Recent Advances of Deep Learning for Computational Histopathology: Principles and Applications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Deep Neural Network
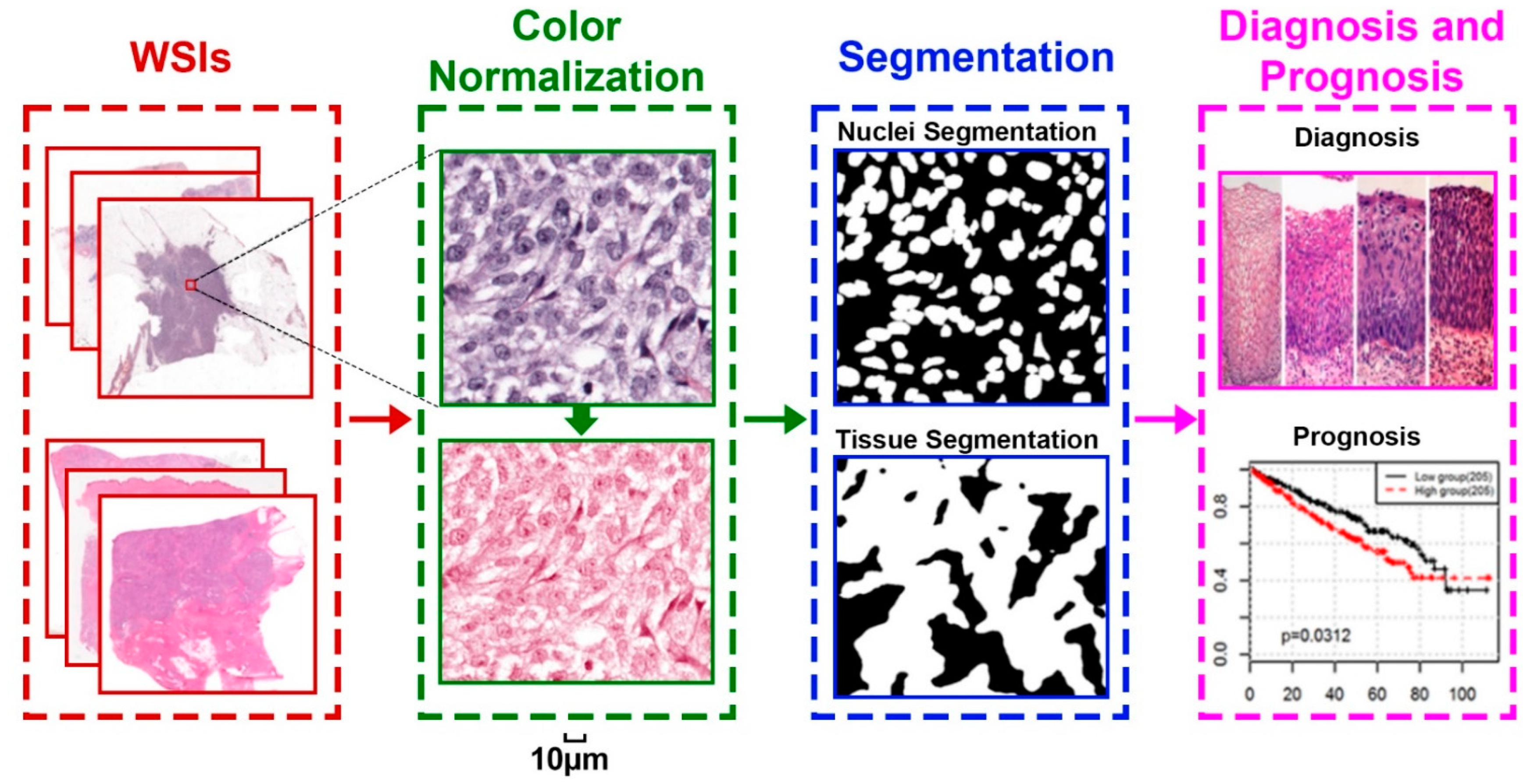
3. Color Normalization
4. Pathology Image Segmentation
4.1. Nuclei-Level Segmentation
4.2. Tissue-Level Segmentation
5. Cancer Diagnosis and Prognosis
5.1. Patch-Level Methods
5.2. WSI-Level Methods
6. Open Resources and Future Work
6.1. Open Resources
6.2. Future Work
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Li, X.; Razmjooy, N.; Ghadimi, N. Breast Cancer Diagnosis by Convolutional Neural Network and Advanced Thermal Exchange Optimization Algorithm. Comput. Math. Methods Med. 2021, 2021, 5595180. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Han, Z.; Cheng, J.; Cheng, L.; Wang, T.; Sun, L.; Lu, Z.; Zhang, J.; Zhang, D.; Huang, K. Integrative Analysis of Pathological Images and Multi-Dimensional Genomic Data for Early-Stage Cancer Prognosis. IEEE Trans. Med. Imaging 2020, 39, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xiang, L.; Liu, Q.; Gilmore, H.; Wu, J.; Tang, J.; Madabhushi, A. Stacked Sparse Autoencoder (SSAE) for Nuclei Detection on Breast Cancer Histopathology Images. IEEE Trans. Med. Imaging 2016, 35, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Stacke, K.; Eilertsen, G.; Unger, J.; Lundström, C. Measuring domain shift for deep learning in histopathology. IEEE J. Biomed. Health Inform. 2020, 25, 325–336. [Google Scholar] [CrossRef]
- Zhu, W.; Xie, L.; Han, J.; Guo, X. The application of deep learning in cancer prognosis prediction. Cancers 2020, 12, 603. [Google Scholar] [CrossRef] [Green Version]
- Peikari, M.; Gangeh, M.J.; Zubovits, J.; Clarke, G.; Martel, A.L. Triaging diagnostically relevant regions from pathology whole slides of breast cancer: A texture based approach. IEEE Trans. Med. Imaging 2015, 35, 307–315. [Google Scholar]
- Anoraganingrum, D. Cell segmentation with median filter and mathematical morphology operation. In Proceedings of the 10th International Conference on Image Analysis and Processing, Venice, Italy, 27–29 September 1999; pp. 1043–1046. [Google Scholar]
- Platt, J. Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines. 1998. Available online: https://www.microsoft.com/en-us/research/publication/sequential-minimal-optimization-a-fast-algorithm-for-training-support-vector-machines/ (accessed on 16 February 2022).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Dudani, S.A. The distance-weighted k-nearest-neighbor rule. IEEE Trans. Syst. Man Cybern. 1976, SMC-6, 325–327. [Google Scholar] [CrossRef]
- Kruk, M.; Kurek, J.; Osowski, S.; Koktysz, R.; Swiderski, B.; Markiewicz, T. Ensemble of classifiers and wavelet transformation for improved recognition of Fuhrman grading in clear-cell renal carcinoma. Biocybern. Biomed. Eng. 2017, 37, 357–364. [Google Scholar] [CrossRef]
- Fuchs, T.J.; Wild, P.J.; Moch, H.; Buhmann, J.M. Computational pathology analysis of tissue microarrays predicts survival of renal clear cell carcinoma patients. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, New York, NY, USA, 6–10 September 2008; pp. 1–8. [Google Scholar]
- Zarella, M.D.; Yeoh, C.; Breen, D.E.; Garcia, F.U. An alternative reference space for H&E color normalization. PLoS ONE 2017, 12, e0174489. [Google Scholar]
- Freitag, D. Information extraction from HTML: Application of a general machine learning approach. In Proceedings of the AAAI/IAAI, Madison, WI, USA, 26–30 July 1998; pp. 517–523. [Google Scholar]
- Cheng, J.; Han, Z.; Mehra, R.; Shao, W.; Cheng, M.; Feng, Q.; Ni, D.; Huang, K.; Cheng, L.; Zhang, J. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11. 2 translocation renal cell carcinoma. Nat. Commun. 2020, 11, 1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, B.; Zhang, L.; Ning, P.; Ding, F.; Wu, F.; Lu, G.; Geng, Y.; Ma, J. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur. Radiol. 2020, 30, 6924–6932. [Google Scholar] [CrossRef] [PubMed]
- Cosatto, E.; Laquerre, P.-F.; Malon, C.; Graf, H.-P.; Saito, A.; Kiyuna, T.; Marugame, A.; Kamijo, K.I. Automated gastric cancer diagnosis on h&e-stained sections; ltraining a classifier on a large scale with multiple instance machine learning. In Proceedings of the Medical Imaging 2013: Digital Pathology, Lake Buena Vista, FL, USA, 29 March 2013; p. 867605. [Google Scholar]
- Komura, D.; Ishikawa, S. Machine learning approaches for pathologic diagnosis. Virchows Arch. 2019, 475, 131–138. [Google Scholar] [CrossRef]
- Jimenez-del-Toro, O.; Otálora, S.; Andersson, M.; Eurén, K.; Hedlund, M.; Rousson, M.; Müller, H.; Atzori, M. Analysis of histopathology images: From traditional machine learning to deep learning. In Biomedical Texture Analysis; Elsevier: Amsterdam, The Netherlands, 2017; pp. 281–314. [Google Scholar]
- Pasquini, G.; Arias, J.E.R.; Schäfer, P.; Busskamp, V. Automated methods for cell type annotation on scRNA-seq data. Comput. Struct. Biotechnol. J. 2021, 19, 961–969. [Google Scholar] [CrossRef]
- van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A new era: Artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef]
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using deep learning to enhance cancer diagnosis and classification. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; pp. 3937–3949. [Google Scholar]
- Zhao, Z.-Q.; Zheng, P.; Xu, S.-T.; Wu, X. Object detection with deep learning: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Li, Y.; Hao, Z.; Lei, H. Survey of convolutional neural network. J. Comput. Appl. 2016, 36, 2508–2515. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 818–833. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Krithiga, R.; Geetha, P. Breast cancer detection, segmentation and classification on histopathology images analysis: A systematic review. Arch. Comput. Methods Eng. 2021, 28, 2607–2619. [Google Scholar] [CrossRef]
- Shaban, M.T.; Baur, C.; Navab, N.; Albarqouni, S. Staingan: Stain style transfer for digital histological images. In Proceedings of the 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 953–956. [Google Scholar]
- de Bel, T.; Hermsen, M.; Kers, J.; van der Laak, J.; Litjens, G. Stain-transforming cycle-consistent generative adversarial networks for improved segmentation of renal histopathology. In Proceedings of the International Conference on Medical Imaging with Deep Learning–Full Paper Track, Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Bentaieb, A.; Hamarneh, G. Adversarial Stain Transfer for Histopathology Image Analysis. IEEE Trans. Med. Imaging 2018, 37, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, D.; Bozorgtabar, B.; Thiran, J.-P.; Shao, L. Structure preserving stain normalization of histopathology images using self supervised semantic guidance. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Online, 4–8 October 2020; pp. 309–319. [Google Scholar]
- Cong, C.; Liu, S.; Di Ieva, A.; Pagnucco, M.; Berkovsky, S.; Song, Y. Texture Enhanced Generative Adversarial Network For Stain Normalisation In Histopathology Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1949–1952. [Google Scholar]
- Patil, A.; Talha, M.; Bhatia, A.; Kurian, N.C.; Mangale, S.; Patel, S.; Sethi, A. Fast, Self Supervised, Fully Convolutional Color Normalization Of H&E Stained Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1563–1567. [Google Scholar]
- Janowczyk, A.; Basavanhally, A.; Madabhushi, A. Stain Normalization using Sparse AutoEncoders (StaNoSA): Application to digital pathology. Comput. Med. Imaging Graph. 2017, 57, 50–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar]
- Reinhard, E.; Ashikhmin, N.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Vahadane, A.; Peng, T.; Sethi, A.; Albarqouni, S.; Wang, L.; Baust, M.; Steiger, K.; Schlitter, A.M.; Esposito, I.; Navab, N. Structure-Preserving Color Normalization and Sparse Stain Separation for Histological Images. IEEE. Trans. Med. Imaging 2016, 35, 1962–1971. [Google Scholar] [CrossRef]
- Bug, D.; Schneider, S.; Grote, A.; Oswald, E.; Feuerhake, F.; Schüler, J.; Merhof, D. Context-based normalization of histological stains using deep convolutional features. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2017; pp. 135–142. [Google Scholar]
- Zanjani, F.G.; Zinger, S.; Bejnordi, B.E.; van der Laak, J.A.; de With, P.H. Stain normalization of histopathology images using generative adversarial networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 573–577. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Chan, L.; Hosseini, M.S.; Rowsell, C.; Plataniotis, K.N.; Damaskinos, S. Histosegnet: Semantic segmentation of histological tissue type in whole slide images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Thessaloniki, Greece, 23–25 September 2019; pp. 10662–10671. [Google Scholar]
- Zhang, H.; Liu, J.; Yu, Z.; Wang, P. MASG-GAN: A multi-view attention superpixel-guided generative adversarial network for efficient and simultaneous histopathology image segmentation and classification. Neurocomputing 2021, 463, 275–291. [Google Scholar] [CrossRef]
- Sucher, R.; Sucher, E. Artificial intelligence is poised to revolutionize human liver allocation and decrease medical costs associated with liver transplantation. HepatoBiliary Surg. Nutr. 2020, 9, 679. [Google Scholar] [CrossRef]
- Chen, H.; Qi, X.; Yu, L.; Dou, Q.; Qin, J.; Heng, P.A. DCAN: Deep contour-aware networks for object instance segmentation from histology images. Med. Image Anal. 2017, 36, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, J.; Polson, J.; Wang, Z.; Speier, W.; Arnold, C. High resolution histopathology image generation and segmentation through adversarial training. Med. Image Anal 2022, 75, 102251. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, J.; Liao, Z.; Verjans, J.; Shen, C.; Xia, Y. Pairwise relation learning for semi-supervised gland segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Online, 4–8 October 2020; pp. 417–427. [Google Scholar]
- Lu, Z.; Zhan, X.; Wu, Y.; Cheng, J.; Shao, W.; Ni, D.; Han, Z.; Zhang, J.; Feng, Q.; Huang, K. BrcaSeg: A Deep Learning Approach for Tissue Quantification and Genomic Correlations of Histopathological Images. Genom. Proteom. Bioinform. 2021. [Google Scholar] [CrossRef]
- Raza, S.E.A.; Cheung, L.; Epstein, D.; Pelengaris, S.; Khan, M.; Rajpoot, N.M. Mimo-net: A multi-input multi-output convolutional neural network for cell segmentation in fluorescence microscopy images. In Proceedings of the 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 337–340. [Google Scholar]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar] [CrossRef] [Green Version]
- Samanta, P.; Raipuria, G.; Singhal, N. Context Aggregation Network For Semantic Labeling In Histopathology Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 673–676. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Smedby, Ö.; Wang, C. A two-stage U-Net algorithm for segmentation of nuclei in H&E-stained tissues. In Proceedings of the European Congress on Digital Pathology, Warwick, UK, 10–13 April 2019; pp. 75–82. [Google Scholar]
- Yang, L.; Ghosh, R.P.; Franklin, J.M.; Chen, S.; You, C.; Narayan, R.R.; Melcher, M.L.; Liphardt, J.T. NuSeT: A deep learning tool for reliably separating and analyzing crowded cells. PLoS Comput. Biol. 2020, 16, e1008193. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, X.; Li, Z.; Yu, Z.; Yao, S.; Yan, L.; Wang, Y.; Liu, Z.; Liang, C.; Han, C. Triple U-net: Hematoxylin-aware nuclei segmentation with progressive dense feature aggregation. Med. Image Anal. 2020, 65, 101786. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, L.; Chen, S.; Ni, D.; Lei, B.; Wang, T. Accurate Segmentation of Cervical Cytoplasm and Nuclei Based on Multiscale Convolutional Network and Graph Partitioning. IEEE Trans. Biomed. Eng. 2015, 62, 2421–2433. [Google Scholar] [CrossRef]
- Zhou, Y.; Chang, H.; Barner, K.E.; Parvin, B. Nuclei segmentation via sparsity constrained convolutional regression. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 1284–1287. [Google Scholar]
- Ciresan, D.; Giusti, A.; Gambardella, L.; Schmidhuber, J. Deep neural networks segment neuronal membranes in electron microscopy images. Adv. Neural Inf Process. Syst. 2012, 25, 2843–2851. [Google Scholar]
- Qi, X.; Xing, F.; Foran, D.J.; Yang, L. Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans. Biomed. Eng. 2012, 59, 754–765. [Google Scholar] [CrossRef] [Green Version]
- Veta, M.; van Diest, P.J.; Willems, S.M.; Wang, H.; Madabhushi, A.; Cruz-Roa, A.; Gonzalez, F.; Larsen, A.B.; Vestergaard, J.S.; Dahl, A.B.; et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015, 20, 237–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oren, A.; Fernandes, J. The Bethesda system for the reporting of cervical/vaginal cytology. J. Am. Osteopath. Assoc. 1991, 91, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, G.; Nie, J.; Tarokh, A.; Zhou, X.; Guo, L.; Malicki, J.; Xia, W.; Wong, S.T. An automated method for cell detection in zebrafish. Neuroinformatics 2008, 6, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Carneiro, G.; Bradley, A.P. An improved joint optimization of multiple level set functions for the segmentation of overlapping cervical cells. IEEE Trans. Image Process. 2015, 24, 1261–1272. [Google Scholar]
- Dorini, L.B.; Minetto, R.; Leite, N.J. Semiautomatic white blood cell segmentation based on multiscale analysis. IEEE J. Biomed. Health Inform. 2012, 17, 250–256. [Google Scholar] [CrossRef]
- Zhang, C.; Yarkony, J.; Hamprecht, F.A. Cell detection and segmentation using correlation clustering. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, MA, USA, 14–18 September 2014; pp. 9–16. [Google Scholar]
- Bergeest, J.-P.; Rohr, K. Efficient globally optimal segmentation of cells in fluorescence microscopy images using level sets and convex energy functionals. Med. Image Anal. 2012, 16, 1436–1444. [Google Scholar] [CrossRef]
- Sahara, K.; Paredes, A.Z.; Tsilimigras, D.I.; Sasaki, K.; Moro, A.; Hyer, J.M.; Mehta, R.; Farooq, S.A.; Wu, L.; Endo, I. Machine learning predicts unpredicted deaths with high accuracy following hepatopancreatic surgery. Hepatobiliary Surg. Nutr. 2021, 10, 20. [Google Scholar] [CrossRef]
- Mahmood, F.; Borders, D.; Chen, R.J.; McKay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 2019, 39, 3257–3267. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Zhang, D.; Song, Y.; Zhang, C.; Zhang, F.; O’Donnell, L.; Cai, W. Nuclei Segmentation via a Deep Panoptic Model with Semantic Feature Fusion. In Proceedings of the 2019 International Joint Conference on Artificial Intelligence, Macao, China, 10–16 August 2019; pp. 861–868. [Google Scholar]
- Moris, D.; Shaw, B.I.; Ong, C.; Connor, A.; Samoylova, M.L.; Kesseli, S.J.; Abraham, N.; Gloria, J.; Schmitz, R.; Fitch, Z.W.; et al. A simple scoring system to estimate perioperative mortality following liver resection for primary liver malignancy—the Hepatectomy Risk Score (HeRS). Hepatobiliary Surg. Nutr. 2021, 10, 315–324. [Google Scholar] [CrossRef]
- Schmitz, R.; Madesta, F.; Nielsen, M.; Krause, J.; Steurer, S.; Werner, R.; Rösch, T. Multi-scale fully convolutional neural networks for histopathology image segmentation: From nuclear aberrations to the global tissue architecture. Med. Image Anal. 2021, 70, 101996. [Google Scholar] [CrossRef]
- Xing, F.; Xie, Y.; Yang, L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans. Med. Imaging 2015, 35, 550–566. [Google Scholar] [CrossRef] [PubMed]
- Settouti, N.; Bechar, M.E.; Daho, M.E.; Chikh, M.A. An optimised pixel-based classification approach for automatic white blood cells segmentation. Int. J. Biomed. Eng. Technol. 2020, 32, 144–160. [Google Scholar] [CrossRef]
- Sahasrabudhe, M.; Christodoulidis, S.; Salgado, R.; Michiels, S.; Loi, S.; André, F.; Paragios, N.; Vakalopoulou, M. Self-supervised nuclei segmentation in histopathological images using attention. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Online, 4–8 October 2020; pp. 393–402. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Muller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana, K.; Pluim, J.P.W.; Chen, H.; Qi, X.; Heng, P.A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Vukicevic, A.M.; Radovic, M.; Zabotti, A.; Milic, V.; Hocevar, A.; Callegher, S.Z.; De Lucia, O.; De Vita, S.; Filipovic, N. Deep learning segmentation of Primary Sjögren’s syndrome affected salivary glands from ultrasonography images. Comput. Biol. Med. 2021, 129, 104154. [Google Scholar] [CrossRef]
- Gunduz-Demir, C.; Kandemir, M.; Tosun, A.B.; Sokmensuer, C. Automatic segmentation of colon glands using object-graphs. Med. Image Anal. 2010, 14, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Salvi, M.; Bosco, M.; Molinaro, L.; Gambella, A.; Papotti, M.; Acharya, U.R.; Molinari, F. A hybrid deep learning approach for gland segmentation in prostate histopathological images. Artif. Intell. Med. 2021, 115, 102076. [Google Scholar] [CrossRef]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [CrossRef]
- Chen, H.; Qi, X.; Yu, L.; Heng, P.-A. DCAN: Deep contour-aware networks for accurate gland segmentation. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Nevada, NV, USA, 26 June–1 July 2016; pp. 2487–2496. [Google Scholar]
- Musulin, J.; Stifanic, D.; Zulijani, A.; Cabov, T.; Dekanic, A.; Car, Z. An Enhanced Histopathology Analysis: An AI-Based System for Multiclass Grading of Oral Squamous Cell Carcinoma and Segmenting of Epithelial and Stromal Tissue. Cancers 2021, 13, 1784. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, J.; Fang, W.; Deng, S. SCAU-Net: Spatial-Channel Attention U-Net for Gland Segmentation. Front. Bioeng. Biotechnol. 2020, 8, 670. [Google Scholar] [CrossRef]
- Yan, Z.; Yang, X.; Cheng, K.T. Enabling a Single Deep Learning Model for Accurate Gland Instance Segmentation: A Shape-Aware Adversarial Learning Framework. IEEE Trans. Med. Imaging 2020, 39, 2176–2189. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Feng, R.; Liu, J.; Li, Y.; Ying, S. GCSBA-Net: Gabor-Based and Cascade Squeeze Bi-Attention Network for Gland Segmentation. IEEE J. Biomed. Health Inform. 2020, 25, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- van Rijthoven, M.; Balkenhol, M.; Siliņa, K.; van der Laak, J.; Ciompi, F. HookNet: Multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. Med. Image Anal. 2021, 68, 101890. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, D.; Poellinger, A.; Shao, L.; Reyes, M. Interpretability-Driven Sample Selection Using Self Supervised Learning for Disease Classification and Segmentation. IEEE Trans. Med. Imaging 2021, 40, 2548–2562. [Google Scholar] [CrossRef]
- Lai, Z.; Wang, C.; Oliveira, L.C.; Dugger, B.N.; Cheung, S.-C.; Chuah, C.-N. Joint Semi-supervised and Active Learning for Segmentation of Gigapixel Pathology Images with Cost-Effective Labeling. In Proceedings of the Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 591–600. [Google Scholar]
- Gupta, L.; Klinkhammer, B.M.; Boor, P.; Merhof, D.; Gadermayr, M. GAN-based image enrichment in digital pathology boosts segmentation accuracy. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 631–639. [Google Scholar]
- Hu, A.; Razmjooy, N. Brain tumor diagnosis based on metaheuristics and deep learning. Int. J. Imaging Syst. Technol. 2021, 31, 657–669. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, H.; Jin, X.; Chen, J.; Yu, Z.; Ramen, K.; Zheng, X.; Wu, X.; Shan, Y.; Bai, J. Development and validation of a machine learning-based nomogram for prediction of intrahepatic cholangiocarcinoma in patients with intrahepatic lithiasis. Hepatobiliary Surg. Nutr. 2021, 10, 749. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef]
- Lau, W.Y.; Wang, K.; Zhang, X.P.; Li, L.Q.; Wen, T.F.; Chen, M.S.; Jia, W.D.; Xu, L.; Shi, J.; Guo, W.X.; et al. A new staging system for hepatocellular carcinoma associated with portal vein tumor thrombus. Hepatobiliary Surg. Nutr. 2021, 10, 782–795. [Google Scholar] [CrossRef]
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Re, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, M.S.; Brawley-Hayes, J.A.; Zhang, Y.; Chan, L.; Plataniotis, K.N.; Damaskinos, S. Focus quality assessment of high-throughput whole slide imaging in digital pathology. IEEE Trans. Med. Imaging 2019, 39, 62–74. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Zhu, X.; Huang, J. Deep multi-instance learning for survival prediction from whole slide images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 496–504. [Google Scholar]
- Yang, H.; Kim, J.-Y.; Kim, H.; Adhikari, S.P. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans. Med. Imaging 2019, 39, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lu, M.Y.; Williamson, D.F.; Chen, T.Y.; Schaumberg, A.J.; Mahmood, F. Fast and Scalable Image Search For Histology. arXiv 2021, arXiv:2107.13587. [Google Scholar]
- Sun, H.; Zeng, X.; Xu, T.; Peng, G.; Ma, Y. Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms. IEEE J. Biomed. Health Inform. 2019, 24, 1664–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Yao, J.; Huang, J. Deep convolutional neural network for survival analysis with pathological images. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 544–547. [Google Scholar]
- Cheng, J.; Mo, X.; Wang, X.; Parwani, A.; Feng, Q.; Huang, K. Identification of topological features in renal tumor microenvironment associated with patient survival. Bioinformatics 2018, 34, 1024–1030. [Google Scholar] [CrossRef]
- Källén, H.; Molin, J.; Heyden, A.; Lundström, C.; Åström, K. Towards grading gleason score using generically trained deep convolutional neural networks. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1163–1167. [Google Scholar]
- Mercan, C.; Aksoy, S.; Mercan, E.; Shapiro, L.G.; Weaver, D.L.; Elmore, J.G. From patch-level to ROI-level deep feature representations for breast histopathology classification. In Proceedings of the Medical Imaging 2019: Digital Pathology, San Diego, CA, USA, 20–21 February 2019; p. 109560H. [Google Scholar]
- Shao, W.; Wang, T.; Huang, Z.; Han, Z.; Zhang, J.; Huang, K. Weakly supervised deep ordinal cox model for survival prediction from whole-slide pathological images. IEEE Trans. Med. Imaging 2021, 40, 3739–3747. [Google Scholar] [CrossRef]
- Yao, J.; Zhu, X.; Jonnagaddala, J.; Hawkins, N.; Huang, J. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med. Image Anal. 2020, 65, 101789. [Google Scholar] [CrossRef]
- Chikontwe, P.; Kim, M.; Nam, S.J.; Go, H.; Park, S.H. Multiple instance learning with center embeddings for histopathology classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 519–528. [Google Scholar]
- Chen, R.J.; Lu, M.Y.; Shaban, M.; Chen, C.; Chen, T.Y.; Williamson, D.F.; Mahmood, F. Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction using Patch-based Graph Convolutional Networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 339–349. [Google Scholar]
- Chen, R.J.; Lu, M.Y.; Weng, W.-H.; Chen, T.Y.; Williamson, D.F.; Manz, T.; Shady, M.; Mahmood, F. Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide Images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 4015–4025. [Google Scholar]
- Xu, Y.; Jia, Z.; Wang, L.B.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E.I. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef] [Green Version]
- Marini, N.; Otalora, S.; Muller, H.; Atzori, M. Semi-supervised training of deep convolutional neural networks with heterogeneous data and few local annotations: An experiment on prostate histopathology image classification. Med. Image Anal. 2021, 73, 102165. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Gan, C.; Lin, H.; Dou, Q.; Tsougenis, E.; Huang, Q.; Cai, M.; Heng, P.A. Weakly Supervised Deep Learning for Whole Slide Lung Cancer Image Analysis. IEEE Trans. Cybern. 2020, 50, 3950–3962. [Google Scholar] [CrossRef]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci. Rep. 2020, 10, 1504. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Yao, J.; Zhu, X.; Li, Y.; Huang, J. Graph CNN for survival analysis on whole slide pathological images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 174–182. [Google Scholar]
- Chen, Z.; Zhang, J.; Che, S.; Huang, J.; Han, X.; Yuan, Y. Diagnose Like A Pathologist: Weakly-Supervised Pathologist-Tree Network for Slide-Level Immunohistochemical Scoring. In Proceedings of the 35th AAAI Conference on Artificial Intelligence (AAAI-21), Online, 2–9 February 2021; pp. 47–54. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Stritt, M.; Stalder, A.K.; Vezzali, E. Orbit image analysis: An open-source whole slide image analysis tool. PLoS Comput. Biol. 2020, 16, e1007313. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [Green Version]
- Goode, A.; Gilbert, B.; Harkes, J.; Jukic, D.; Satyanarayanan, M. OpenSlide: A vendor-neutral software foundation for digital pathology. J. Pathol. Inform. 2013, 4, 27. [Google Scholar] [CrossRef]
- Della Mea, V.; Baroni, G.L.; Pilutti, D.; Di Loreto, C. SlideJ: An ImageJ plugin for automated processing of whole slide images. PLoS ONE 2017, 12, e0180540. [Google Scholar] [CrossRef] [Green Version]
- Marée, R.; Rollus, L.; Stévens, B.; Hoyoux, R.; Louppe, G.; Vandaele, R.; Begon, J.-M.; Kainz, P.; Geurts, P.; Wehenkel, L. Cytomine: An open-source software for collaborative analysis of whole-slide images. Diagn. Pathol. 2016, 1, 8. [Google Scholar] [CrossRef]
- Bao, G.; Wang, X.; Xu, R.; Loh, C.; Adeyinka, O.D.; Pieris, D.A.; Cherepanoff, S.; Gracie, G.; Lee, M.; McDonald, K.L.; et al. PathoFusion: An Open-Source AI Framework for Recognition of Pathomorphological Features and Mapping of Immunohistochemical Data. Cancers 2021, 13, 617. [Google Scholar] [CrossRef]
- Sheng, V.S.; Zhang, J. Machine learning with crowdsourcing: A brief summary of the past research and future directions. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 9837–9843. [Google Scholar]
- Alaghehbandan, R.; Perez Montiel, D.; Luis, A.S.; Hes, O. Molecular genetics of renal cell tumors: A practical diagnostic approach. Cancers 2020, 12, 85. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.J.; Lu, M.Y.; Wang, J.; Williamson, D.F.; Rodig, S.J.; Lindeman, N.I.; Mahmood, F. Pathomic fusion: An integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans. Med. Imaging 2020, 1. [Google Scholar] [CrossRef]







| Model | AlexNet [28] | ZFNet [29] | VGGNet-19 [30] | GoogLeNet [31] | ResNet-152 [32] | SENET [33] |
|---|---|---|---|---|---|---|
| Input size | 227 × 227 | 224 × 224 | 224 × 224 | 224 × 224 | 224 × 224 | 224 × 224 |
| Top-5 error(%) | 15.3 | 11.2 | 7.50 | 6.67 | 3.57 | 97.75 |
| Layer number | 8 | 8 | 19 | 22 | 152 | 152 |
| Convolution layer number | 5 | 5 | 16 | 21 | 151 | 151 |
| Kernel size | 11, 5, 3 | 7, 5, 3 | 3 | 7, 1, 3, 5 | 7, 1, 3, 5 | 7, 1, 3, 5 |
| Full connected layer number | 3 | 3 | 3 | 1 | 1 | 1 |
| Model size | 60 M | 140 M | 144 M | 500 M | 60 M | 64 M |
| Calculation speed | 727 M | 1.6 G | 20 G | 2 G | 11 G | 21 G |
| Dropout | √ | √ | √ | √ | √ | √ |
| Batch Normalization | × | × | × | × | √ | √ |
| ID | Cancer Types | Images/Cases | Link |
|---|---|---|---|
| Color normalization | |||
| NIA Lymphoma 2009 | lymphoma | 375 | https://www.nia.nih.gov (accessed on 17 January 2022) |
| UCSB | Breast | 58 | http://iridl.ldeo.columbia.edu/SOURCES/.UCSB/ (accessed on 17 January 2022) |
| CAMELYON16 2016 | Breast | 400 | https://camelyon16.grand-challenge.org/ (accessed on 17 January 2022) |
| CAMELYON17 2017 | Breast | 1000 | https://camelyon17.grand-challenge.org/ (accessed on 17 January 2022) |
| Pathology image segmentation | |||
| Nuclei segmentation | |||
| MoNuSeg 2018 | Multi-tissue | 44 | https://monuseg.grand-challenge.org/Home/ (accessed on 17 January 2022) |
| TNBC 2018 | Breast | 50 | https://github.com/PeterJackNaylor/DRFNS (accessed on 17 January 2022) |
| Gland segmentation | |||
| GLAS 2015 | Colon | 165 | https://warwick.ac.uk/fac/sci/dcs/research/tia/glascontest/ (accessed on 17 January 2022) |
| CRAG 2019 | Colon | 213 | https://warwick.ac.uk/fac/cross_fac/tia/data/mildnet/ (accessed on 17 January 2022) |
| Diagnosis and prognosis | |||
| Diagnosis | |||
| ICPR 2014 | Breast | 2112 | https://mitos-atypia-14.grand-challenge.org/ (accessed on 17 January 2022) |
| BreakHis 2016 | Breast | 82 | https://mitos-atypia-14.grand-challenge.org/ (accessed on 17 January 2022) |
| HER2 Scoring 2016 | Breast | 86 | https://warwick.ac.uk/fac/sci/dcs/research/tia/her2contest/ (accessed on 17 January 2022) |
| BACH 2018 | Breast | 500 | https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/ (accessed on 17 January 2022) |
| Prognosis | |||
| CRCHisto 2016 | Colon | 100 | https://warwick.ac.uk/fac/cross_fac/tia/data/crchistolabelednucleihe/ (accessed on 17 January 2022) |
| NCT-CRC-HE-100k 2019 | Colon | 100,000 | https://zenodo.org/record/1214456#.YeV8MnpByUl (accessed on 17 January 2022) |
| ACDC-LungHP 2019 | Lung | 200 | https://acdc-lunghp.grand-challenge.org/ (accessed on 17 January 2022) |
| CoNSeP 2019 | Colon | 41 | https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/ (accessed on 17 January 2022) |
| Multiple | |||
| TCIA | Multi-cancer | - | https://www.cancerimagingarchive.net/ (accessed on 17 January 2022) |
| Tool Name | Language | View | Color Normalization | Segmentation | Diagnosis /Prognosis | Link | Reference |
|---|---|---|---|---|---|---|---|
| Qupath | Java | √ | √ | √ | √ | https://qupath.github.io/ (accessed on 17 January 2022) | [123] |
| Cytomine | Java, web | √ | × | √ | √ | https://cytomine.be/ (accessed on 17 January 2022) | [128] |
| Orbit | Java, Scala, Python, R, and SQL | √ | √ | √ | √ | https://www.orbit.bio/ (accessed on 17 January 2022) | [124] |
| ASAP | Python | √ | × | × | × | https://computationalpathologygroup.github.io/ASAP/ (accessed on 17 January 2022) | \ |
| Openslide | C, Java | √ | √ | √ | √ | https://openslide.org/demo/ (accessed on 17 January 2022) | [126] |
| ImageJ | Java | √ | √ | √ | √ | https://imagej.net/plugins/slidej (accessed on 17 January 2022) | [127] |
| PMA.start | Web | √ | √ | √ | √ | https://free.pathomation.com/ (accessed on 17 January 2022) | \ |
| CellProfiler | Python | √ | √ | √ | √ | https://cellprofiler.org/ (accessed on 17 January 2022) | [125] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Cheng, M.; Huang, S.; Pei, Z.; Zuo, Y.; Liu, J.; Yang, K.; Zhu, Q.; Zhang, J.; Hong, H.; et al. Recent Advances of Deep Learning for Computational Histopathology: Principles and Applications. Cancers 2022, 14, 1199. https://doi.org/10.3390/cancers14051199
Wu Y, Cheng M, Huang S, Pei Z, Zuo Y, Liu J, Yang K, Zhu Q, Zhang J, Hong H, et al. Recent Advances of Deep Learning for Computational Histopathology: Principles and Applications. Cancers. 2022; 14(5):1199. https://doi.org/10.3390/cancers14051199
Chicago/Turabian StyleWu, Yawen, Michael Cheng, Shuo Huang, Zongxiang Pei, Yingli Zuo, Jianxin Liu, Kai Yang, Qi Zhu, Jie Zhang, Honghai Hong, and et al. 2022. "Recent Advances of Deep Learning for Computational Histopathology: Principles and Applications" Cancers 14, no. 5: 1199. https://doi.org/10.3390/cancers14051199
APA StyleWu, Y., Cheng, M., Huang, S., Pei, Z., Zuo, Y., Liu, J., Yang, K., Zhu, Q., Zhang, J., Hong, H., Zhang, D., Huang, K., Cheng, L., & Shao, W. (2022). Recent Advances of Deep Learning for Computational Histopathology: Principles and Applications. Cancers, 14(5), 1199. https://doi.org/10.3390/cancers14051199









