diagnosis and Treatment of Hypopharynx and Larynx Cancer: Are Longer Delays Associated with Higher Discrepancy between Clinical and Pathological Staging? Int. Arch. Otorhinolaryngol. 2021, 25, e108–e114. [Google Scholar] [CrossRef]
- Baird, B.J.; Sung, C.K.; Beadle, B.M.; Divi, V. Treatment of Early-Stage Laryngeal Cancer: A Comparison of Treatment Options. Oral Oncol. 2018, 87, 8–16. [Google Scholar] [CrossRef]
- García Lorenzo, J.; Montoro Martínez, V.; Rigo Quera, A.; Codina Aroca, A.; López Vilas, M.; Quer Agustí, M.; León Vintró, X. Modifications in the Treatment of Advanced Laryngeal Cancer throughout the Last 30 Years. Eur. Arch. OtoRhinoLaryngol. 2017, 274, 3449–3455. [Google Scholar] [CrossRef]
- Forastiere, A.A.; Ismaila, N.; Lewin, J.S.; Nathan, C.A.; Adelstein, D.J.; Eisbruch, A.; Fass, G.; Fisher, S.G.; Laurie, S.A.; Le, Q.-T.; et al. Use of Larynx-Preservation Strategies in the Treatment of Laryngeal Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 1143–1169. [Google Scholar] [CrossRef]
- Singh, A.; Qayyumi, B.; Chaturvedi, P. An Update on Surgical Margins in the Head Neck Squamous Cell Carcinoma: Assessment, Clinical Outcome, and Future Directions. Curr. Oncol. Rep. 2020, 22, 82. [Google Scholar] [CrossRef]
- Beibei, Y.; Rong, Y.; Yunfei, Y.; Wenchao, Z. Research Progress Regarding Surgical Margins, Molecular Margins, and Prognosis of Laryngeal Carcinoma. Ear Nose Throat J. 2021, 100, 597–603. [Google Scholar] [CrossRef]
- Holliday, E.B.; Smith, B.D.; Gross, N.D.; Fuller, C.D.; Rosenthal, D.I. Larynx Cancer. In The American Cancer Society’s Oncology in Practice; The American Cancer Society, Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 201–210. ISBN 978-1-118-59216-8. [Google Scholar]
- Nocini, R.; Molteni, G.; Mattiuzzi, C.; Lippi, G. Updates on Larynx Cancer Epidemiology. Chin. J. Cancer Res. 2020, 32, 18–25. [Google Scholar] [CrossRef]
- Pezzuto, F.; Buonaguro, L.; Caponigro, F.; Ionna, F.; Starita, N.; Annunziata, C.; Buonaguro, F.M.; Tornesello, M.L. Update on Head and Neck Cancer: Current Knowledge on Epidemiology, Risk Factors, Molecular Features and Novel Therapies. Oncology 2015, 89, 125–136. [Google Scholar] [CrossRef]
- Chen, X.; Gao, L.; Sturgis, E.M.; Liang, Z.; Zhu, Y.; Xia, X.; Zhu, X.; Chen, X.; Li, G.; Gao, Z. HPV16 DNA and Integration in Normal and Malignant Epithelium: Implications for the Etiology of Laryngeal Squamous Cell Carcinoma. Ann. Oncol. 2017, 28, 1105–1110. [Google Scholar] [CrossRef]
- Lifsics, A.; Groma, V.; Cistjakovs, M.; Skuja, S.; Deksnis, R.; Murovska, M. Identification of High-Risk Human Papillomavirus DNA, P16, and E6/E7 Oncoproteins in Laryngeal and Hypopharyngeal Squamous Cell Carcinomas. Viruses 2021, 13, 1008. [Google Scholar] [CrossRef]
- Thompson, L.D.R. Laryngeal Dysplasia, Squamous Cell Carcinoma, and Variants. Surg. Pathol. Clin. 2017, 10, 15–33. [Google Scholar] [CrossRef]
- García, J.J.; Richardson, M.S. Common Lesions of the Larynx and Hypopharynx. Surg. Pathol. Clin. 2011, 4, 1153–1175. [Google Scholar] [CrossRef]
- Ciolofan, M.S.; Vlăescu, A.N. Clinical, Histological and Immunohistochemical Evaluation of Larynx Cancer. Curr. Health Sci. J. 2017, 43, 367–375. [Google Scholar] [CrossRef]
- Marioni, G.; Marchese-Ragona, R.; Cartei, G.; Marchese, F.; Staffieri, A. Current Opinion in Diagnosis and Treatment of Laryngeal Carcinoma. Cancer Treat. Rev. 2006, 32, 504–515. [Google Scholar] [CrossRef]
- Dispenza, F.; De Stefano, A.; Marchese, D.; Martines, F.; Dispenza, C. Management of Laryngeal Precancerous Lesions. Auris. Nasus. Larynx 2012, 39, 280–283. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primer 2020, 6, 92. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, H. MicroRNA-10a-5p and MicroRNA-34c-5p in Laryngeal Epithelial Premalignant Lesions: Differential Expression and Clinicopathological Correlation. Eur. Arch. OtoRhinoLaryngol. 2015, 272, 391–399. [Google Scholar] [CrossRef]
- Tuncturk, F.R.; Akalin, I.; Uzun, L.; Zenginkinet, T. Comparison of MiRNA Expressions among Benign, Premalignant and Malignant Lesions of the Larynx: Could They Be Transformation Biomarkers? J. Otolaryngol. Head Neck Surg. 2021, 50, 14. [Google Scholar] [CrossRef]
- Ying, X.; Kai, W.; Wei, G.; Chunming, Z.; Fuhui, H.; Shuxin, W.; Binquan, W. MicroRNA-106b Regulates the Tumor Suppressor RUNX3 in Laryngeal Carcinoma Cells. FEBS Lett. 2013, 587, 3166–3174. [Google Scholar] [CrossRef]
- Daquan, W.; Tian, W.; Shen, N.; Danzheng, L.; Xinsheng, H. Decrement of Prognostic Nutrition Index in Laryngeal Diseases: From Precancerous Lesion to Squamous Cell Carcinoma. Acta OtoLaryngol. 2021, 141, 1070–1074. [Google Scholar] [CrossRef]
- Lan, L.; Cao, H.; Chi, W.; Meng, W.; Zhao, L.; Cui, W.; Wang, B. Aberrant DNA Hyper-Methylation-Silenced LINC00886 Gene Accelerates Malignant Progression of Laryngeal Carcinoma. Pathol. Res. Pract. 2020, 216, 152877. [Google Scholar] [CrossRef]
- Manterola, L.; Aguirre, P.; Larrea, E.; Arestín, M.; Gaafar, A.; Elorriaga, K.; Goicoechea, I.; Armesto, M.; Fernández-Mercado, M.; Zabalza, I.; et al. Mutational Profiling Can Identify Laryngeal Dysplasia at Risk of Progression to Invasive Carcinoma. Sci. Rep. 2018, 8, 6613. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, Z.; Myers, J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell. Biochem. 2016, 117, 2682–2692. [Google Scholar] [CrossRef] [PubMed]
- Poeta, M.L.; Manola, J.; Goldwasser, M.A.; Forastiere, A.; Benoit, N.; Califano, J.A.; Ridge, J.A.; Goodwin, J.; Kenady, D.; Saunders, J.; et al. TP53 Mutations and Survival in Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2007, 357, 2552–2561. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Braakhuis, B.J.M.; Brakenhoff, R.H. The Molecular Biology of Head and Neck Cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Chrysovergis, A.; Papanikolaou, V.; Tsiambas, E.; Stavraka, C.; Ragos, V.; Peschos, D.; Psyrri, A.; Mastronikolis, N.; Kyrodimos, E. P53/MDM2 Co-Expression in Laryngeal Squamous Cell Carcinoma Based on Digital Image Analysis. Anticancer Res. 2019, 39, 4137–4142. [Google Scholar] [CrossRef]
- Osman, I.; Sherman, E.; Singh, B.; Venkatraman, E.; Zelefsky, M.; Bosl, G.; Scher, H.; Shah, J.; Shaha, A.; Kraus, D.; et al. Alteration of P53 Pathway in Squamous Cell Carcinoma of the Head and Neck: Impact on Treatment Outcome in Patients Treated with Larynx Preservation Intent. J. Clin. Oncol. 2002, 20, 2980–2987. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Liu, S.; Ouyang, X.; Liang, C. The relationship of p53 gene mutation to cell differentiation and metastasis of laryngeal squamous cell carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi Chin. J. Med. Genet. 2002, 19, 61–63. [Google Scholar]
- Pruneri, G.; Pignataro, L.; Manzotti, M.; Carboni, N.; Ronchetti, D.; Neri, A.; Cesana, B.M.; Viale, G. P63 in Laryngeal Squamous Cell Carcinoma: Evidence for a Role of TA-P63 down-Regulation in Tumorigenesis and Lack of Prognostic Implications of P63 Immunoreactivity. Lab. Investig. J. Tech. Methods Pathol. 2002, 82, 1327–1334. [Google Scholar] [CrossRef]
- Marcos, C.Á.; Alonso-Guervós, M.; Prado, N.R.; Gimeno, T.S.; Iglesias, F.D.; Hermsen, M.; Llorente, J.L. Genetic Model of Transformation and Neoplastic Progression in Laryngeal Epithelium. Head Neck 2011, 33, 216–224. [Google Scholar] [CrossRef]
- Pruneri, G.; Pignataro, L.; Carboni, N.; Ronchetti, D.; Cesana, B.M.; Ottaviani, A.; Neri, A.; Buffa, R. Clinical Relevance of P53 and Bcl-2 Protein over-Expression in Laryngeal Squamous-Cell Carcinoma. Int. J. Cancer 1998, 79, 263–268. [Google Scholar] [CrossRef]
- Giotakis, A.I.; Lazaris, A.C.; Kataki, A.; Kontos, C.K.; Giotakis, E.I. Positive BCL2L12 Expression Predicts Favorable Prognosis in Patients with Laryngeal Squamous Cell Carcinoma. Cancer Biomark. 2019, 25, 141–149. [Google Scholar] [CrossRef]
- Chrysovergis, A.; Papanikolaou, V.S.; Tsiambas, E.; Ragos, V.; Peschos, D.; Kyrodimos, E. Digital Analysis of BCL2 Expression in Laryngeal Squamous Cell Carcinoma. Anticancer Res. 2019, 39, 1253–1257. [Google Scholar] [CrossRef]
- Giotakis, A.I.; Kontos, C.K.; Manolopoulos, L.D.; Sismanis, A.; Konstadoulakis, M.M.; Scorilas, A. High BAX/BCL2 MRNA Ratio Predicts Favorable Prognosis in Laryngeal Squamous Cell Carcinoma, Particularly in Patients with Negative Lymph Nodes at the Time of Diagnosis. Clin. Biochem. 2016, 49, 890–896. [Google Scholar] [CrossRef]
- Jovanovic, I.P.; Radosavljevic, G.D.; Simovic-Markovic, B.J.; Stojanovic, S.P.; Stefanovic, S.M.; Pejnovic, N.N.; Arsenijevic, N.N. Clinical Significance of Cyclin D1, FGF3 and P21 Protein Expression in Laryngeal Squamous Cell Carcinoma. J. BUON Off. J. Balk. Union Oncol. 2014, 19, 944–952. [Google Scholar]
- Pruneri, G.; Pignataro, L.; Carboni, N.; Buffa, R.; Di Finizio, D.; Cesana, B.M.; Neri, A. Clinical Relevance of Expression of the CIP/KIP Cell-Cycle Inhibitors P21 and P27 in Laryngeal Cancer. J. Clin. Oncol. 1999, 17, 3150–3159. [Google Scholar] [CrossRef]
- Fan, G.K.; Fujieda, S.; Sunaga, H.; Tsuzuki, H.; Ito, N.; Saito, H. Expression of Protein P27 Is Associated with Progression and Prognosis in Laryngeal Cancer. Laryngoscope 1999, 109, 815–820. [Google Scholar] [CrossRef]
- Tamura, N.; Dong, Y.; Sui, L.; Tai, Y.; Sugimoto, K.; Nagahata, S.; Tokuda, M. Cyclin-Dependent Kinase Inhibitor P27 Is Related to Cell Proliferation and Prognosis in Laryngeal Squamous Cell Carcinomas. J. Laryngol. Otol. 2001, 115, 400–406. [Google Scholar] [CrossRef]
- Peschos, D.; Tsanou, E.; Stefanou, D.; Damala, C.; Vougiouklakis, T.; Mitselou, A.; Agnantis, N.J. Expression of Cyclin-Dependent Kinases Inhibitors P21(WAF1) and P27(KIP1) in Benign, Premalignant and Malignant Laryngeal Lesions. Correlation with Cell Cycle Regulatory Proteins. Vivo Athens Greece 2004, 18, 719–724. [Google Scholar]
- Politi, A.; Tsiambas, E.; Mastronikolis, N.S.; Peschos, D.; Asproudis, I.; Kyrodimos, E.; Armata, I.E.; Chrysovergis, A.; Asimakopoulos, A.; Papanikolaou, V.S.; et al. Combined EGFR/ALK Expression Analysis in Laryngeal Squamous Cell Carcinoma. Vivo Athens Greece 2019, 33, 815–819. [Google Scholar] [CrossRef]
- Cercelaru, L.; Stepan, A.E.; Mărgăritescu, C.; Osman, A.; Popa, I.-C.; Simionescu, C.E.; Mărgăritescu, O. EGFR Immunoexpression in Laryngeal Squamous Cell Carcinoma. Curr. Health Sci. J. 2017, 43, 340–344. [Google Scholar] [CrossRef]
- Lin, X.; Wen, G.; Wang, S.; Lu, H.; Li, C.; Wang, X. Expression and Role of EGFR, Cyclin D1 and KRAS in Laryngocarcinoma Tissues. Exp. Ther. Med. 2019, 17, 782–790. [Google Scholar] [CrossRef]
- Jung, A.R.; Jung, C.-H.; Noh, J.K.; Lee, Y.C.; Eun, Y.-G. Epithelial-Mesenchymal Transition Gene Signature Is Associated with Prognosis and Tumor Microenvironment in Head and Neck Squamous Cell Carcinoma. Sci. Rep. 2020, 10, 3652. [Google Scholar] [CrossRef] [PubMed]
- Larizadeh, M.H.; Damghani, M.A.; Tabrizchi, H.; Mirshekari, T.R. Expression of E-Cadherin in Squamous Cell Carcinoma of the Larynx and its Correlation with Clinicopathological Features. J. Med. Sci. 2009, 9, 41–45. [Google Scholar] [CrossRef][Green Version]
- Ahmed, R.A.; Shawky, A.E.-A.; Hamed, R.H. Prognostic Significance of Cyclin D1 and E-Cadherin Expression in Laryngeal Squamous Cell Carcinoma. Pathol. Oncol. Res. 2014, 20, 625–633. [Google Scholar] [CrossRef]
- Nardi, C.E.; Dedivitis, R.A.; de Almeida, R.C.; de Matos, L.L.; Cernea, C.R. The Role of E-Cadherin and β-Catenin in Laryngeal Cancer. Oncotarget 2018, 9, 30199–30209. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, G.-J.; Song, P.-P.; Zhou, H.; Shen, X.-H.; Wang, J.-G.; Ma, X.-F.; Gu, Y.-J.; Liu, D.-D.; Feng, A.-N.; Qian, X.-Y.; et al. Role of Epithelial-Mesenchymal Transition Markers E-Cadherin, N-Cadherin, β-Catenin and ZEB2 in Laryngeal Squamous Cell Carcinoma. Oncol. Lett. 2018, 15, 3472–3481. [Google Scholar] [CrossRef] [PubMed]
- Fanjul-Fernández, M.; Quesada, V.; Cabanillas, R.; Cadiñanos, J.; Fontanil, T.; Obaya, A.; Ramsay, A.J.; Llorente, J.L.; Astudillo, A.; Cal, S.; et al. Cell-Cell Adhesion Genes CTNNA2 and CTNNA3 Are Tumour Suppressors Frequently Mutated in Laryngeal Carcinomas. Nat. Commun. 2013, 4, 2531. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Gaykalova, D.A.; Ochs, M.F.; Mambo, E.; Arnaoutakis, D.; Liu, Y.; Loyo, M.; Agrawal, N.; Howard, J.; Li, R.; et al. Activation of the NOTCH Pathway in Head and Neck Cancer. Cancer Res. 2014, 74, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Fukusumi, T.; Califano, J.A. The NOTCH Pathway in Head and Neck Squamous Cell Carcinoma. J. Dent. Res. 2018, 97, 645–653. [Google Scholar] [CrossRef]
- Li, D.; Dong, P.; Wu, C.; Cao, P.; Zhou, L. Notch1 Overexpression Associates with Poor Prognosis in Human Laryngeal Squamous Cell Carcinoma. Ann. Otol. Rhinol. Laryngol. 2014, 123, 705–710. [Google Scholar] [CrossRef]
- Dai, M.-Y.; Fang, F.; Zou, Y.; Yi, X.; Ding, Y.-J.; Chen, C.; Tao, Z.-Z.; Chen, S.-M. Downregulation of Notch1 Induces Apoptosis and Inhibits Cell Proliferation and Metastasis in Laryngeal Squamous Cell Carcinoma. Oncol. Rep. 2015, 34, 3111–3119. [Google Scholar] [CrossRef]
- Zou, Y.; Fang, F.; Ding, Y.-J.; Dai, M.-Y.; Yi, X.; Chen, C.; Tao, Z.-Z.; Chen, S.-M. Notch 2 Signaling Contributes to Cell Growth, Anti-Apoptosis and Metastasis in Laryngeal Squamous Cell Carcinoma. Mol. Med. Rep. 2016, 14, 3517–3524. [Google Scholar] [CrossRef]
- Henning, S.; Cascorbi, I.; Münchow, B.; Jahnke, V.; Roots, I. Association of Arylamine N-Acetyltransferases NAT1 and NAT2 Genotypes to Laryngeal Cancer Risk. Pharmacogenetics 1999, 9, 103–112. [Google Scholar] [CrossRef]
- Varzim, G.; Monteiro, E.; Silva, R.; Pinheiro, C.; Lopes, C. Polymorphisms of Arylamine N-Acetyltransferase (NAT1 and NAT2) and Larynx Cancer Susceptibility. J. Oto-Rhino-Laryngol. Its Relat. Spec. 2002, 64, 206–212. [Google Scholar] [CrossRef]
- Mahjabeen, I.; Masood, N.; Baig, R.M.; Sabir, M.; Inayat, U.; Malik, F.A.; Kayani, M.A. Novel Mutations of OGG1 Base Excision Repair Pathway Gene in Laryngeal Cancer Patients. Fam. Cancer 2012, 11, 587–593. [Google Scholar] [CrossRef]
- Mastronikolis, N.S.; Tsiambas, E.; Papadas, T.A.; Karameris, A.; Ragos, V.; Peschos, D.; Mastronikolis, S.N.; Papadas, A.T.; Liatsos, C.; Armata, I.E.; et al. Deregulation of PTEN Expression in Laryngeal Squamous Cell Carcinoma Based on Tissue Microarray Digital Analysis. Anticancer Res. 2017, 37, 5521–5524. [Google Scholar] [CrossRef]
- Zhu, X.-L.; Wang, Z.-F.; Lei, W.-B.; Zhuang, H.-W.; Hou, W.-J.; Wen, Y.-H.; Wen, W.-P. Tumorigenesis Role and Clinical Significance of DJ-1, a Negative Regulator of PTEN, in Supraglottic Squamous Cell Carcinoma. J. Exp. Clin. Cancer Res. 2012, 31, 94. [Google Scholar] [CrossRef]
- Kim, R.H.; Peters, M.; Jang, Y.; Shi, W.; Pintilie, M.; Fletcher, G.C.; DeLuca, C.; Liepa, J.; Zhou, L.; Snow, B.; et al. DJ-1, a Novel Regulator of the Tumor Suppressor PTEN. Cancer Cell 2005, 7, 263–273. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, Z.; Liu, C.; Gong, Z.; Yang, Y.; Kang, H.; Li, Y.; Hu, G. TrkB Promotes Laryngeal Cancer Metastasis via Activation PI3K/AKT Pathway. Oncotarget 2017, 8, 108726–108737. [Google Scholar] [CrossRef]
- Yilmaz, T.; Jiffar, T.; de la Garza, G.; Lin, H.; Milas, Z.; Takahashi, Y.; Hanna, E.; MacIntyre, T.; Brown, J.L.; Myers, J.N.; et al. Theraputic Targeting of Trk Supresses Tumor Proliferation and Enhances Cisplatin Activity in HNSCC. Cancer Biol. Ther. 2010, 10, 644–653. [Google Scholar] [CrossRef]
- Reddy, K.B. MicroRNA (MiRNA) in Cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef]
- Yu, X.; Li, Z. The Role of MicroRNAs Expression in Laryngeal Cancer. Oncotarget 2015, 6, 23297–23305. [Google Scholar] [CrossRef]
- Bouyssou, J.M.C.; Manier, S.; Huynh, D.; Issa, S.; Roccaro, A.M.; Ghobrial, I.M. Regulation of MicroRNAs in Cancer Metastasis. Biochim. Biophys. Acta 2014, 1845, 255–265. [Google Scholar] [CrossRef]
- Chen, L.; Sun, D.-Z.; Fu, Y.-G.; Yang, P.-Z.; Lv, H.-Q.; Gao, Y.; Zhang, X.-Y. Upregulation of MicroRNA-141 Suppresses Epithelial-Mesenchymal Transition and Lymph Node Metastasis in Laryngeal Cancer through HOXC6-Dependent TGF-β Signaling Pathway. Cell. Signal. 2020, 66, 109444. [Google Scholar] [CrossRef]
- Gao, S.; Wang, J.; Xie, J.; Zhang, T.; Dong, P. Role of MiR-138 in the Regulation of Larynx Carcinoma Cell Metastases. Tumor Biology 2015, 37, 15601–15606. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, W.; Ji, W. MYO5A Inhibition by MiR-145 Acts as a Predictive Marker of Occult Neck Lymph Node Metastasis in Human Laryngeal Squamous Cell Carcinoma. OncoTargets Ther. 2018, 11, 3619–3635. [Google Scholar] [CrossRef]
- Tian, L.; Li, M.; Ge, J.; Guo, Y.; Sun, Y.; Liu, M.; Xiao, H. MiR-203 Is Downregulated in Laryngeal Squamous Cell Carcinoma and Can Suppress Proliferation and Induce Apoptosis of Tumours. Tumour Biol. 2014, 35, 5953–5963. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Wu, Y.; He, X.; Zhang, C.; Zhu, M.; Chen, B.; Liu, Q.; Qu, X.; Li, W.; Wen, S.; et al. MicroRNA-204-5p Inhibits Invasion and Metastasis of Laryngeal Squamous Cell Carcinoma by Suppressing Forkhead Box C1. J. Cancer 2017, 8, 2356–2368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cao, H. MicroRNA-143-3p Suppresses Cell Growth and Invasion in Laryngeal Squamous Cell Carcinoma via Targeting the K-Ras/Raf/MEK/ERK Signaling Pathway. Int. J. Oncol. 2019, 54, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tian, L.; Ren, H.; Chen, X.; Wang, Y.; Ge, J.; Wu, S.; Sun, Y.; Liu, M.; Xiao, H. MicroRNA-101 Is a Potential Prognostic Indicator of Laryngeal Squamous Cell Carcinoma and Modulates CDK8. J. Transl. Med. 2015, 13, 271. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, Y.-P.; Yang, D.; Zhang, G.; Zhou, H.-F. Clinical Significance of MiR-149 in the Survival of Patients with Laryngeal Squamous Cell Carcinoma. BioMed Res. Int. 2016, 2016, 8561251. [Google Scholar] [CrossRef]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A New Weapon against Cancer? Mol. Ther. Nucleic Acids 2014, 3, e194. [Google Scholar] [CrossRef]
- Shen, Z.; Zhan, G.; Ye, D.; Ren, Y.; Cheng, L.; Wu, Z.; Guo, J. MicroRNA-34a Affects the Occurrence of Laryngeal Squamous Cell Carcinoma by Targeting the Antiapoptotic Gene Survivin. Med. Oncol. Northwood Lond. Engl. 2012, 29, 2473–2480. [Google Scholar] [CrossRef]
- Shuang, Y.; Li, C.; Zhou, X.; Huang, Y.; Zhang, L. MicroRNA-195 Inhibits Growth and Inva-sion of Laryngeal Carcinoma Cells by Directly Targeting DCUN1D1. Oncol. Rep. 2017, 38, 2155–2165. [Google Scholar] [CrossRef]
- Li, J.Z.-H.; Gao, W.; Lei, W.-B.; Zhao, J.; Chan, J.Y.-W.; Wei, W.I.; Ho, W.-K.; Wong, T.-S. MicroRNA 744-3p Promotes MMP-9-Mediated Metastasis by Simultaneously Suppressing PDCD4 and PTEN in Laryngeal Squamous Cell Carcinoma. Oncotarget 2016, 7, 58218–58233. [Google Scholar] [CrossRef]
- Ren, J.; Zhu, D.; Liu, M.; Sun, Y.; Tian, L. Downregulation of MiR-21 Modulates Ras Expression to Promote Apoptosis and Suppress Invasion of Laryngeal Squamous Cell Carcinoma. Eur. J. Cancer 2010, 46, 3409–3416. [Google Scholar] [CrossRef]
- Li, M.; Tian, L.; Wang, L.; Yao, H.; Zhang, J.; Lu, J.; Sun, Y.; Gao, X.; Xiao, H.; Liu, M. Down-Regulation of MiR-129-5p Inhibits Growth and Induces Apoptosis in Laryngeal Squamous Cell Carcinoma by Targeting APC. PLoS ONE 2013, 8, e77829. [Google Scholar] [CrossRef]
- Ricciardiello, F.; Capasso, R.; Kawasaki, H.; Abate, T.; Oliva, F.; Lombardi, A.; Misso, G.; Ingrosso, D.; Leone, C.A.; Iengo, M.; et al. A MiRNA Signature Suggestive of Nodal Metastases from Laryngeal Carcinoma. Acta Otorhinolaryngol. Ital. 2017, 37, 467–474. [Google Scholar] [CrossRef]
- Kawasaki, H.; Takeuchi, T.; Ricciardiello, F.; Lombardi, A.; Biganzoli, E.; Fornili, M.; De Bortoli, D.; Mesolella, M.; Cossu, A.M.; Scrima, M.; et al. Definition of MiRNA Signatures of Nodal Metastasis in LCa: MiR-449a Targets Notch Genes and Suppresses Cell Migration and Invasion. Mol. Ther. Nucleic Acids 2020, 20, 711–724. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Cheng, G. Circulating MiRNAs: Roles in Cancer Diagnosis, Prognosis and Therapy. Adv. Drug Deliv. Rev. 2015, 81, 75–93. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined Detection of Serum Exosomal MiR-21 and HOTAIR as Diagnostic and Prognostic Biomarkers for Laryngeal Squamous Cell Carcinoma. Med. Oncol. Northwood Lond. Engl. 2014, 31, 148. [Google Scholar] [CrossRef]
- Wang, J.L.; Wang, X.; Yang, D.; Shi, W.J. The Expression of MicroRNA-155 in Plasma and Tissue Is Matched in Human Laryngeal Squamous Cell Carcinoma. Yonsei Med. J. 2016, 57, 298–305. [Google Scholar] [CrossRef]
- Cao, Y.-C.; Song, L.-Q.; Xu, W.-W.; Qi, J.-J.; Wang, X.-Y.; Su, Y. Serum MiR-632 Is a Potential Marker for the Diagnosis and Prognosis in Laryngeal Squamous Cell Carcinoma. Acta Oto-laryngol. 2020, 140, 418–421. [Google Scholar] [CrossRef]
- Cao, Z.; Zhao, K.; Jose, I.; Hoogenraad, N.J.; Osellame, L.D. Biomarkers for Cancer Cachexia: A Mini Review. Int. J. Mol. Sci. 2021, 22, 4501. [Google Scholar] [CrossRef]
- Powrózek, T.; Mlak, R.; Brzozowska, A.; Mazurek, M.; Gołębiowski, P.; Małecka-Massalska, T. MiRNA-130a Significantly Improves Accuracy of SGA Nutritional Assessment Tool in Prediction of Malnutrition and Cachexia in Radiotherapy-Treated Head and Neck Cancer Patients. Cancers 2018, 10, 294. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Qu, L.; Jin, M.; Yang, L.; Sun, C.; Wang, P.; Li, Y.; Tian, L.; Liu, M.; Sun, Y. Expression of Long Non-Coding RNA HOXA11-AS Is Correlated with Progression of Laryngeal Squamous Cell Carcinoma. Am. J. Transl. Res. 2018, 10, 573–580. [Google Scholar]
- Xu, Z.; Xi, K. LncRNA RGMB-AS1 Promotes Laryngeal Squamous Cell Carcinoma Cells Progression via Sponging MiR-22/NLRP3 Axis. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 118, 109222. [Google Scholar] [CrossRef]
- Yang, T.; Li, S.; Liu, J.; Yin, D.; Yang, X.; Tang, Q. LncRNA-NKILA/NF-ΚB Feedback Loop Modulates Laryngeal Cancer Cell Proliferation, Invasion, and Radioresistance. Cancer Med. 2018, 7, 2048–2063. [Google Scholar] [CrossRef]
- Wu, T.; Qu, L.; He, G.; Tian, L.; Li, L.; Zhou, H.; Jin, Q.; Ren, J.; Wang, Y.; Wang, J.; et al. Regulation of Laryngeal Squamous Cell Cancer Progression by the LncRNA H19/MiR-148a-3p/DNMT1 Axis. Oncotarget 2016, 7, 11553–11566. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Cao, S.; Han, X.; Wang, Z.; Zhao, X.; Liu, X.; Li, G.; Pan, X.; Lei, D. The Long Noncoding RNA TUG1 Promotes Laryngeal Cancer Proliferation and Migration. Cell. Physiol. Biochem. Cell. Physiol. Biochem. 2018, 49, 2511–2520. [Google Scholar] [CrossRef]
- Wang, P.; Wu, T.; Zhou, H.; Jin, Q.; He, G.; Yu, H.; Xuan, L.; Wang, X.; Tian, L.; Sun, Y.; et al. Long Noncoding RNA NEAT1 Promotes Laryngeal Squamous Cell Cancer through Regulating MiR-107/CDK6 Pathway. J. Exp. Clin. Cancer Res. 2016, 35, 22. [Google Scholar] [CrossRef]
- Qin, H.; Xu, J.; Gong, L.; Jiang, B.; Zhao, W. The Long Noncoding RNA ST7-AS1 Promotes Laryngeal Squamous Cell Carcinoma by Stabilizing CARM1. Biochem. Biophys. Res. Commun. 2019, 512, 34–40. [Google Scholar] [CrossRef]
- Xiao, D.; Cui, X.; Wang, X. Long Noncoding RNA XIST Increases the Aggressiveness of Laryngeal Squamous Cell Carcinoma by Regulating MiR-124-3p/EZH2. Exp. Cell Res. 2019, 381, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Gong, C.; Yuan, K. LncRNA UCA1 Promotes Cell Proliferation, Invasion and Migration of Laryngeal Squamous Cell Carcinoma Cells by Activating Wnt/β-Catenin Signaling Pathway. Exp. Ther. Med. 2019, 17, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Yang, Y.; Zha, D.; Yue, B.; Qiu, J.; Zhang, C. Overexpression of LncRNA SnaR Is Correlated with Progression and Predicts Poor Survival of Laryngeal Squamous Cell Carcinoma. J. Cell. Biochem. 2018, 120, 8492–8498. [Google Scholar] [CrossRef] [PubMed]
- Mäbert, K.; Cojoc, M.; Peitzsch, C.; Kurth, I.; Souchelnytskyi, S.; Dubrovska, A. Cancer Bi-omarker Discovery: Current Status and Future Perspectives. Int. J. Radiat. Biol. 2014, 90, 659–677. [Google Scholar] [CrossRef]
- Yap, T.A.; Lorente, D.; Omlin, A.; Olmos, D.; de Bono, J.S. Circulating Tumor Cells: A Multi-Functional Biomarker. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 2553–2568. [Google Scholar] [CrossRef]
- Yong, E. Cancer Biomarkers: Written in Blood. Nature 2014, 511, 524–526. [Google Scholar] [CrossRef]
- Chan, K.C.A.; Jiang, P.; Zheng, Y.W.L.; Liao, G.J.W.; Sun, H.; Wong, J.; Siu, S.S.N.; Chan, W.C.; Chan, S.L.; Chan, A.T.C.; et al. Cancer Genome Scanning in Plasma: Detection of Tumor-Associated Copy Number Aberrations, Single-Nucleotide Variants, and Tumoral Heterogeneity by Massively Parallel Sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef]
- Balgkouranidou, I.; Chimonidou, M.; Milaki, G.; Tsarouxa, E.G.; Kakolyris, S.; Welch, D.R.; Georgoulias, V.; Lianidou, E.S. Breast Cancer Metastasis Suppressor-1 Promoter Methylation in Cell-Free DNA Provides Prognostic Information in Non-Small Cell Lung Cancer. Br. J. Cancer 2014, 110, 2054–2062. [Google Scholar] [CrossRef]
- Sanchez-Cespedes, M.; Esteller, M.; Wu, L.; Nawroz-Danish, H.; Yoo, G.H.; Koch, W.M.; Jen, J.; Herman, J.G.; Sidransky, D. Gene Promoter Hypermethylation in Tumors and Serum of Head and Neck Cancer Patients. Cancer Res. 2000, 60, 892–895. [Google Scholar]
- Schröck, A.; Leisse, A.; de Vos, L.; Gevensleben, H.; Dröge, F.; Franzen, A.; Wachendörfer, M.; Schröck, F.; Ellinger, J.; Teschke, M.; et al. Free-Circulating Methylated DNA in Blood for Diagnosis, Staging, Prognosis, and Monitoring of Head and Neck Squamous Cell Carcinoma Patients: An Observational Prospective Cohort Study. Clin. Chem. 2017, 63, 1288–1296. [Google Scholar] [CrossRef]
- Chan, K.C.A.; Hung, E.C.W.; Woo, J.K.S.; Chan, P.K.S.; Leung, S.-F.; Lai, F.P.T.; Cheng, A.S.M.; Yeung, S.W.; Chan, Y.W.; Tsui, T.K.C.; et al. Early Detection of Nasopharyngeal Carcinoma by Plasma Epstein-Barr Virus DNA Analysis in a Surveillance Program. Cancer 2013, 119, 1838–1844. [Google Scholar] [CrossRef]
- Campitelli, M.; Jeannot, E.; Peter, M.; Lappartient, E.; Saada, S.; de la Rochefordière, A.; Fourchotte, V.; Alran, S.; Petrow, P.; Cottu, P.; et al. Human Papillomavirus Mutational Insertion: Specific Marker of Circulating Tumor DNA in Cervical Cancer Patients. PLoS ONE 2012, 7, e43393. [Google Scholar] [CrossRef]
- Stroun, M.; Lyautey, J.; Lederrey, C.; Olson-Sand, A.; Anker, P. About the Possible Origin and Mechanism of Circulating DNA Apoptosis and Active DNA Release. Clin. Chim. Acta Int. J. Clin. Chem. 2001, 313, 139–142. [Google Scholar] [CrossRef]
- van der Vaart, M.; Pretorius, P.J. The Origin of Circulating Free DNA. Clin. Chem. 2007, 53, 2215. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Payne, K.; Spruce, R.; Beggs, A.; Sharma, N.; Kong, A.; Martin, T.; Parmar, S.; Praveen, P.; Nankivell, P.; Mehanna, H. Circulating Tumor DNA as a Biomarker and Liquid Biopsy in Head and Neck Squamous Cell Carcinoma. Head Neck 2018, 40, 1598–1604. [Google Scholar] [CrossRef]
- Terry, S.; Savagner, P.; Ortiz-Cuaran, S.; Mahjoubi, L.; Saintigny, P.; Thiery, J.-P.; Chouaib, S. New Insights into the Role of EMT in Tumor Immune Escape. Mol. Oncol. 2017, 11, 824–846. [Google Scholar] [CrossRef]
- Mazurek, A.M.; Rutkowski, T.; Fiszer-Kierzkowska, A.; Małusecka, E.; Składowski, K. Assessment of the Total CfDNA and HPV16/18 Detection in Plasma Samples of Head and Neck Squamous Cell Carcinoma Patients. Oral Oncol. 2016, 54, 36–41. [Google Scholar] [CrossRef]
- Khandelwal, A.R.; Greer, A.H.; Hamiter, M.; Fermin, J.M.; McMullen, T.; Moore-Medlin, T.; Mills, G.; Flores, J.M.; Yin, H.; Nathan, C.-A.O. Comparing Cell-Free Circulating Tumor DNA Mutational Profiles of Disease-Free and Nonresponders Patients with Oropharyngeal Squamous Cell Carcinoma. Laryngoscope Investig. Otolaryngol. 2020, 5, 868–878. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of Somatic Mutations and HPV in the Saliva and Plasma of Patients with Head and Neck Squamous Cell Carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- McMullen, H.; Greer, A.; Khandelwal, A.R.; Mickie, H.; Ma, X.; Moore-Medlin, T.; Hong, Y.; Mills, G.; Nathan, C.-A.O. Abstract 06: Comparing Mutational Profiles between Cell-Free Circu-lating Tumor DNA and Tumor DNA in Laryngeal Carcinoma Patients. Clin. Cancer Res. 2017, 23, 6. [Google Scholar] [CrossRef]
- Kawada, T.; Takahashi, H.; Sakakura, K.; Ida, S.; Mito, I.; Toyoda, M.; Chikamatsu, K. Circulating Tumor Cells in Patients with Head and Neck Squamous Cell Carcinoma: Feasibility of Detection and Quantitation. Head Neck 2017, 39, 2180–2186. [Google Scholar] [CrossRef]
- Nichols, A.C.; Lowes, L.E.; Szeto, C.C.T.; Basmaji, J.; Dhaliwal, S.; Chapeskie, C.; Todorovic, B.; Read, N.; Venkatesan, V.; Hammond, A.; et al. Detection of Circulating Tumor Cells in Advanced Head and Neck Cancer Using the CellSearch System. Head Neck 2012, 34, 1440–1444. [Google Scholar] [CrossRef]
- Rizzo, M.I.; Ralli, M.; Nicolazzo, C.; Gradilone, A.; Carletti, R.; Di Gioia, C.; De Vincentiis, M.; Greco, A. Detection of Circulating Tumor Cells in Patients with Laryngeal Cancer Using ScreenCell: Comparative Pre- and Post-Operative Analysis and Association with Prognosis. Oncol. Lett. 2020, 19, 4183–4188. [Google Scholar] [CrossRef]
- Kanwal, R.; Gupta, S. Epigenetic Modifications in Cancer. Clin. Genet. 2012, 81, 303–311. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, M.; Ni, S.; Li, Q.; Ye, D.; Li, J.; Shen, Z.; Deng, H. DNA Methylation Biomarkers for Head and Neck Squamous Cell Carcinoma. Epigenetics 2018, 13, 398–409. [Google Scholar] [CrossRef]
- Chen, Z.; Riggs, A.D. DNA Methylation and Demethylation in Mammals. J. Biol. Chem. 2011, 286, 18347–18353. [Google Scholar] [CrossRef] [PubMed]
- Witte, T.; Plass, C.; Gerhauser, C. Pan-Cancer Patterns of DNA Methylation. Genome Med. 2014, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Rosas, S.L.; Koch, W.; da Costa Carvalho, M.G.; Wu, L.; Califano, J.; Westra, W.; Jen, J.; Sidransky, D. Promoter Hypermethylation Patterns of P16, O6-Methylguanine-DNA-Methyltransferase, and Death-Associated Protein Kinase in Tumors and Saliva of Head and Neck Cancer Patients. Cancer Res. 2001, 61, 939–942. [Google Scholar] [PubMed]
- Wong, T.-S.; Gao, W.; Li, Z.-H.; Chan, J.Y.-W.; Ho, W.-K. Epigenetic Dysregulation in Laryngeal Squamous Cell Carcinoma. J. Oncol. 2012, 2012, 739461. [Google Scholar] [CrossRef]
- Smigiel, R.; Sasiadek, M.; Krecicki, T.; Ramsey, D.; Jagielski, J.; Blin, N. Inactivation of the Cyclin-Dependent Kinase Inhibitor 2A (CDKN2A) Gene in Squamous Cell Carcinoma of the Larynx. Mol. Carcinog. 2004, 39, 147–154. [Google Scholar] [CrossRef]
- Padhi, S.S.; Roy, S.; Kar, M.; Saha, A.; Roy, S.; Adhya, A.; Baisakh, M.; Banerjee, B. Role of CDKN2A/P16 Expression in the Prognostication of Oral Squamous Cell Carcinoma. Oral Oncol. 2017, 73, 27–35. [Google Scholar] [CrossRef]
- Todorova, T.A.; Jordanov, S.H.; Stancheva, G.S.; Chalakov, I.J.; Melnicharov, M.B.; Kunev, K.V.; Mitev, V.I.; Kaneva, R.P.; Goranova, T.E. Mutational Status of CDKN2A and TP53 Genes in Laryngeal Squamous Cell Carcinoma. Pathol. Oncol. Res. 2015, 21, 413–421. [Google Scholar] [CrossRef]
- Pietruszewska, W.; Rieske, P.; Murlewska, A.; Kobos, J.; Gryczyński, M. Expression of p16 gene and protein in the evaluation of dynamics of laryngeal cancer growth. Otolaryngol. Pol. Pol. Otolaryngol. 2004, 58, 173–180. [Google Scholar]
- Temam, S.; Bénard, J.; Dugas, C.; Trassard, M.; Gormally, E.; Soria, J.-C.; Faivre, S.; Luboinski, B.; Marandas, P.; Hainaut, P.; et al. Molecular Detection of Early-Stage Laryngopharyn-geal Squamous Cell Carcinomas. Clin. Cancer Res. 2005, 11, 2547–2551. [Google Scholar] [CrossRef]
- Giefing, M.; Zemke, N.; Brauze, D.; Kostrzewska-Poczekaj, M.; Luczak, M.; Szaumkessel, M.; Pelinska, K.; Kiwerska, K.; Tönnies, H.; Grenman, R.; et al. High Resolution ArrayCGH and Expression Profiling Identifies PTPRD and PCDH17/PCH68 as Tumor Suppressor Gene Candidates in Laryngeal Squamous Cell Carcinoma. Genes Chromosomes Cancer 2011, 50, 154–166. [Google Scholar] [CrossRef]
- Byzia, E.; Soloch, N.; Bodnar, M.; Szaumkessel, M.; Kiwerska, K.; Kostrzewska-Poczekaj, M.; Jarmuz-Szymczak, M.; Szylberg, L.; Wierzbicka, M.; Bartochowska, A.; et al. Recurrent Transcriptional Loss of the PCDH17 Tumor Suppressor in Laryngeal Squamous Cell Carcinoma Is Partially Mediated by Aberrant Promoter DNA Methylation. Mol. Carcinog. 2018, 57, 878–885. [Google Scholar] [CrossRef]
- Shen, Z.; Lin, L.; Cao, B.; Zhou, C.; Hao, W.; Ye, D. LZTS2 Promoter Hypermethylation: A Potential Biomarker for the Diagnosis and Prognosis of Laryngeal Squamous Cell Carcinoma. World J. Surg. Oncol. 2018, 16, 42. [Google Scholar] [CrossRef]
- Starska, K.; Forma, E.; Lewy-Trenda, I.; Papiez, P.; Wos, J.; Brys, M. Diagnostic impact of promoter methylation and E-cadherin gene and protein expression levels in laryngeal carcinoma. Contemp. Oncol. 2013, 17, 263–271. [Google Scholar] [CrossRef]
- Calmon, M.F.; Colombo, J.; Carvalho, F.; Souza, F.P.; Filho, J.F.G.; Fukuyama, E.E.; Camargo, A.A.; Caballero, O.L.S.; Tajara, E.H.; Cordeiro, J.A.; et al. Methylation Profile of Genes CDKN2A (P14 and P16), DAPK1, CDH1, and ADAM23 in Head and Neck Cancer. Cancer Genet. Cytogenet. 2007, 173, 31–37. [Google Scholar] [CrossRef]
- Bouras, E.; Karakioulaki, M.; Bougioukas, K.I.; Aivaliotis, M.; Tzimagiorgis, G.; Chourdakis, M. Gene Promoter Methylation and Cancer: An Umbrella Review. Gene 2019, 710, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Paluszczak, J.; Misiak, P.; Wierzbicka, M.; Woźniak, A.; Baer-Dubowska, W. Frequent Hy-permethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in Laryngeal Squamous Cell Carcinomas and Adjacent Normal Mucosa. Oral Oncol. 2011, 47, 104–107. [Google Scholar] [CrossRef]
- Pierini, S.; Jordanov, S.H.; Mitkova, A.V.; Chalakov, I.J.; Melnicharov, M.B.; Kunev, K.V.; Mitev, V.I.; Kaneva, R.P.; Goranova, T.E. Promoter Hypermethylation of CDKN2A, MGMT, MLH1, and DAPK Genes in Laryngeal Squamous Cell Carcinoma and Their Associations with Clinical Profiles of the Patients. Head Neck 2014, 36, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Dikshit, R.P.; Gillio-Tos, A.; Brennan, P.; De Marco, L.; Fiano, V.; Martinez-Peñuela, J.M.; Boffetta, P.; Merletti, F. Hypermethylation, Risk Factors, Clinical Characteristics, and Survival in 235 Patients with Laryngeal and Hypopharyngeal Cancers. Cancer 2007, 110, 1745–1751. [Google Scholar] [CrossRef]
- Xu, S.; Li, Y.; Lu, Y.; Huang, J.; Ren, J.; Zhang, S.; Yin, Z.; Huang, K.; Wu, G.; Yang, K. LZTS2 Inhibits PI3K/AKT Activation and Radioresistance in Nasopharyngeal Carcinoma by Interacting with P85. Cancer Lett. 2018, 420, 38–48. [Google Scholar] [CrossRef]
- Azarschab, P.; Stembalska, A.; Loncar, M.B.; Pfister, M.; Sasiadek, M.M.; Blin, N. Epigenetic Control of E-Cadherin (CDH1) by CpG Methylation in Metastasising Laryngeal Cancer. Oncol. Rep. 2003, 10, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Ma, X.; Zhou, H.; Yu, C.; Zhang, Y.; Yang, X.; Shen, X.; Gao, X. Expression and Prognostic Value of E-Cadherin in Laryngeal Cancer. Acta OtoLaryngol. 2016, 136, 722–728. [Google Scholar] [CrossRef]
- Paksoy, M.; Hardal, U.; Caglar, C. Expression of Cathepsin D and E-Cadherin in Primary Laryngeal Cancers Correlation with Neck Lymph Node Involvement. J. Cancer Res. Clin. Oncol. 2011, 137, 1371–1377. [Google Scholar] [CrossRef]
- Rodrigo, J.P.; Domínguez, F.; Alvarez, C.; Manrique, C.; Herrero, A.; Suárez, C. Expression of E-Cadherin in Squamous Cell Carcinomas of the Supraglottic Larynx with Correlations to Clinicopathological Features. Eur J Cancer. 2002, 38, 1059–1064. [Google Scholar] [CrossRef]
- Galera-Ruiz, H.; Ríos-Moreno, M.J.; González-Cámpora, R.; Ortega, I.; Fernández, A.; García-Escudero, A.; Galera-Davidson, H. The Cadherin-Catenin Complex in Laryngeal Squamous Cell Carcinoma. Eur. Arch. OtoRhinoLaryngol. 2012, 269, 1183–1188. [Google Scholar] [CrossRef]
- Eriksen, J.G.; Buffa, F.M.; Alsner, J.; Steiniche, T.; Bentzen, S.M.; Overgaard, J. Molecular Profiles as Predictive Marker for the Effect of Overall Treatment Time of Radiotherapy in Supraglottic Larynx Squamous Cell Carcinomas. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2004, 72, 275–282. [Google Scholar] [CrossRef]
- Farag, A.K.; Roh, E.J. Death-Associated Protein Kinase (DAPK) Family Modulators: Current and Future Therapeutic Outcomes. Med. Res. Rev. 2019, 39, 349–385. [Google Scholar] [CrossRef]
- Onerci Celebi, O.; Tezel, G.G.; Hosal, A.S.; Cengiz, M.; Gullu, I.H.; Hayran, M. Detection of O6-Methylguanine-DNA Methyltransferase Gene Promoter Region Methylation Pattern Using Pyrosequencing and the Effect of Methylation Pattern on Survival, Recurrence, and Chemotherapy Sensitivity in Patients with Laryngeal Cancer. Pathol. Res. Pract. 2016, 212, 456–462. [Google Scholar] [CrossRef]
- Paluszczak, J.; Hemmerling, D.; Kostrzewska-Poczekaj, M.; Jarmuż-Szymczak, M.; Grenman, R.; Wierzbicka, M.; Baer-Dubowska, W. Frequent Hypermethylation of WNT Pathway Genes in Laryngeal Squamous Cell Carcinomas. J. Oral Pathol. Med. 2014, 43, 652–657. [Google Scholar] [CrossRef]
- Langevin, S.M.; Stone, R.A.; Bunker, C.H.; Lyons-Weiler, M.A.; LaFramboise, W.A.; Kelly, L.; Seethala, R.R.; Grandis, J.R.; Sobol, R.W.; Taioli, E. MicroRNA-137 Promoter Methylation Is Associated with Poorer Overall Survival in Patients with Squamous Cell Carcinoma of the Head and Neck. Cancer 2011, 117, 1454–1462. [Google Scholar] [CrossRef]
- Shen, Z.; Zhou, C.; Li, J.; Ye, D.; Li, Q.; Wang, J.; Cui, X.; Chen, X.; Bao, T.; Duan, S. Promoter Hypermethylation of MiR-34a Contributes to the Risk, Progression, Metastasis and Poor Survival of Laryngeal Squamous Cell Carcinoma. Gene 2016, 593, 272–276. [Google Scholar] [CrossRef]
- Gaudet, F.; Hodgson, J.G.; Eden, A.; Jackson-Grusby, L.; Dausman, J.; Gray, J.W.; Leonhardt, H.; Jaenisch, R. Induction of Tumors in Mice by Genomic Hypomethylation. Science 2003, 300, 489–492. [Google Scholar] [CrossRef]
- Liu, J.; Xu, Z.-M.; Qiu, G.-B.; Zheng, Z.-H.; Sun, K.-L.; Fu, W.-N. S100A4 Is Upregulated via the Binding of C-Myb in Methylation-Free Laryngeal Cancer Cells. Oncol. Rep. 2014, 31, 442–449. [Google Scholar] [CrossRef]
- Liu, J.; Guo, Y.; Fu, S.; Yang, M.; Sun, K.-L.; Fu, W.-N. Hypomethylation-Induced Expression of S100A4 Increases the Invasiveness of Laryngeal Squamous Cell Carcinoma. Oncol. Rep. 2010, 23, 1101–1107. [Google Scholar]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug Resistance in Cancer: An Overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, J.; Ma, W.; Zhang, W.; Ke, Z.; Ma, L. Anti-Tumor Effect of HOTAIR–MiR-613-SNAI2 Axis through Suppressing EMT and Drug Resistance in Laryngeal Squamous Cell Carcinoma. RSC Adv. 2018, 8, 29879–29889. [Google Scholar] [CrossRef]
- Yuan, Z.; Xiu, C.; Liu, D.; Zhou, G.; Yang, H.; Pei, R.; Ding, C.; Cui, X.; Sun, J.; Song, K. Long Noncoding RNA LINC-PINT Regulates Laryngeal Carcinoma Cell Stemness and Chemoresistance through MiR-425-5p/PTCH1/SHH Axis. J. Cell. Physiol. 2019, 234, 23111–23122. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, S.; Zhan, J.; Li, X.; Liu, W.; Sheng, X.; Lu, Z.; Zhong, R.; Chen, L.; Luo, X.; et al. Long Noncoding RNA FOXD2-AS1 Enhances Chemotherapeutic Resistance of Laryngeal Squamous Cell Carcinoma via STAT3 Activation. Cell Death Dis. 2020, 11, 41. [Google Scholar] [CrossRef]
- Yuan, Z.; Xiu, C.; Song, K.; Pei, R.; Miao, S.; Mao, X.; Sun, J.; Jia, S. Long Non-Coding RNA AFAP1-AS1/MiR-320a/RBPJ Axis Regulates Laryngeal Carcinoma Cell Stemness and Chemoresistance. J. Cell. Mol. Med. 2018, 22, 4253–4262. [Google Scholar] [CrossRef]
- Jiang, Q.; Liu, S.; Hou, L.; Guan, Y.; Yang, S.; Luo, Z. The Implication of LncRNA MALAT1 in Promoting Chemo-Resistance of Laryngeal Squamous Cell Carcinoma Cells. J. Clin. Lab. Anal. 2020, 34, e23116. [Google Scholar] [CrossRef]
- Liu, J.; Tang, Q.; Li, S.; Yang, X. Inhibition of HAX-1 by MiR-125a Reverses Cisplatin Resistance in Laryngeal Cancer Stem Cells. Oncotarget 2016, 7, 86446–86456. [Google Scholar] [CrossRef]
- Tian, L.; Zhang, J.; Ren, X.; Liu, X.; Gao, W.; Zhang, C.; Sun, Y.; Liu, M. Overexpression of MiR-26b Decreases the Cisplatin-Resistance in Laryngeal Cancer by Targeting ATF2. Oncotarget 2017, 8, 79023–79033. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, W.; Zhao, X.; Wang, P. MiR-133a Enhances the Sensitivity of Hep-2 Cells and Vincristine-Resistant Hep-2v Cells to Cisplatin by Downregulating ATP7B Expression. Int. J. Mol. Med. 2016, 37, 1636–1642. [Google Scholar] [CrossRef]
- Lin, X.-J.; Liu, H.; Li, P.; Wang, H.-F.; Yang, A.-K.; Di, J.-M.; Jiang, Q.-W.; Yang, Y.; Huang, J.-R.; Yuan, M.-L.; et al. MiR-936 Suppresses Cell Proliferation, Invasion, and Drug Resistance of Laryngeal Squamous Cell Carcinoma and Targets GPR78. Front. Oncol. 2020, 10, 60. [Google Scholar] [CrossRef]
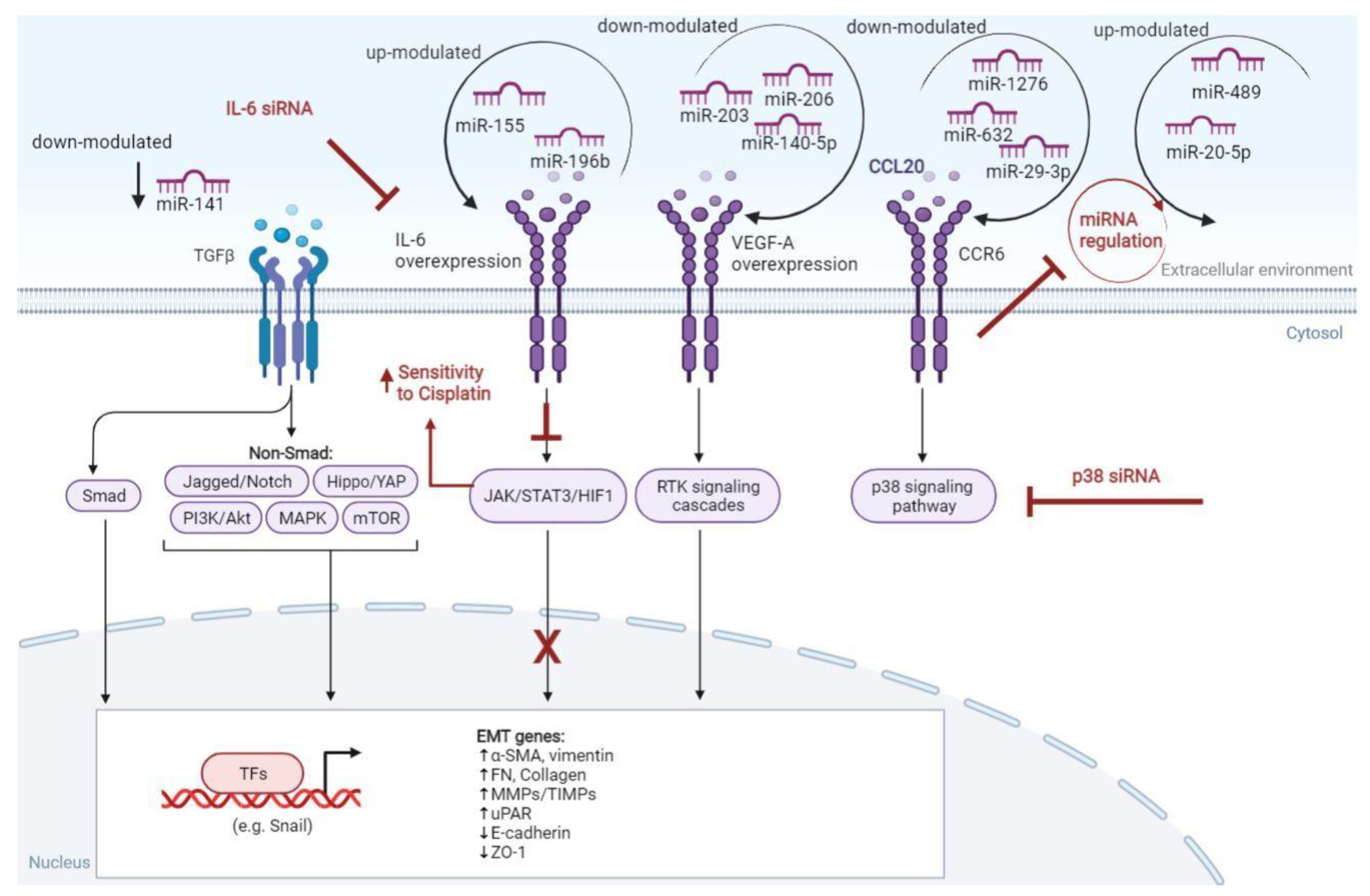
- Fu, Q.; Liu, P.; Sun, X.; Huang, S.; Han, F.; Zhang, L.; Xu, Y.; Liu, T. Ribonucleic Acid Inter-ference Knockdown of IL-6 Enhances the Efficacy of Cisplatin in Laryngeal Cancer Stem Cells by down-Regulating the IL-6/STAT3/HIF1 Pathway. Cancer Cell Int. 2017, 17, 79. [Google Scholar] [CrossRef]
- Sheng, X.; Li, Y.; Li, Y.; Liu, W.; Lu, Z.; Zhan, J.; Xu, M.; Chen, L.; Luo, X.; Cai, G.; et al. PLOD2 Contributes to Drug Resistance in Laryngeal Cancer by Promoting Cancer Stem Cell-like Characteristics. BMC Cancer 2019, 19, 840. [Google Scholar] [CrossRef]
- Zhu, M.; Yin, F.; Yang, L.; Chen, S.; Chen, R.; Zhou, X.; Jing, W.; Fan, X.; Jia, R.; Wang, H.; et al. Contribution of TIP30 to Chemoresistance in Laryngeal Carcinoma. Cell Death Dis. 2014, 5, e1468. [Google Scholar] [CrossRef]
- Li, G.; Hu, X.; Sun, L.; Li, X.; Li, J.; Li, T.; Zhang, X. C-Fos Upregulates P-Glycoprotein, Contributing to the Development of Multidrug Resistance in HEp-2 Laryngeal Cancer Cells with VCR-Induced Resistance. Cell. Mol. Biol. Lett. 2018, 23, 6. [Google Scholar] [CrossRef]
- Wang, L.; Sun, J.; Gao, P.; Su, K.; Wu, H.; Li, J.; Lou, W. Wnt1-Inducible Signaling Protein 1 Regulates Laryngeal Squamous Cell Carcinoma Glycolysis and Chemoresistance via the YAP1/TEAD1/GLUT1 Pathway. J. Cell. Physiol. 2019, 234, 15941–15950. [Google Scholar] [CrossRef]
- Xu, D.; Li, D.W.; Xie, J.; Chen, X.W. Effect and Mechanism of Survivin on Hypoxia-Induced Multidrug Resistance of Human Laryngeal Carcinoma Cells. BioMed Res. Int. 2019, 2019, e5696801. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and Cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Dvorak, H.F. Tumors: Wounds That Do Not Heal. Similarities between Tumor Stroma Generation and Wound Healing. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar] [CrossRef]
- Xu, L.; Shen, B.; Chen, T.; Dong, P. MiR-203 Is Involved in the Laryngeal Carcinoma Pathogenesis via Targeting VEGFA and Cox-2. OncoTargets Ther. 2016, 9, 4629–4637. [Google Scholar] [CrossRef]
- Kyzas, P.A.; Stefanou, D.; Agnantis, N.J. COX-2 Expression Correlates with VEGF-C and Lymph Node Metastases in Patients with Head and Neck Squamous Cell Carcinoma. Mod. Pathol. 2005, 18, 153–160. [Google Scholar] [CrossRef]
- Morita, Y.; Hata, K.; Nakanishi, M.; Nishisho, T.; Yura, Y.; Yoneda, T. Cyclooxygenase-2 Promotes Tumor Lymphangiogenesis and Lymph Node Metastasis in Oral Squamous Cell Carcinoma. Int. J. Oncol. 2012, 41, 885–892. [Google Scholar] [CrossRef]
- Zhang, J.-R.; Zhu, R.-H.; Han, X.-P. MiR-140-5p Inhibits Larynx Carcinoma Invasion and Angiogenesis by Targeting VEGF-A. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5994–6001. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, M.; Wang, C.; Lin, C.; Sun, Y.; Jin, D. Down-Regulation of MiR-206 Promotes Proliferation and Invasion of Laryngeal Cancer by Regulating VEGF Expression. Anticancer Res. 2011, 31, 3859–3863. [Google Scholar]
- Ribatti, D. Epithelial-Mesenchymal Transition in Morphogenesis, Cancer Progression and Angiogenesis. Exp. Cell Res. 2017, 353, 1–5. [Google Scholar] [CrossRef]
- Mojtahedi, Z.; Khademi, B.; Hashemi, S.B.; Abtahi, S.M.B.; Ghasemi, M.A.; Fattahi, M.J.; Ghaderi, A. Serum Interleukine-6 Concentration, but Not Interleukine-18, Is Associated with Head and Neck Squamous Cell Carcinoma Progression. Pathol. Oncol. Res. POR 2011, 17, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.A.; Taylor, J.M.G.; Terrell, J.E.; Islam, M.; Li, Y.; Fowler, K.E.; Wolf, G.T.; Teknos, T.N. Interleukin-6 Predicts Recurrence and Survival among Head and Neck Cancer Patients. Cancer 2008, 113, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Kumar, B.; Datta, J.; Teknos, T.N.; Kumar, P. IL-6 Promotes Head and Neck Tu-mor Metastasis by Inducing Epithelial-Mesenchymal Transition via the JAK-STAT3-SNAIL Signaling Pathway. Mol. Cancer Res. 2011, 9, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Zhu, Y.; Zhou, H. Prognostic Value of Interleukin-6 and Interleukin-8 in Larynge-al Squamous Cell Cancer. Med. Oncol. 2013, 30, 333. [Google Scholar] [CrossRef]
- Nikakhlagh, S.; Ranjbari, N.; Khorami, E.; Saki, N. Association between Serum Levels of Interleukin-6 and Stage of Laryngeal Cancer. Iran. J. Otorhinolaryngol. 2015, 27, 199–205. [Google Scholar]
- Weng, Y.-S.; Tseng, H.-Y.; Chen, Y.-A.; Shen, P.-C.; Al Haq, A.T.; Chen, L.-M.; Tung, Y.-C.; Hsu, H.-L. MCT-1/MiR-34a/IL-6/IL-6R Signaling Axis Promotes EMT Progression, Cancer Stemness and M2 Macrophage Polarization in Triple-Negative Breast Cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef]
- Rokavec, M.; Öner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/MiR-34a Feedback Loop Promotes EMT-Mediated Colorectal Cancer Invasion and Metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, W.; Chang, H.; Han, Z.; Yu, X.; Zhang, T. Reciprocal Regulation of MiR-206 and IL-6/STAT3 Pathway Mediates IL6-Induced Gefitinib Resistance in EGFR-Mutant Lung Cancer Cells. J. Cell. Mol. Med. 2019, 23, 7331–7341. [Google Scholar] [CrossRef]
- Li, F.; Gu, C.; Tian, F.; Jia, Z.; Meng, Z.; Ding, Y.; Yang, J. MiR-218 Impedes IL-6-Induced Prostate Cancer Cell Proliferation and Invasion via Suppression of LGR4 Expression. Oncol. Rep. 2016, 35, 2859–2865. [Google Scholar] [CrossRef]
- Misso, G.; Zarone, M.R.; Lombardi, A.; Grimaldi, A.; Cossu, A.M.; Ferri, C.; Russo, M.; Vuoso, D.C.; Luce, A.; Kawasaki, H.; et al. MiR-125b Upregulates MiR-34a and Sequentially Activates Stress Adaption and Cell Death Mechanisms in Multiple Myeloma. Mol. Ther. Nucleic Acids 2019, 16, 391–406. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, W.; Liang, H.; Ji, W. Overexpression of MiR-155 Promotes Proliferation and Invasion of Human Laryngeal Squamous Cell Carcinoma via Targeting SOCS1 and STAT3. PLoS ONE 2013, 8, e56395. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, W.; Ji, W. MiR-196b Is a Prognostic Factor of Human Laryngeal Squamous Cell Carcinoma and Promotes Tumor Progression by Targeting SOCS2. Biochem. Biophys. Res. Commun. 2018, 501, 584–592. [Google Scholar] [CrossRef]
- Wang, B.; Lv, K.; Chen, W.; Zhao, J.; Luo, J.; Wu, J.; Li, Z.; Qin, H.; Wong, T.-S.; Yang, W.; et al. MiR-375 and MiR-205 Regulate the Invasion and Migration of Laryngeal Squamous Cell Carcinoma Synergistically via AKT-Mediated EMT. BioMed Res. Int. 2016, 2016, 9652789. [Google Scholar] [CrossRef]
- Fiorella, R.; Assennato, G.; Di Nicola, V.; Troia, M.; Colucci, G.A.; Resta, L. Multivariate Analysis of Metastasis Risk in Laryngeal Carcinoma. II. Immune Response. Boll. Della Soc. Ital. Biol. Sper. 1991, 67, 199–205. [Google Scholar]
- Stockmann, C.; Schadendorf, D.; Klose, R.; Helfrich, I. The Impact of the Immune System on Tumor: Angiogenesis and Vascular Remodeling. Front. Oncol. 2014, 4, 69. [Google Scholar] [CrossRef]
- Davoine, F.; Lacy, P. Eosinophil Cytokines, Chemokines, and Growth Factors: Emerging Roles in Immunity. Front. Immunol. 2014, 5, 570. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the Immune System in Cancer: From Tumor Initiation to Metastatic Progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, B.; Tan, H.; Su, H.; He, W. Relationship among lymphatic metastasis, pericancerous lymphocytic reaction and dendritic cell infiltration in laryngeal carcinoma cells. Zhonghua Er Bi Yan Hou Ke Za Zhi 2001, 36, 264–266. [Google Scholar]
- Remedi, M.M.; Donadio, A.C.; Chiabrando, G.A. Polymorphonuclear Cells Stimulate the Migration and Metastatic Potential of Rat Sarcoma Cells. Int. J. Exp. Pathol. 2009, 90, 44–51. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-Beta: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Burkholder, B.; Huang, R.-Y.; Burgess, R.; Luo, S.; Jones, V.S.; Zhang, W.; Lv, Z.-Q.; Gao, C.-Y.; Wang, B.-L.; Zhang, Y.-M.; et al. Tumor-Induced Perturbations of Cytokines and Immune Cell Networks. Biochim. Biophys. Acta 2014, 1845, 182–201. [Google Scholar] [CrossRef]
- Wang, R.; Guo, Y.; Ma, H.; Feng, L.; Wang, Q.; Chen, X.; Lian, M.; Wang, H.; Fang, J. Tumor Necrosis Factor Superfamily Member 13 Is a Novel Biomarker for Diagnosis and Prognosis and Promotes Cancer Cell Proliferation in Laryngeal Squamous Cell Carcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 2635–2645. [Google Scholar] [CrossRef]
- Chen, L.; Guan, H.; Gu, C.; Cao, Y.; Shao, J.; Wang, F. MiR-383 Inhibits Hepatocellular Carcinoma Cell Proliferation via Targeting APRIL. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 2497–2507. [Google Scholar] [CrossRef]
- Kara, M.; Uysal, S.; Altinişik, U.; Cevizci, S.; Güçlü, O.; Dereköy, F.S. The Pre-Treatment Neutrophil-to-Lymphocyte Ratio, Platelet-to-Lymphocyte Ratio, and Red Cell Distribution Width Predict Prognosis in Patients with Laryngeal Carcinoma. Eur. Arch. OtoRhinoLaryngol. 2017, 274, 535–542. [Google Scholar] [CrossRef]
- Eskiizmir, G.; Uz, U.; Onur, E.; Ozyurt, B.; Karaca Cikrikci, G.; Sahin, N.; Oran, A.; Celik, O. The Evaluation of Pretreatment Neutrophil-Lymphocyte Ratio and Derived Neutrophil-Lymphocyte Ratio in Patients with Laryngeal Neoplasms. Braz. J. Otorhinolaryngol. 2019, 85, 578–587. [Google Scholar] [CrossRef]
- Marchi, F.; Missale, F.; Incandela, F.; Filauro, M.; Mazzola, F.; Mora, F.; Paderno, A.; Parrinello, G.; Piazza, C.; Peretti, G. Prognostic Significance of Peripheral T-Cell Subsets in Laryngeal Squamous Cell Carcinoma. Laryngoscope Investig. Otolaryngol. 2019, 4, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Drennan, S.; Stafford, N.D.; Greenman, J.; Green, V.L. Increased Frequency and Suppressive Activity of CD127low/− Regulatory T Cells in the Peripheral Circulation of Patients with Head and Neck Squamous Cell Carcinoma Are Associated with Advanced Stage and Nodal Involvement. Immunology 2013, 140, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, D.; Zhou, J.; Li, Q.; Zhou, L.; Li, S.-M.; Zhu, L.; Chou, K.-Y.; Zhou, L.; Tao, L.; et al. High CCR6/CCR7 Expression and Foxp3+ Treg Cell Number Are Positively Related to the Progression of Laryngeal Squamous Cell Carcinoma. Oncol. Rep. 2013, 30, 1380–1390. [Google Scholar] [CrossRef] [PubMed]
- Lu, E.; Su, J.; Zhou, Y.; Zhang, C.; Wang, Y. CCL20/CCR6 Promotes Cell Proliferation and Metastasis in Laryngeal Cancer by Activating P38 Pathway. Biomed. Pharmacother. Biomedecine Pharmacother. 2017, 85, 486–492. [Google Scholar] [CrossRef]
| Gene | Mutation | Regulation | Biological Role | Clinical-Pathological and Prognostic Implications | Refs. |
|---|---|---|---|---|---|
| TP53 | Disruptive (exons 2 through 11) | Down (wild type isoform) | Tumor suppressor | Decreased overall survival HR (95% CI):1.4 (1.1–1.8) (p = 0.009) Primary tumor recurrence (p = 0.001) | [31,32,33,34,35,37] |
| TP63 | - | Abnormal Expression | Tumor suppressor/ oncogene | Cancer development Tumor recurrence Higher death rate | [36] |
| IL1A | - | Down | Anti-tumor immune response | Primary tumor recurrence (p = 0.004) | [37] |
| RB1 | - | Down | Tumor suppressor | Primary tumor recurrence (p = 0.031) | [37] |
| STK1 | - | Up | Tumor suppressor | Primary tumor recurrence (p = 0.031) | [37] |
| LMNA | - | Down | Transcriptional signaling | Lymph node metastases | [37] |
| RECQL4 | - | Down | DNA replication | Lymph node metastases | [37] |
| IGF1R | - | Down | Receptor | Lymph node metastases | [37] |
| N33 | - | Up | Tumor suppressor | Lymph node metastases | [37] |
| CDKN2D | - | Up | Signal transduction | Lymph node metastases | [37] |
| Cyclin D1 | - | Up | Oncogene | Lymph node metastases Tumor staging Decreased overall survival | [49] |
| P21 | - | Up | Tumor suppressor | - | [42] |
| FGF3 | - | Up | Signal transduction | - | [42] |
| P16 | - | Up | Cell cycle regulator | - | [42] |
| P27 | - | Down | Tumor suppressor | Shorter 5-year overall survival (p = 0.045) Advanced tumor size (p = 0.0023) Lymph node metastasis (p = 0.0016) General metastasis occurrence (p = 0.0097) Advanced clinical stage (p = 0.0008) | [43,44,45] |
| NOTCH1 | - | Up (wild type isoform) Down (mutated isoform) | Oncogene/tumor suppressor | Higher clinical stage incidence of lymph node metastasis Histological grade (p < 0.05) Shorter overall survival and disease-free survival p < 0.01 | [56,58,59] |
| NOTCH2 | - | Up | Oncogene | Tumorigenesis Lymph node metastasis | [60] |
| EGFR | Exons Deletions (19-del) Insertions (20-ins), Point Missense Mutations (L858R, T790M, G719X, T790M and L858R, L861Q, G719X and L861Q) | Up | Oncogene | Tumor aggressiveness Lymph node metastasis (p = 0.534) Decreased overall survival | [48] |
| BCL2 | - | Up | Oncogene | Tumor recurrence Reduced survival Chemo-resistance | [38,41] |
| BCL2L12 | - | Up | Tumor suppressor | Increased overall survival | [39] |
| OGG1 | Silent and missense mutations | Aberrant expression | DNA structure regulation | Higher cancer risk T3–T4 stage (p < 0.04) Laryngeal cancer (p < 0.02) | [63] |
| E-cadherin | - | Down | Tumor suppressor | Tumor staging Degree of differentiation Lymph node metastasis Supraglottic localization Poor differentiation | [53] |
| FGFR3 | Non-synonymous mutations (S249C) | Aberrant expression | Tumor suppressor/ oncogene | Progressing dysplasia Carcinoma progression | [29] |
| PIK3CA | Non-synonymous mutations (E542K) | Aberrant expression | Oncogene | Progressing dysplasia Carcinoma progression | [29] |
| TRKB | - | Up | Oncogene | - | [67,68] |
| NAT1/NAT2 | Polymorphism | Aberrant expression | Oncogene | Increased risk of laryngeal cancer | [62] |
| PARK7 | - | Up | Oncogene | Cancer cells proliferation Lymph node metastasis Laryngeal cancer localization Clinical stage | [65] |
| β-catenin | - | Up | Oncogene | Cervical metastasis Later tumor (T) stage Decreased tumor differentiation Reduced overall survival (p = 0.002) | [53,54] |
| ZEB2 | - | Up | Oncogene | Later tumor (T) stage Decreased tumor differentiation Reduced overall survival (p = 0.0003) | [54] |
| N.cadherin | - | Up | Oncogene | Later tumor (T) stage Decreased tumor differentiation Reduced overall survival (p = 0.003) | [54] |
| CTNNA2 | Point mutations Missense mutations Nonsense mutations | Up (mutated isoform) Down (wild type isoform) | Tumor suppressor | Worse prognosis | [55] |
| CTNNA3 | Point mutations Missense mutations Nonsense mutations | Up (mutated isoform) Down (wild type isoform) | Tumor suppressor | Worse prognosis | [55] |
| JAK3 | Non-synonymous mutations c.2164G > A | Aberrant expression | Oncogene | Non-progressing dysplasia Carcinoma progression | [29] |
| MET | Non-synonymous mutations c.2962C > T | Aberrant expression | Oncogene | Non-progressing dysplasia Carcinoma progression | [29] |
| FWXB7 | Non-synonymous mutations c.1273C > A | Aberrant expression | Oncogene | Non-progressing dysplasia Carcinoma progression | [29] |
| PTEN | Point mutations LOH Epigenetic silencing (abnormal methylation) | Aberrant expression | Tumor suppressor | Shorter overall survival Glottic localization Advanced tumor grade (when downregulated) | [64] |
| KRAS | - | Up | Oncogene | Tumorigenesis Invasion Lymph node metastasis Recurrence Decreased overall survival | [49] |
| Non-Coding RNA | Regulation | Sample Type | Biological Role | Target | Clinic-Pathologic and Prognostic Implications | Ref. |
|---|---|---|---|---|---|---|
| miRNA | ||||||
| miR-141 | Down | Tissue | Tumor supressor | HOXC6 | TNM stage Differentiation degree Lymph node metastasis (p < 0.001) | [72] |
| miR-138 | Down | Tissue | Tumor supressor | ZEB2 | Distal metastases of primary LC Poor prognosis (p < 0.05) | [73] |
| miR-145 | Down | Tissue | Tumor supressor | MYO5A | T stage Cell differentiation Cervical metastatic state (p < 0.05) | [74] |
| miR-203 | Down | Tissue | Tumor supressor | ASAP1 | Advanced T stage (p < 0.002) Differentiation (p < 0.001) Lymph node metastasis (p = 0.044) Decreased 5-year overall survival (p = 0.002) | [75] |
| miR-204-5p | Down | Tissue | Tumor supressor | FOXC1 | Cervical lymph node (p = 0.019) Clinical stage (p = 0.005) | [76] |
| miR-143-3p | Down | Tissue | Tumor supressor | KRAS | T classification Differentiation Lymph node metastasis Clinical stage (p < 0.05) | [77] |
| miR-101 | Down | Tissue | Tumor supressor | CDK8 | T classification (p = 0.015) Lymph node metastasis (p = 0.044) Clinical stage (p = 0.004) | [78] |
| miR-744-3p | Up | Tissue | Oncogene | PDCD4 | Lymph node metastasis (p = 0.007) | [83] |
| miR-21 | Up | Tissue | Oncogene | Ras | T classification (p = 0.0001) Differentiation (p = 0.004) Lymph node metastasis (p = 0.0008) Clinical stage (p < 0.001) | [84] |
| miR-149 | Down | Tissue | Tumor supressor | - | T Stage (p = 0.022) Lymph node metastasis (p = 0.018) Differentiation (p = 0.036) Shorter overall survival (median survival of 48 months, 95% CI of ratio 0.2536 to 1.385; p = 0.0405) | [79] |
| miR-618, miR-542-5p, let7b, miR-135a, miR-20b, miR-324-3p, miR-886-5p | Up | Tissue | - | - | Lymph node metastasis | [86] |
| miR486-3p, miR-328, miR-376a, miR-493 | Down | Tissue | - | - | Lymph node metastasis | [86] |
| miR-129-5p | Up | Tissue | Oncogene | APC | T classification (p = 0.04) Lymph node metastasis (p = 0.02) Clinical stage (p = 0.01) | [85] |
| miR-34a | Down | Tissue | Tumor supressor | Survivin | Histological Differentiation (p < 0.0001) Lymphatic metastasis (p = 0.0022) TNM stage (p = 0.0111) | [81] |
| miR-195 | Down | Tissue | Tumor supressor | DCUN1D1 | Shorter overall survival (p = 0.029) T stage (p < 0.001) Lymph node metastases (p = 0.035) Clinical stage (p < 0.001) | [82] |
| miR-21 | Up | Serum | - | - | - | [89] |
| miR-155 | Up | Plasma | Oncogene | - | T stage (p = 0.001) Lymph node metastases (p = 0.007) Tumor size (p = 0.033) | [91] |
| miR-632 | Up | Serum | - | - | T stage (p = 0.014) Lymph node metastases (p = 0.020) Histological grade (p = 0.001) | [92] |
| miR-449a | Down | Tissue | Oncogene | - | Lymph node metastases (p < 0.01) (ROC sensitivity = 0.55, specificity = 0.76, AUC = 0.67) | [87] |
| miR-130a | Down | Plasma | - | Grade I of cachexia (p = 0.044) Cachexia grade I vs. grade II Sensitivity of 63.6% and specificity of 64.7% (AUC = 0.663; p < 0.05) Shorter overall survival (p = 0.087 HR = 2.582) | [94] | |
| lncRNA | ||||||
| HOXA11-AS | Up | Tissue | Oncogene | - | T stage (p = 0.011) Differentiation (p = 0.014) Lymph node metastasis (p = 0.026) Clinical stage (p = 0.001) | [96] |
| RGMB-AS1 | Up | Tissue | Oncogene | miR-22/NLRP3 axis | T stage (p < 0.05) Lymph node metastasis (p < 0.05) Shorter Overall Survival and Disease Free Survival (p < 0.05) | [97] |
| NKILA | Down | Tissue | Tumor-supressor | NF-kB pathway | T stage (p = 0.002) Lymph node metastasis Clinical stage (p < 0.001) Shorter overall survival | [98] |
| H19 | Up | Tissue | Oncogene | miR-148a-3p/DNMT1 axis | T stage (p < 0.01) Lymph node metastasis (p < 0.01) Differentiation (p < 0.017) Clinical stage (p < 0.01) Shorter overall survival (p = 0.003) | [99] |
| TUG1 | Up | Tissue | Oncogene | - | T stage (p = 0.025) Lymph node metastasis (p = 0.014) Clinical stage (p = 0.003) | [100] |
| MIR155HG | Up | Tissue | Oncogene | miR-155/ SOX10 axis | T stage (p < 0.001) Lymph node metastases (p < 0.01) Differentiation (p < 0.05) | [91] |
| NEAT1 | Up | Tissue | Oncogene | miR-107/CDK6 axis | T stage (p < 0.001) Lymph node metastases (p < 0.05) Histological grade (p < 0.001) | [101] |
| ST7-AS1 | Up | Tissue | Oncogene | CARM1/ SOX2 axis | T stage (p < 0.001) Lymph node metastases (p < 0.001) Shorter overall survival (p = 0.0023) | [102] |
| XIST | Up | Tissue | Oncogene | miR-124/EZH2 axis | T stage (p < 0.05) | [103] |
| HOTAIR | Up | Serum | - | - | T stage (p < 0.0038) Lymph node metastases (p = 0.0003) Clinical stage (p = 0.0061) | [90] |
| UCA1 | Up | Serum | - | Wnt/β-catenin pathway | Distant metastasis Shorter overall survival (p = 0.032) | [104] |
| snaR | Up | Plasma | Oncogene | TGF-β1 | AJCC stages (p < 0.05) Shorter overall survival (p < 0.05) | [105] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falco, M.; Tammaro, C.; Takeuchi, T.; Cossu, A.M.; Scafuro, G.; Zappavigna, S.; Itro, A.; Addeo, R.; Scrima, M.; Lombardi, A.; et al. Overview on Molecular Biomarkers for Laryngeal Cancer: Looking for New Answers to an Old Problem. Cancers 2022, 14, 1716. https://doi.org/10.3390/cancers14071716
Falco M, Tammaro C, Takeuchi T, Cossu AM, Scafuro G, Zappavigna S, Itro A, Addeo R, Scrima M, Lombardi A, et al. Overview on Molecular Biomarkers for Laryngeal Cancer: Looking for New Answers to an Old Problem. Cancers. 2022; 14(7):1716. https://doi.org/10.3390/cancers14071716
Chicago/Turabian StyleFalco, Michela, Chiara Tammaro, Takashi Takeuchi, Alessia Maria Cossu, Giuseppe Scafuro, Silvia Zappavigna, Annalisa Itro, Raffaele Addeo, Marianna Scrima, Angela Lombardi, and et al. 2022. "Overview on Molecular Biomarkers for Laryngeal Cancer: Looking for New Answers to an Old Problem" Cancers 14, no. 7: 1716. https://doi.org/10.3390/cancers14071716
APA StyleFalco, M., Tammaro, C., Takeuchi, T., Cossu, A. M., Scafuro, G., Zappavigna, S., Itro, A., Addeo, R., Scrima, M., Lombardi, A., Ricciardiello, F., Irace, C., Caraglia, M., & Misso, G. (2022). Overview on Molecular Biomarkers for Laryngeal Cancer: Looking for New Answers to an Old Problem. Cancers, 14(7), 1716. https://doi.org/10.3390/cancers14071716









