Immunologic Gene Signature Analysis Correlates Myeloid Cells and M2 Macrophages with Time to Trabectedin Failure in Sarcoma Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Data Collection
2.2. Multiplex Immunohistochemistry
2.3. NanoString
3. Results
3.1. Patient Demographics
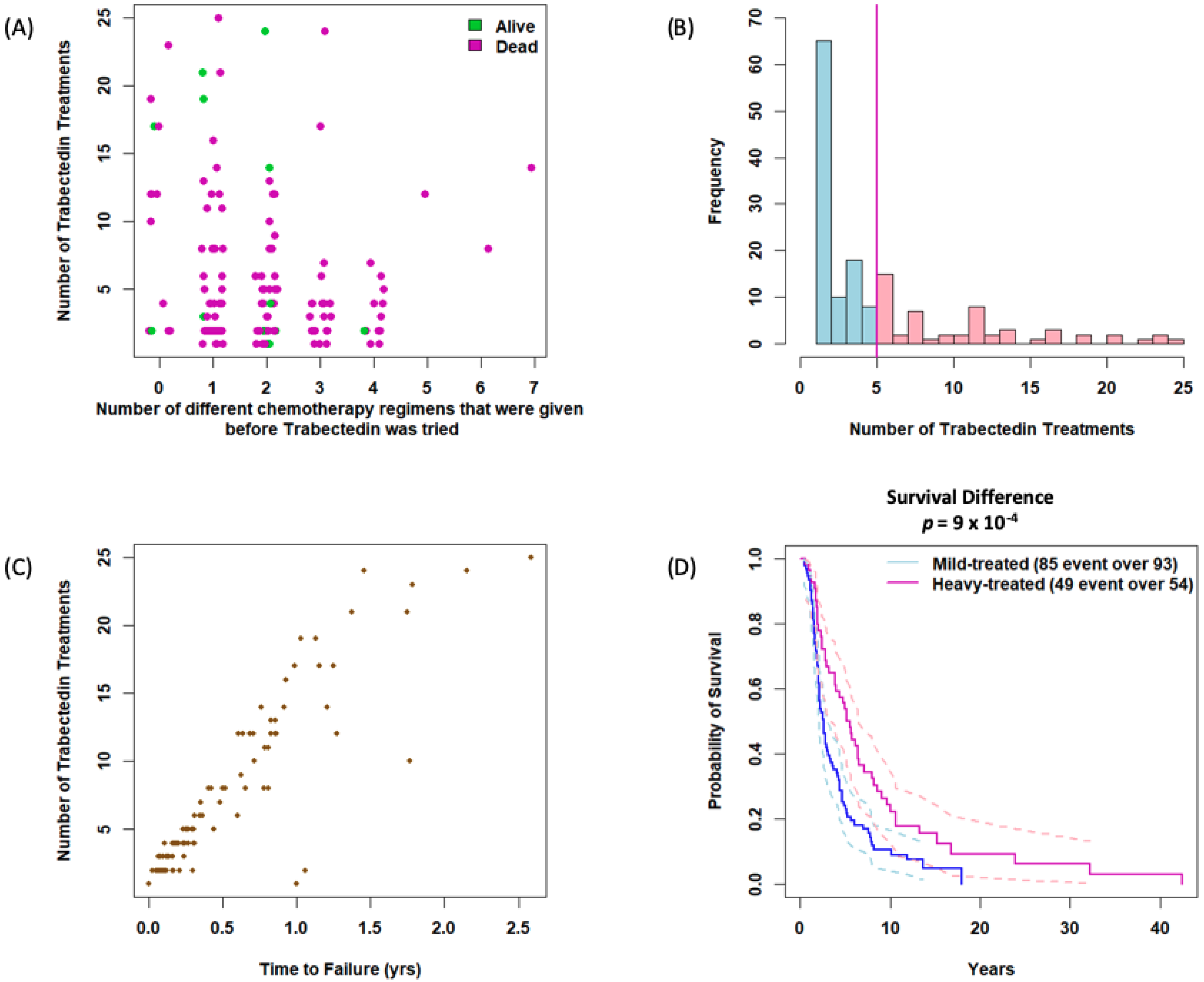
3.2. Impact of Prior Treatment and Trabectedin Duration on Outcomes
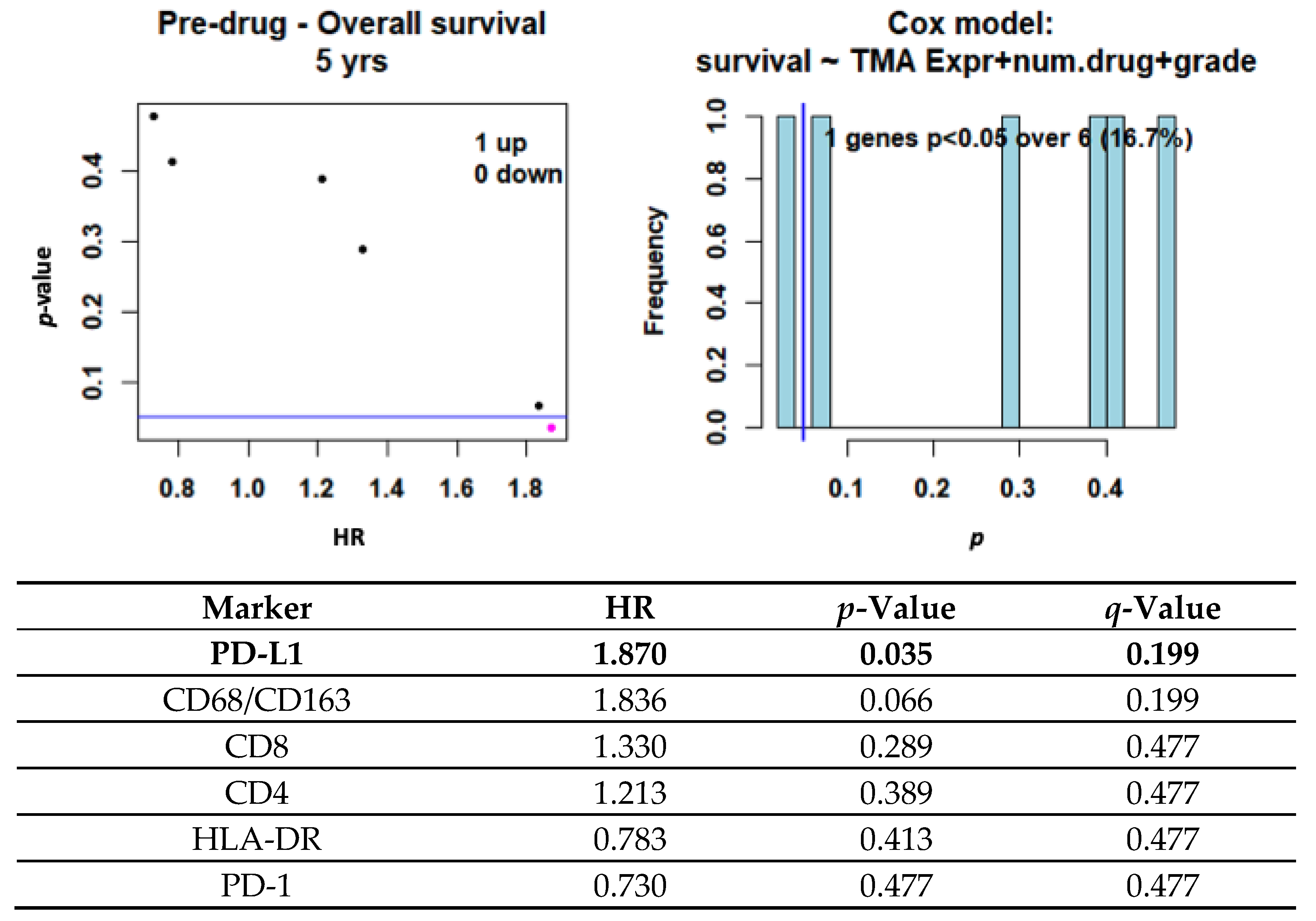
3.3. Impact of PD-L1 and Immune Infiltration on Clinical Outcome
3.4. NanoString Data
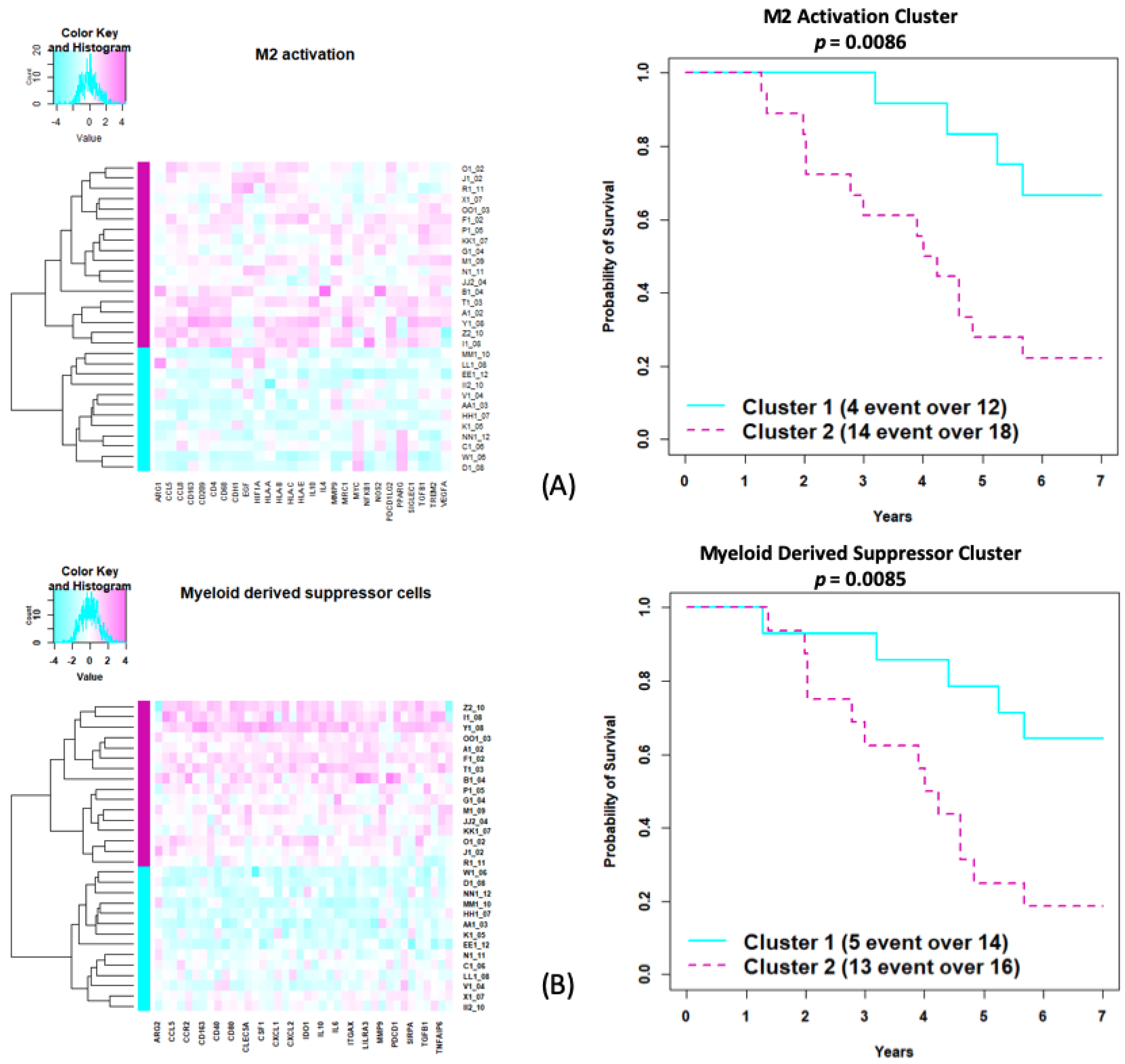
3.5. Exploratory Immunologic Analysis Using MiRKAT-S
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Demetri, G.D.; von Mehren, M.; Jones, R.L.; Hensley, M.L.; Schuetze, S.M.; Staddon, A.; Milhem, M.; Elias, A.; Ganjoo, K.; Tawbi, H.; et al. Efficacy and Safety of Trabectedin or Dacarbazine for Metastatic Liposarcoma or Leiomyosarcoma After Failure of Conventional Chemotherapy: Results of a Phase III Randomized Multicenter Clinical Trial. J. Clin. Oncol. 2016, 34, 786–793. [Google Scholar] [CrossRef]
- Kawai, A.; Araki, N.; Sugiura, H.; Ueda, T.; Yonemoto, T.; Takahashi, M.; Morioka, H.; Hiraga, H.; Hiruma, T.; Kunisada, T.; et al. Trabectedin monotherapy after standard chemotherapy versus best supportive care in patients with advanced, translocation-related sarcoma: A randomised, open-label, phase 2 study. Lancet Oncol. 2015, 16, 406–416. [Google Scholar] [CrossRef]
- Cuevas, C.; Francesch, A. Development of Yondelis (trabectedin, ET-743). A semisynthetic process solves the supply problem. Nat. Prod. Rep. 2009, 26, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.K.; Galmarini, C.M.; D’Incalci, M. Unique features of trabectedin mechanism of action. Cancer Chemother. Pharmacol. 2016, 77, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Di Giandomenico, S.; Frapolli, R.; Bello, E.; Uboldi, S.; Licandro, S.A.; Marchini, S.; Beltrame, L.; Brich, S.; Mauro, V.; Tamborini, E.; et al. Mode of action of trabectedin in myxoid liposarcomas. Oncogene 2014, 33, 5201–5210. [Google Scholar] [CrossRef] [Green Version]
- Germano, G.; Frapolli, R.; Simone, M.; Tavecchio, M.; Erba, E.; Pesce, S.; Pasqualini, F.; Grosso, F.; Sanfilippo, R.; Casali, P.G.; et al. Antitumor and anti-inflammatory effects of trabectedin on human myxoid liposarcoma cells. Cancer Res. 2010, 70, 2235–2244. [Google Scholar] [CrossRef] [Green Version]
- Germano, G.; Frapolli, R.; Belgiovine, C.; Anselmo, A.; Pesce, S.; Liguori, M.; Erba, E.; Uboldi, S.; Zucchetti, M.; Pasqualini; et al. Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell 2013, 23, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Blay, J.; Italiano, A.; Ray-Coquard, I.; Le Cesne, A.; Duffaud, F.; Rios, M.; Collard, O.; Bertucci, F.; Bompas, E.; Isambert, N.; et al. Long-term outcome and effect of maintenance therapy in patients with advanced sarcoma treated with trabectedin: An analysis of 181 patients of the French ATU compassionate use program. BMC Cancer 2013, 64, 13. [Google Scholar] [CrossRef] [Green Version]
- Grosso, F.; Jones, R.L.; Demetri, G.D.; Judson, I.R.; Blay, J.Y.; Le Cesne, A.; Sanfilippo, R.; Casieri, P.; Collini, P.; Dileo, P.; et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: A retrospective study. Lancet Oncol. 2007, 8, 595–602. [Google Scholar] [CrossRef]
- Plantinga, A.; Zhan, X.; Zhao, N.; Chen, J.; Jeng, R.R.; Wu, M.C. MiRKAT-S: A community level test of association between the microbiota and survival times. Microbiome 2017, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Perry Fell, H.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Payton, J.E.; Grieselhuber, N.R.; Chang, L.W.; Murakami, M.; Geiss, G.K.; Link, D.C.; Nagarajan, R.; Watson, M.A.; Ley, T.J. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J. Clin. Investig. 2009, 119, 1714–1726. [Google Scholar] [CrossRef]
- Kim, S. ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients. Commun. Stat. Appl. Methods 2015, 22, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollack, S.M.; He, Q.; Yearley, J.H.; Emerson, R.; Vignali, M.; Zhang, Y.; Redman, M.W.; Baker, K.K.; Cooper, S.; Donahue, B.; et al. T cell infiltration and clonality correlate with programmed cell death protein 1 and programmed death-ligand 1 expression in patients with soft tissue sarcomas. Cancer 2017, 123, 3291–3304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollack, S.M.; Junbluth, A.A.; Hoch, B.L.; Farrar, E.A.; Bleakley, M.; Schenider, D.J.; Loggers, E.T.; Rodler, E.; Eary, J.F.; Conrad, E.U.; et al. NY-ESO-1 is a ubiquitous immunotherapeutic target antigen for patients with myxoid/round cell liposarcoma. Cancer 2012, 118, 4564–4570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jungbluth, A.A.; Antonescu, C.R.; Busam, K.J.; Iverson, K.; Kolb, D.; Coplan, K.; Chen, Y.T.; Stockert, E.; Ladanyi, M.; Old, L.J. Monophasic and biphasic synovial sarcomas abundantly express cancer/testis antigen NY-ESO-1 but not MAGE-A1 or CT7. Int. J. Cancer 2001, 94, 252–256. [Google Scholar] [CrossRef] [Green Version]
- Allavena, P.; Signorelli, M.; Chieppa, M.; Erba, E.; Bianchi, G.; Marchesi, F.; Olimpio, C.O.; Bonardi, C.; Garbi, A.; Lissoni, A.; et al. Anti-inflammatory properties of the novel antitumor agent Yondelis (trabectedin): Inhibition of macrophage differentiation and cytokine production. Cancer Res. 2005, 65, 2964–2971. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Zhang, R.; Ivan, C.; Galletti, G.; Clise-Dwyer, K.; Barbaglio, F.; Sacrfo, L.; Aracil, M.; Klein, C.; Wierda, W.; et al. Trabectedin reveals a strategy of immunomodulation in chronic lymphocytic leukemia. Cancer Immunol. Res. 2019, 7, 2036–2051. [Google Scholar] [CrossRef]
- Schroeder, B.A.; LaFranzo, N.A.; LaFleur, B.J.; Gittelman, R.M.; Vignali, M.; Zhang, S.; Flanagan, K.C.; Rytlewski, J.; Riolobos, L.; Schulte, B.C.; et al. CD4+ T cell and M2 macrophage infiltration predict dedifferentiate liposarcoma patient outcomes. J. Immunother. Cancer 2021, 9, e002812. [Google Scholar] [CrossRef]
- Tseng, W.W.; Maul, S.; Zhang, M.; Chen, J.; Sim, G.C.; Wei, W.; Ingram, D.; Somaiah, N.; Lev, D.C.; Pollock, R.E.; et al. Analysis of the intratumoral adaptive immune response in well differentiated and dedifferentiated retroperitoneal liposarcoma. Sarcoma 2015, 2015, 547460. [Google Scholar] [CrossRef]
- Conley, A.P.; Wang, W.L.; Livingston, J.A.; Ravi, V.; Tsai, J.W.; Ali, A.; Ingram, D.R.; Lowery, C.D.; Roland, C.L.; Somaiah, N.; et al. MAGE-A3 is a clinically relevant target in undifferentiated pleopmorphic sarcoma/myxofibrosarcoma. Cancers 2019, 11, 677. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Mild-Treated (N = 93) | Heavy-Treated (N = 54) | Total (N = 147) | p-Value | |
|---|---|---|---|---|
| Age | 0.35 | |||
| 20–30 | 8 (8.6%) | 4 (7.4%) | 12 (8.2%) | |
| 30–40 | 5 (5.4%) | 6 (11.1%) | 11 (7.5%) | |
| 40–50 | 16 (17.2%) | 6 (11.1%) | 22 (15.0%) | |
| 50–60 | 33 (35.5%) | 17 (31.5%) | 50 (34.0%) | |
| 60–70 | 21 (22.6%) | 14 (25.9%) | 35 (23.8%) | |
| 70–80 | 6 (6.5%) | 7 (13.0%) | 13 (8.8%) | |
| 80–90 | 4 (4.3%) | 0 (0.0%) | 4 (2.7%) | |
| Gender | 0.027 | |||
| Female | 50 (53.8%) | 39 (72.2%) | 89 (60.5%) | |
| Male | 43 (46.2%) | 15 (27.8%) | 58 (39.5%) | |
| Tumor Grade | 0.047 | |||
| Unknown | 3 (3.2%) | 7 (13.0%) | 10 (6.8%) | |
| High | 70 (75.3%) | 31 (57.4%) | 101 (68.7%) | |
| Intermediate | 15 (16.1%) | 10 (18.5%) | 25 (17.0%) | |
| Low | 5 (5.4%) | 6 (11.1%) | 11 (7.5%) | |
| Num Trabectedin Treatments | <0.001 | |||
| Mean (SD) | 2.323 (0.980) | 11.296 (5.732) | 5.619 (5.601) | |
| Range | 1–4 | 5–25 | 1–25 | |
| Num Previous Chemo Regimens | 0.21 | |||
| Mean (SD) | 2.032 (1.137) | 1.759 (1.466) | 1.932 (1.270) | |
| Range | 0–4 | 0–7 | 0–7 | |
| Vital Status | 0.892 | |||
| Alive | 8 (8.6%) | 5 (9.3%) | 13 (8.8%) | |
| Death | 85 (91.4%) | 49 (90.7%) | 134 (91.2%) | |
| Follow-Up Time (years) | <0.001 | |||
| N-Miss | 1 | 0 | 1 | |
| Mean (SD) | 3.826 (3.405) | 7.249 (7.621) | 5.092 (5.591) | |
| Range | 0.47–17.95 | 0.54–42.46 | 0.47–42.46 | |
| Trabectedin Time to Failure (years) | <0.001 | |||
| Mean (SD) | 0.116 (0.157) | 0.798 (0.512) | 0.366 (0.469) | |
| Range | 0–1.06 | 0.23–2.59 | 0–2.59 | |
| Pathology | 0.344 | |||
| Leiomyosarcoma, Nonuterine | 14 (15.5%) | 10 (18.5%) | 24 (16.3%) | |
| Leiomyosarcoma, Uterine | 9 (9.7%) | 2 (3.7%) | 11 (7.5%) | |
| Myxoid/Round Cell Liposarcoma | 2 (2.2%) | 4 (7.4%) | 6 (4.1%) | |
| Synovial Sarcoma | 16 (17.2%) | 6 (11.1%) | 22 (15.0%) | |
| UPS/Spindle Cell Sarcoma | 17 (18.3%) | 8 (14.8%) | 25 (17.0%) | |
| Other | 35 (37.6%) | 26 (48.1%) | 59 (40.1%) | |
| Name | Number of Genes from Original | Number of Genes in Data | Genes in Data | p-Value | q-Value |
|---|---|---|---|---|---|
| Merck 18-gene | 18 | 17 | CCL5, CD27, CD274, CD276, CMKLR1, CXCL9, CXCR6, HLA-DQA1, HLA-DRB1, HLA-E, IDO1, LAG3, NKG7, PDCD1LG2, PSMB10, STAT1, TIGIT | 0.041 | 0.058 |
| 6-gene IFNg | 6 | 6 | CXCL10, CXCL9, HLA-DRA, IDO1, IFNG, STAT1 | 0.061 | 0.061 |
| 10-gene IFNg | 10 | 10 | CCR5, CXCL10, CXCL11, CXCL9, GZMA, HLA-DRA, IDO1, IFNG, PRF1, STAT1 | 0.048 | 0.058 |
| TCR signaling | 12 | 9 | CCL5, CD27, CD3D, CD3G, CD4, CD74, IL2, LCK, TIGIT | 0.048 | 0.058 |
| M1 activation | 28 | 24 | CCL19, CCL5, CCL8, CD38, CD40, CXCL10, CXCL11, CXCL13, CXCL9, HLA-DPB1, HLA-DQA1, HLA-DRA, HLA-DRB1, IDO1, IFNG, IL2RA, IL6, LAG3, LILRA3, RSAD2, SIGLEC1, STAT1, TNF, TNFAIP6 | 0.039 | 0.058 |
| M2 activation | 35 | 27 | ARG1, CCL5, CCL8, CD163, CD209, CD4, CD68, CDH1, EGF, HIF1A, HLA-A, HLA-B, HLA-C, HLA-E, IL10, IL4, MMP9, MRC1, MYC, NFKB1, NOS2, PDCD1LG2, PPARG, SIGLEC1, TGFB1, TREM2, VEGFA | 0.03 | 0.058 |
| T cell activation CD8 | 48 | 35 | CCL5, CD2, CD247, CD27, CD3D, CD3E, CD3G, CD6, CD69, CD7, CD8B, CD96, DPP4, GNLY GZMA, GZMB, GZMK, ICOS, IL7R, IRF8, KLRB1, KLRD1, KLRK1, LAG3, LCK, LTB, LY9, NKG7, PDCD1, PDCD1LG2, PRF1, PVRIG, SH2D1A, TRAT1, ZAP70 | 0.052 | 0.058 |
| Regulatory T cells | 33 | 23 | CD2, CD247, CD27, CD28, CD3D, CD3E, CD3G, CD4, CD5, CD6, CD70, CD96, CTLA4, DPP4, FOXP3, ICOS, IL2RA, IL2RB, LCK, LTB, SH2D1A, TRAT1, ZAP70 | 0.051 | 0.058 |
| Myeloid-derived suppressor cells | 55 | 40 | ARG2, BTLA, CCL5, CCL8, CCR2, CD14, CD163, CD274, CD40, CD44, CD80, CD86, CLEC5A, CLEC7A, CSF1, CSF1R, CXCL1, CXCL10, CXCL2, HLA-DPB1, IDO1, IFNG, IL10, IL10RA, IL6, ITGAM, ITGAX, LILRA1, LILRA3, LILRA5, MMP9, NOS2, PDCD1, S100A8, SIRPA, STAT1, TGFB1, TGFBR2, TNFAIP6, VEGFA | 0.023 | 0.058 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schroeder, B.A.; Zhang, Y.; Smythe, K.S.; Desai, P.; Thomas, A.; Viveiros, P.; Alexiev, B.A.; Obeidin, F.; Chen, E.Y.; Cranmer, L.D.; et al. Immunologic Gene Signature Analysis Correlates Myeloid Cells and M2 Macrophages with Time to Trabectedin Failure in Sarcoma Patients. Cancers 2022, 14, 1290. https://doi.org/10.3390/cancers14051290
Schroeder BA, Zhang Y, Smythe KS, Desai P, Thomas A, Viveiros P, Alexiev BA, Obeidin F, Chen EY, Cranmer LD, et al. Immunologic Gene Signature Analysis Correlates Myeloid Cells and M2 Macrophages with Time to Trabectedin Failure in Sarcoma Patients. Cancers. 2022; 14(5):1290. https://doi.org/10.3390/cancers14051290
Chicago/Turabian StyleSchroeder, Brett A., Yuzheng Zhang, Kimberly S. Smythe, Parth Desai, Anish Thomas, Pedro Viveiros, Borislav A. Alexiev, Farres Obeidin, Eleanor Y. Chen, Lee D. Cranmer, and et al. 2022. "Immunologic Gene Signature Analysis Correlates Myeloid Cells and M2 Macrophages with Time to Trabectedin Failure in Sarcoma Patients" Cancers 14, no. 5: 1290. https://doi.org/10.3390/cancers14051290






