Targeting IGF2BP2 Promotes Differentiation of Radioiodine Refractory Papillary Thyroid Cancer via Destabilizing RUNX2 mRNA
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Tissue Specimen and Patient Information
2.3. Cell Culture and Transfection
2.4. 125I Uptake Assay
2.5. 131I Clonogenic Assay
2.6. Real-Time RT-qPCR Analysis
2.7. RNA Stability Assay
2.8. Western Blot Assay
2.9. RNA Immunoprecipitation (RIP) Assay
2.10. Immunohistochemistry (IHC)
2.11. Immunofluorescence Test
2.12. Statistical Analysis
3. Results
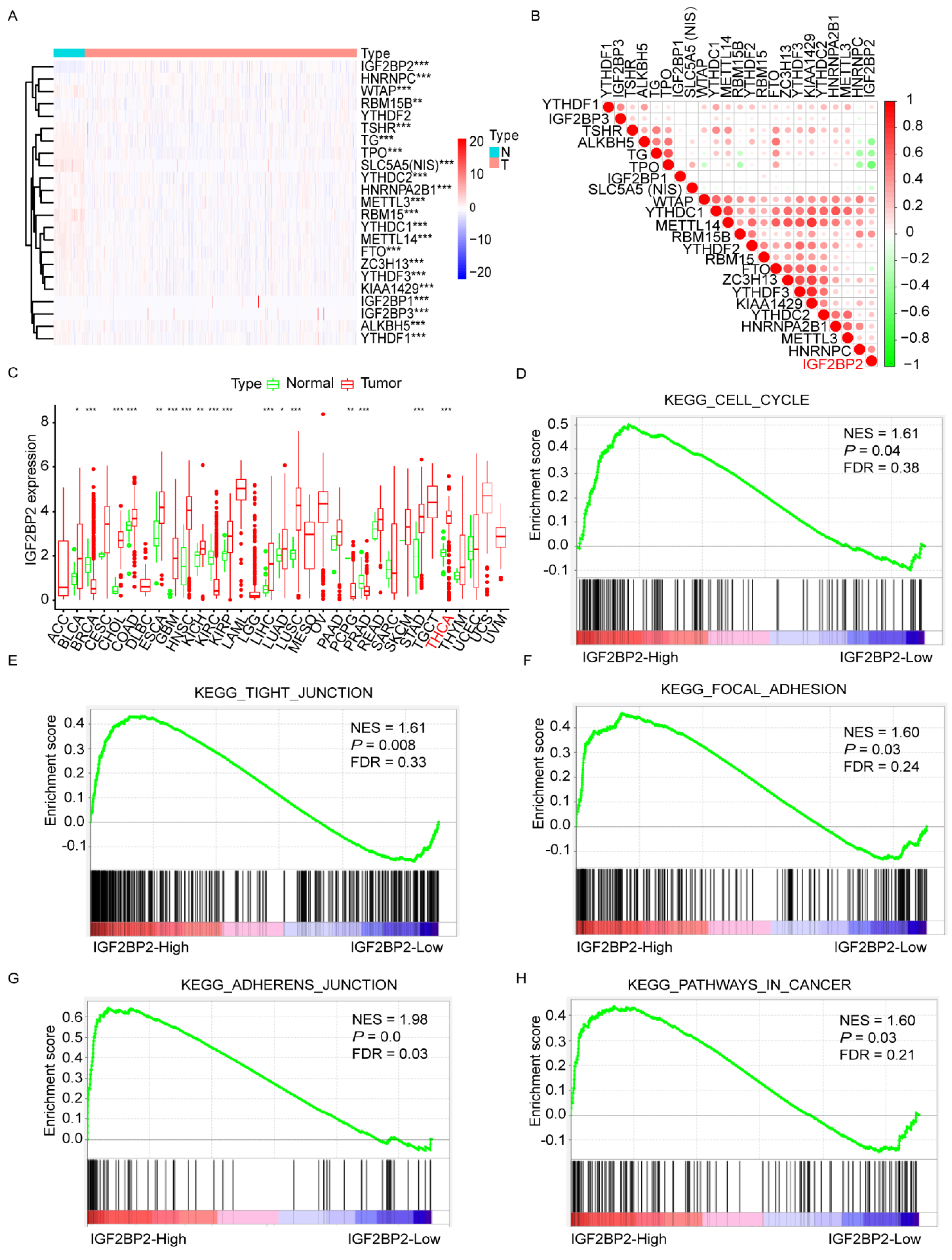
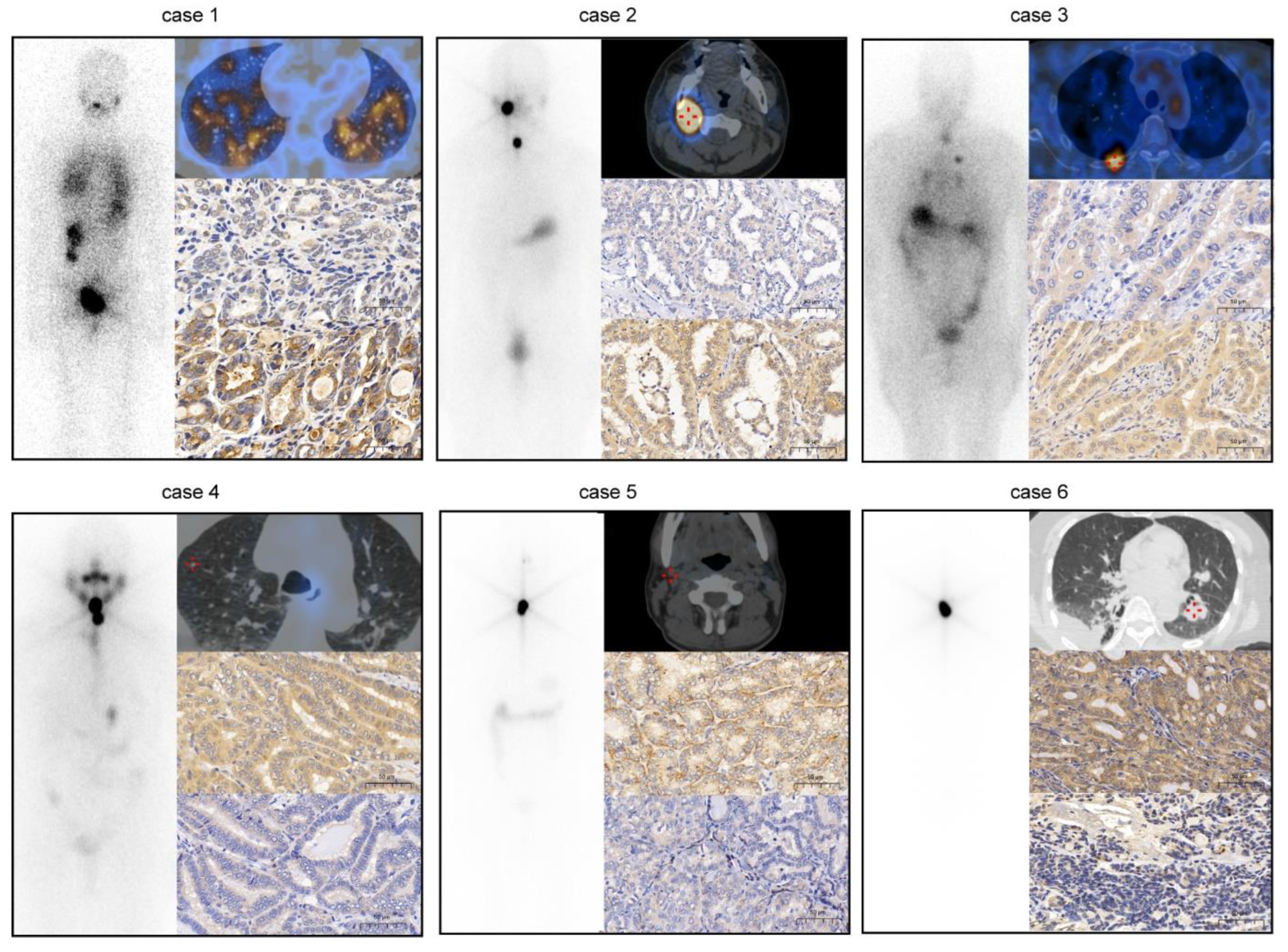
3.1. Negative Association between Expression of IGF2BP2 and Differentiation Level of RR-PTC
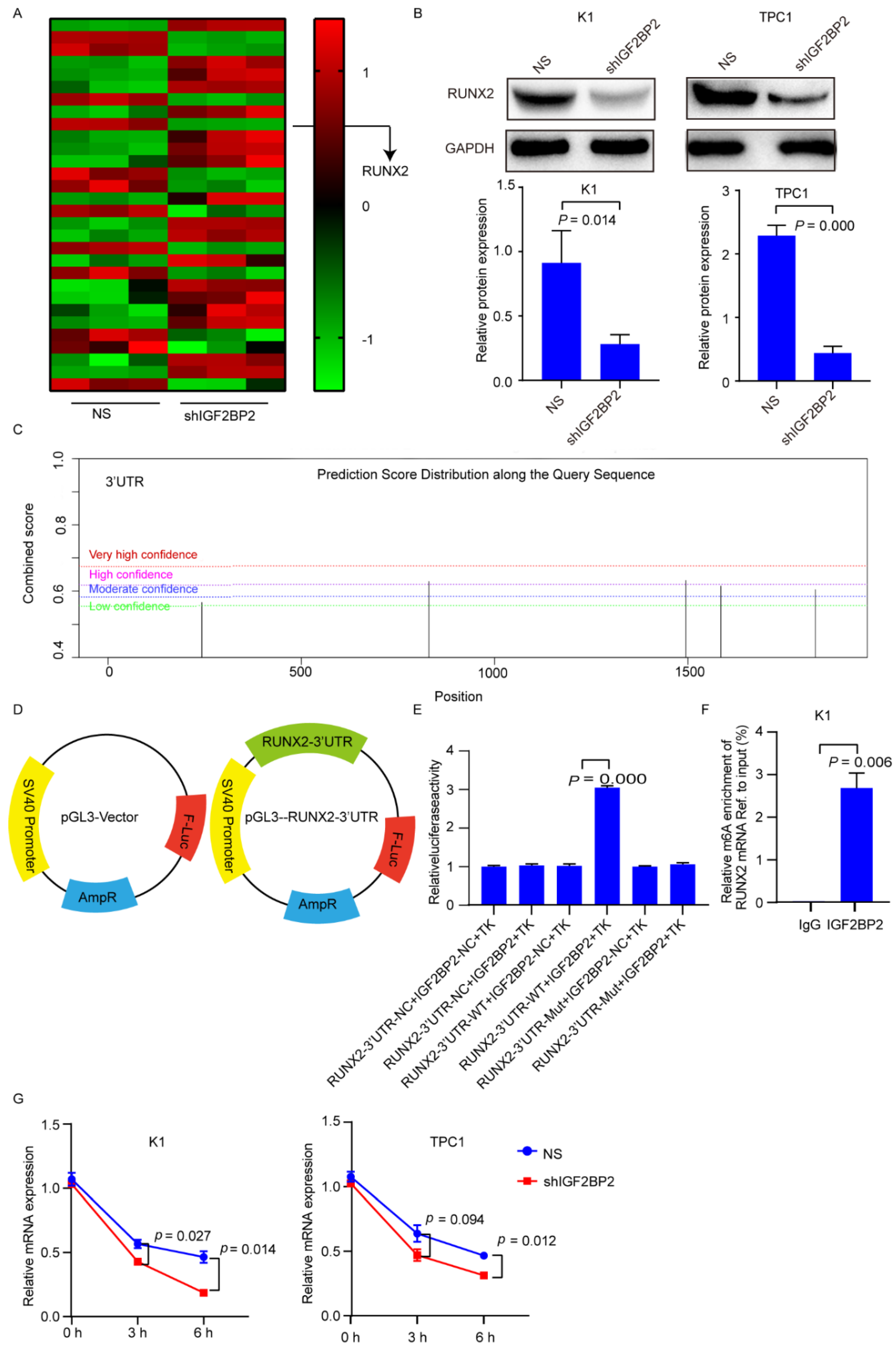
3.2. Knockdown of IGF2BP2 Enhances Differentiation of RR-PTC Cells
3.3. IGF2BP2 Stabilizes RUNX2 mRNA via Binding to 3′-Untranslated Region (UTR)
3.4. Inhibiting RUNX2 Promotes Differentiation of RR-PTC
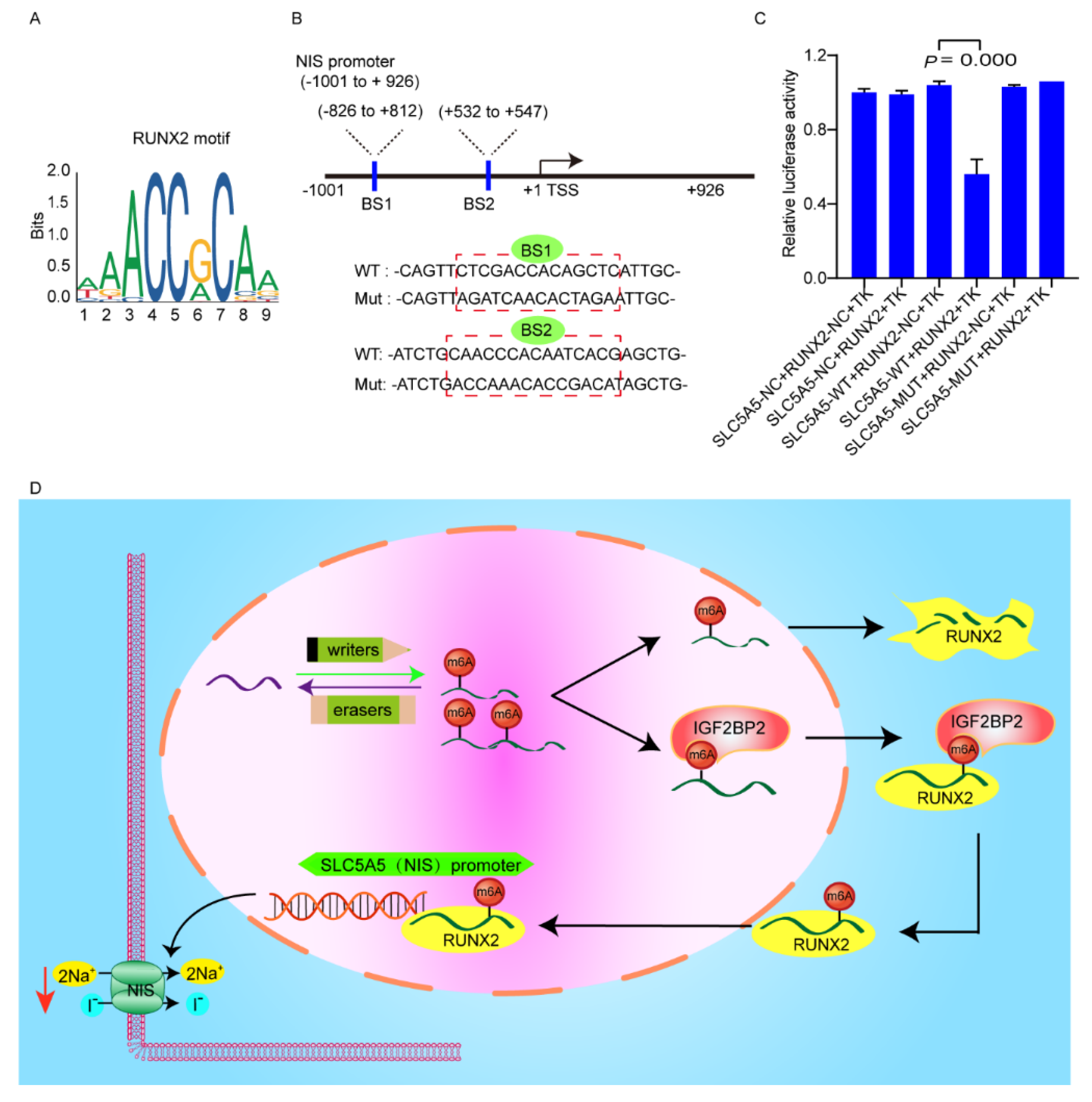
3.5. RUNX2 Downregulates NIS Expression via Binding to Promoter of NIS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Schlumberger, M.; Brose, M.; Elisei, R.; Leboulleux, S.; Luster, M.; Pitoia, F.; Pacini, F. Definition and management of radioactive iodine-refractory differentiated thyroid cancer. Lancet Diabetes Endocrinol. 2014, 2, 356–358. [Google Scholar] [CrossRef]
- Aashiq, M.; Silverman, D.A.; Na’ara, S.; Takahashi, H.; Amit, M. Radioiodine-Refractory Thyroid Cancer: Molecular Basis of Redifferentiation Therapies, Management, and Novel Therapies. Cancers 2019, 11, 1382. [Google Scholar] [CrossRef] [Green Version]
- Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [CrossRef] [Green Version]
- Fröhlich, E.; Wahl, R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat. Rev. 2014, 40, 665–674. [Google Scholar] [CrossRef]
- Qing, Y.; Su, R.; Chen, J. RNA modifications in hematopoietic malignancies: A new research frontier. Blood 2021, 138, 637–648. [Google Scholar] [CrossRef]
- Li, J.; Pei, Y.; Zhou, R.; Tang, Z.; Yang, Y. Regulation of RNA N(6)-methyladenosine modification and its emerging roles in skeletal muscle development. Int. J. Biol. Sci. 2021, 17, 1682–1692. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Bell, J.L.; Wang, J.Y.; Hüttelmaier, S.; Zhang, X.D.; Zhang, L.; Liu, T. The Emerging Roles of RNA m(6)A Methylation and Demethylation as Critical Regulators of Tumorigenesis, Drug Sensitivity, and Resistance. Cancer Res. 2021, 81, 3431–3440. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, C.; Luo, Y.; Zhang, G.; Wu, P.; Sun, N.; He, J. RNA N(6) -methyladenosine modification in the lethal teamwork of cancer stem cells and the tumor immune microenvironment: Current landscape and therapeutic potential. Clin. Transl. Med. 2021, 11, e525. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, S.; Xiang, D.; Liu, J.; Sun, W.; Cui, X.; Ning, B.; Li, X.; Cheng, Z.; Jiang, W.; et al. m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct. Target. Ther. 2020, 5, 296. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Saiselet, M.; Floor, S.; Tarabichi, M.; Dom, G.; Hébrant, A.; van Staveren, W.C.; Maenhaut, C. Thyroid cancer cell lines: An overview. Front. Endocrinol. 2012, 3, 133. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.; Wang, Z.; Zhou, N.; Li, G.; Kou, Y.; Luo, Y.; Wang, Y.; Yang, J.; Tian, F. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 2019, 18, 168. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Yu, Y.; Zong, K.; Lv, P.; Gu, Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 497. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Mu, Q.; Huang, H. The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Stem Cells Int. 2018, 2018, 4217259. [Google Scholar] [CrossRef] [Green Version]
- Su, T.S.; Liu, W.Y.; Han, S.H.; Jansen, M.; Yang-Fen, T.L.; P’Eng, F.K.; Chou, C.K. Transcripts of the insulin-like growth factors I and II in human hepatoma. Cancer Res. 1989, 49, 1773–1777. [Google Scholar]
- Cleynen, I.; Brants, J.R.; Peeters, K.; Deckers, R.; Debiec-Rychter, M.; Sciot, R.; Van de Ven, W.J.; Petit, M.M. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol. Cancer Res. 2007, 5, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Janiszewska, M.; Suvà, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clément-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Fu, X.; Zhang, J.; Xiong, C.; Zhang, S.; Lv, Y. Identification and validation of m(6)A RNA methylation regulators with clinical prognostic value in Papillary thyroid cancer. Cancer Cell Int. 2020, 20, 203. [Google Scholar] [CrossRef]
- Xu, N.; Chen, J.; He, G.; Gao, L.; Zhang, D. Prognostic values of m6A RNA methylation regulators in differentiated Thyroid Carcinoma. J. Cancer 2020, 11, 5187–5197. [Google Scholar] [CrossRef]
- Fu, R.; Jiang, X.; Li, G.; Zhu, Y.; Zhang, H. Junctional complexes in epithelial cells: Sentinels for extracellular insults and intracellular homeostasis. FEBS J. 2021. [Google Scholar] [CrossRef]
- Pedroza-Garcia, J.A.; Xiang, Y.; De Veylder, L. Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. Cell Mol. Biol. 2021, 109, 490–507. [Google Scholar] [CrossRef]
- Miller, A.E.; Hu, P.; Barker, T.H. Feeling Things Out: Bidirectional Signaling of the Cell-ECM Interface, Implications in the Mechanobiology of Cell Spreading, Migration, Proliferation, and Differentiation. Adv. Healthc. Mater. 2020, 9, e1901445. [Google Scholar] [CrossRef]
- Christiansen, J.; Kolte, A.M.; Hansen, T.; Nielsen, F.C. IGF2 mRNA-binding protein 2: Biological function and putative role in type 2 diabetes. J. Mol. Endocrinol. 2009, 43, 187–195. [Google Scholar] [CrossRef]
- Atlas, R.; Behar, L.; Elliott, E.; Ginzburg, I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 2004, 89, 613–626. [Google Scholar] [CrossRef]
- Lang, C.; Yin, C.; Lin, K.; Li, Y.; Yang, Q.; Wu, Z.; Du, H.; Ren, D.; Dai, Y.; Peng, X. m(6)A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 2021, 11, e426. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, H.; Chen, Y.; Shen, Z.; Qiu, W.; Qian, C.; Zhang, J. ZEB1-induced LINC01559 expedites cell proliferation, migration and EMT process in gastric cancer through recruiting IGF2BP2 to stabilize ZEB1 expression. Cell Death Dis. 2021, 12, 349. [Google Scholar] [CrossRef]
- Hou, P.; Meng, S.; Li, M.; Lin, T.; Chu, S.; Li, Z.; Zheng, J.; Gu, Y.; Bai, J. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J. Exp. Clin. Cancer Res. 2021, 40, 52. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Komori, T. Regulation of skeletal development by the Runx family of transcription factors. J. Cell. Biochem. 2005, 95, 445–453. [Google Scholar] [CrossRef]
- Sun, S.S.; Zhang, L.; Yang, J.; Zhou, X. Role of runt-related transcription factor 2 in signal network of tumors as an inter-mediator. Cancer Lett. 2015, 361, 1–7. [Google Scholar] [CrossRef]
- Niu, D.F.; Kondo, T.; Nakazawa, T.; Oishi, N.; Kawasaki, T.; Mochizuki, K.; Yamane, T.; Katoh, R. Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab. Investig. A J. Tech. Methods Pathol. 2012, 92, 1181–1190. [Google Scholar] [CrossRef] [Green Version]
- Sancisi, V.; Borettini, G.; Maramotti, S.; Ragazzi, M.; Tamagnini, I.; Nicoli, D.; Piana, S.; Ciarrocchi, A. Runx2 isoform I controls a panel of proinvasive genes driving aggressiveness of papillary thyroid carcinomas. J. Clin. Endocrinol. Metab. 2012, 97, E2006–E2015. [Google Scholar] [CrossRef] [Green Version]


| Characteristic | High (%) | Low (%) | p-Value |
|---|---|---|---|
| Age | 0.689 | ||
| ≥55 years | 22 (56.4%) | 16 (51.6%) | |
| <55 years | 17 (43.6%) | 15 (48.4%) | |
| Sex | 0.313 | ||
| Female | 26 (66.7%) | 17 (54.8%) | |
| Male | 13 (33.3%) | 14 (45.2%) | |
| Size | 0.023 * | ||
| ≤2.0 cm | 8 (20.5%) | 11 (35.5%) | |
| 2.0~4.0 cm | 11 (28.2%) | 14 (45.2%) | |
| >4.0 cm | 20 (51.3%) | 6 (19.3%) | |
| Metastatic site(s) | 0.233 | ||
| Lymph node-only | 8 (20.5%) | 10 (32.3%) | |
| Lung-only | 11 (28.2%) | 3 (9.6%) | |
| Bone-only | 6 (15.4%) | 7 (22.6%) | |
| Others or combined | 14 (35.9%) | 11 (35.5%) | |
| Rx-WBS | 0.015 * | ||
| 131I-avid | 14 (35.9%) | 21 (67.7%) | |
| 131I-nonavid | 25 (64.1%) | 10 (32.3%) | |
| NIS expression | 0.032 * | ||
| Low | 23 (59.0%) | 10 (32.3%) | |
| High | 16 (41.0%) | 21 (67.7%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sa, R.; Liang, R.; Qiu, X.; He, Z.; Liu, Z.; Chen, L. Targeting IGF2BP2 Promotes Differentiation of Radioiodine Refractory Papillary Thyroid Cancer via Destabilizing RUNX2 mRNA. Cancers 2022, 14, 1268. https://doi.org/10.3390/cancers14051268
Sa R, Liang R, Qiu X, He Z, Liu Z, Chen L. Targeting IGF2BP2 Promotes Differentiation of Radioiodine Refractory Papillary Thyroid Cancer via Destabilizing RUNX2 mRNA. Cancers. 2022; 14(5):1268. https://doi.org/10.3390/cancers14051268
Chicago/Turabian StyleSa, Ri, Rui Liang, Xian Qiu, Ziyan He, Zhiyan Liu, and Libo Chen. 2022. "Targeting IGF2BP2 Promotes Differentiation of Radioiodine Refractory Papillary Thyroid Cancer via Destabilizing RUNX2 mRNA" Cancers 14, no. 5: 1268. https://doi.org/10.3390/cancers14051268
APA StyleSa, R., Liang, R., Qiu, X., He, Z., Liu, Z., & Chen, L. (2022). Targeting IGF2BP2 Promotes Differentiation of Radioiodine Refractory Papillary Thyroid Cancer via Destabilizing RUNX2 mRNA. Cancers, 14(5), 1268. https://doi.org/10.3390/cancers14051268







