Comprehensive Analysis of the Prognosis and Drug Sensitivity of Differentiation-Related lncRNAs in Papillary Thyroid Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Data Acquisition
2.2. Protein–Protein Interaction (PPI) Network
2.3. Construction of the Prognostic Risk Assessment Model
2.4. GO and KEGG Pathway Enrichment Functional Analysis
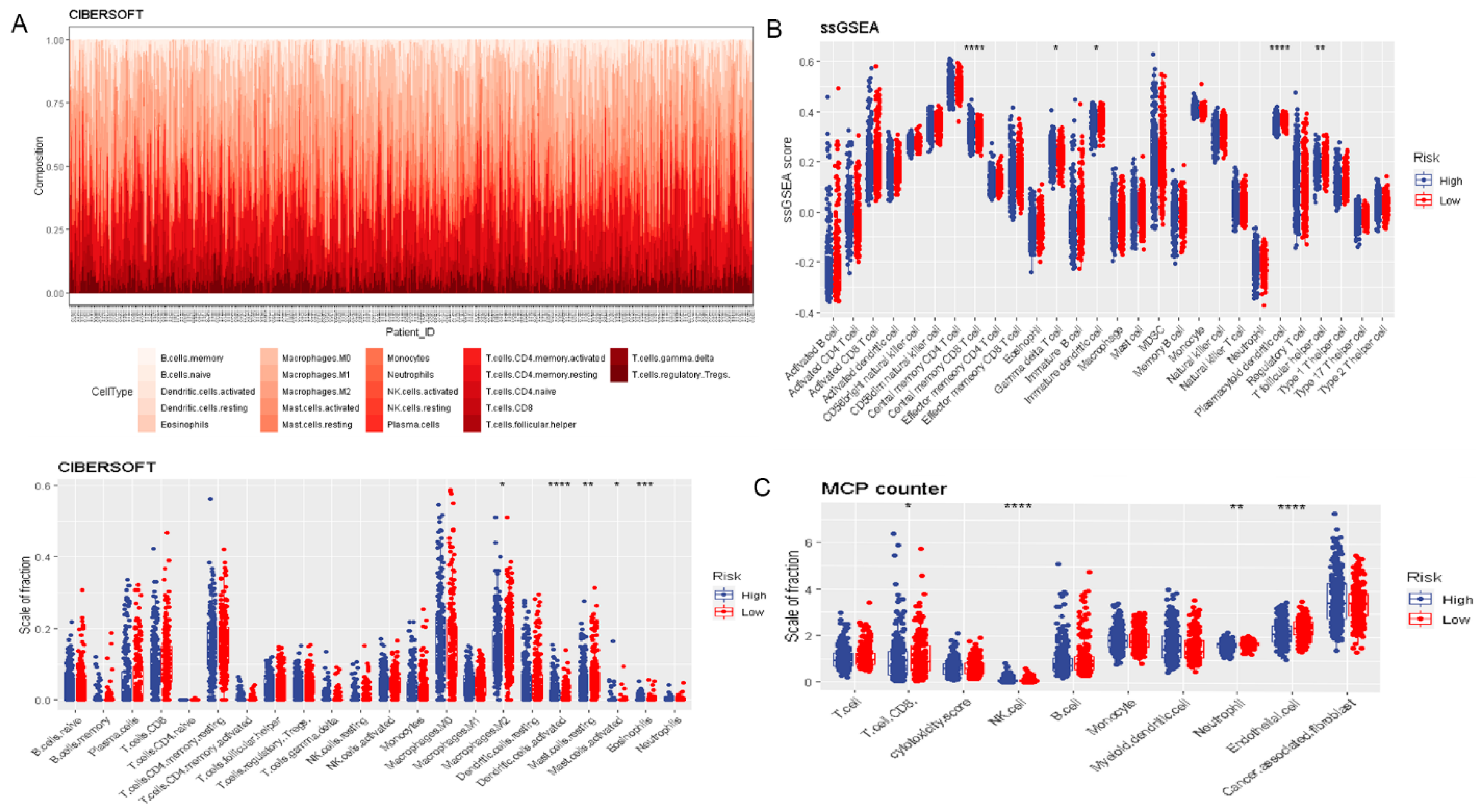
2.5. Evaluation of Immune Infiltration and the Expression of Immune Checkpoints
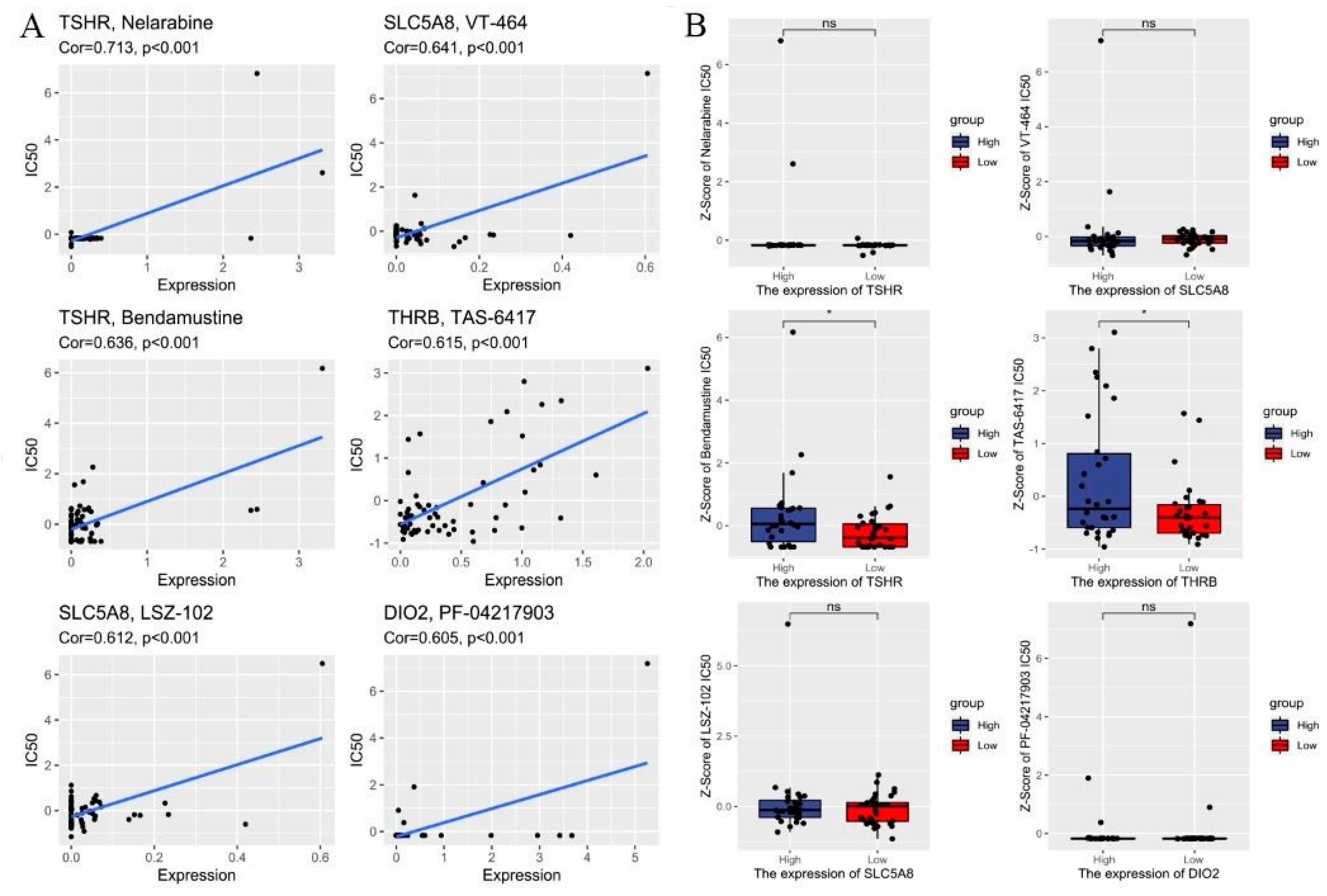
2.6. Drug Susceptibility Prediction
2.7. Tissue, Cell Lines, and Cell Transfection
2.8. RNA Extraction and Quantitative Reverse Transcription-PCR (qRT-PCR)
2.9. Western Blotting
2.10. Cell Counting Kit (CCK)-8 Assay
2.11. 5-Ethynyl-2-deoxyuridine (EdU) Assay
2.12. Transwell Migration and Invasion Assay
2.13. Statistical Analysis
3. Results
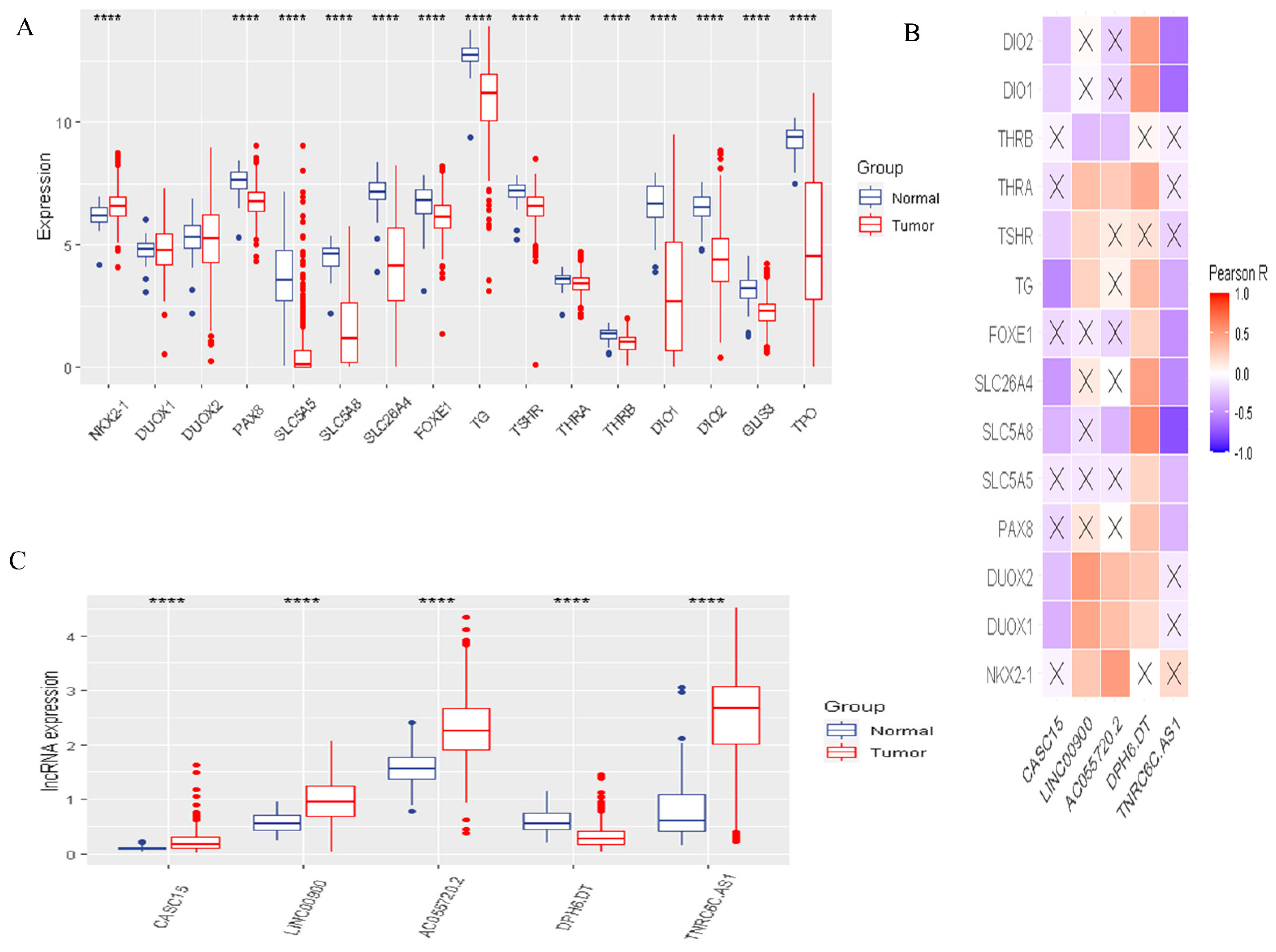
3.1. The Landscape of DR-lncRNA Regulators
3.2. Risk Score Was Associated with Prognosis and Tumor Immune Microenvironment
3.3. Functional and Pathway Enrichment Analysis
3.4. Drug Sensitivity Analysis
3.5. Construction of the DR-lncRNAs Prognosis Nomogram
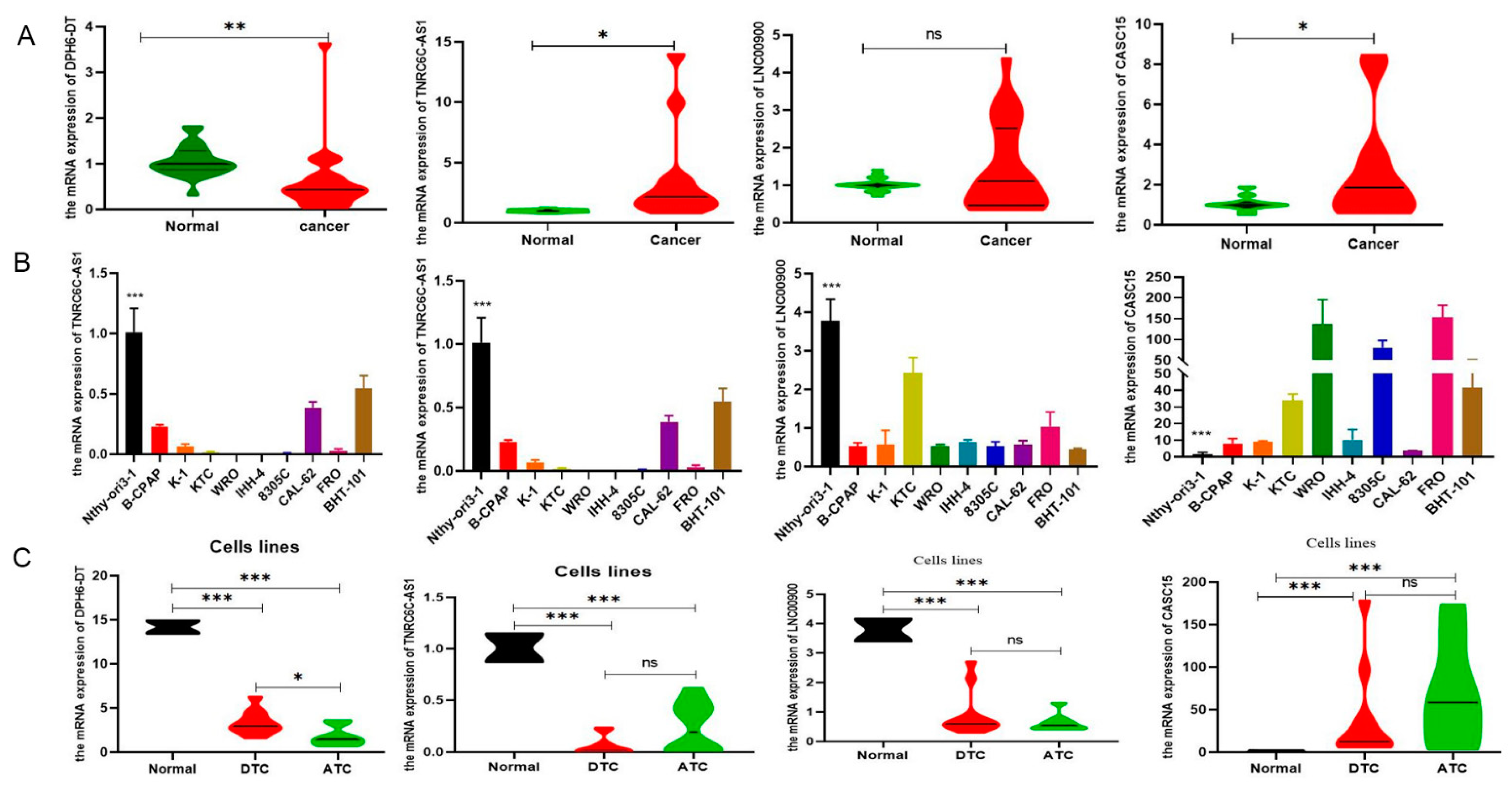
3.6. Validation of the Expression of DR-lncRNAs
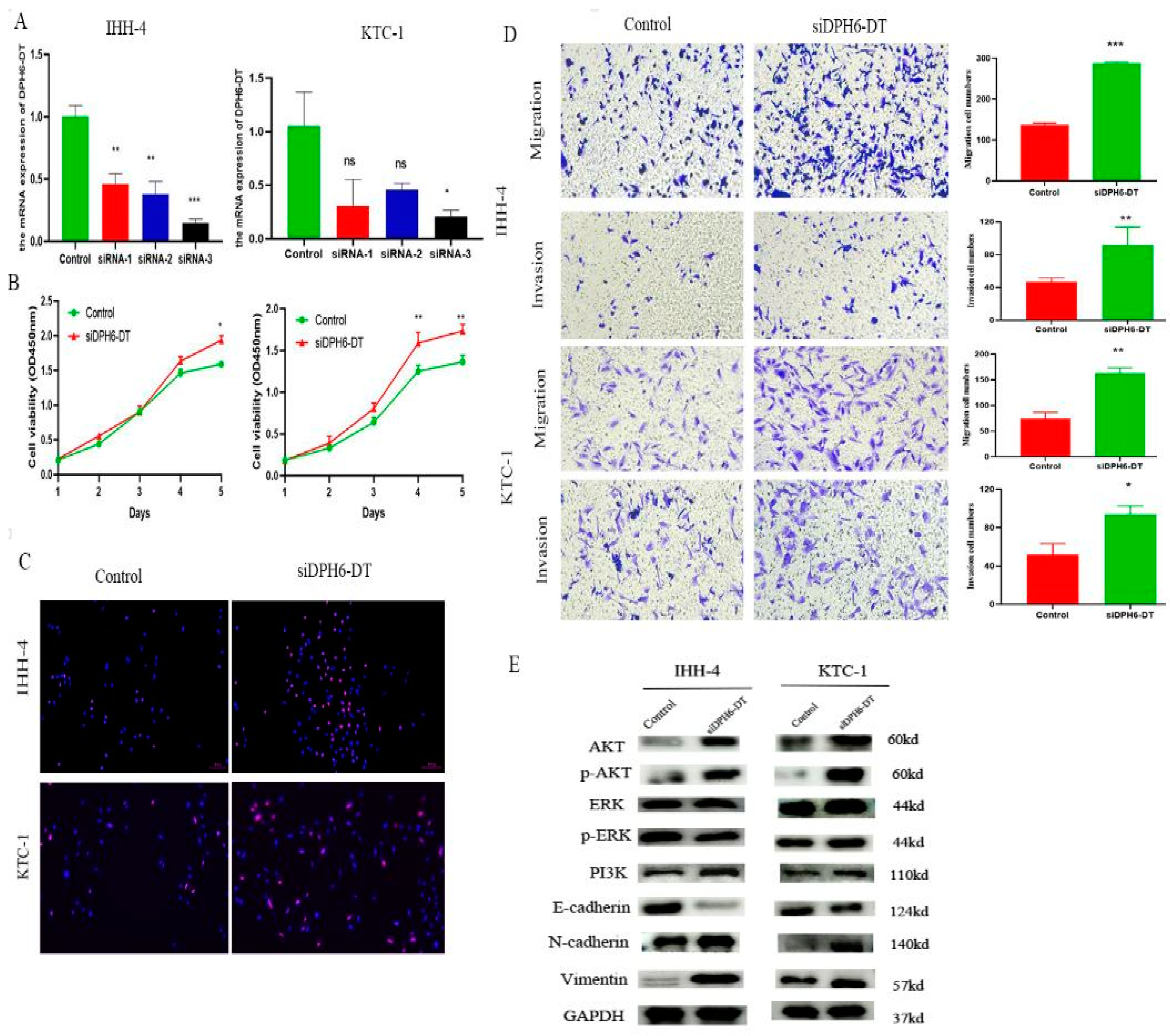
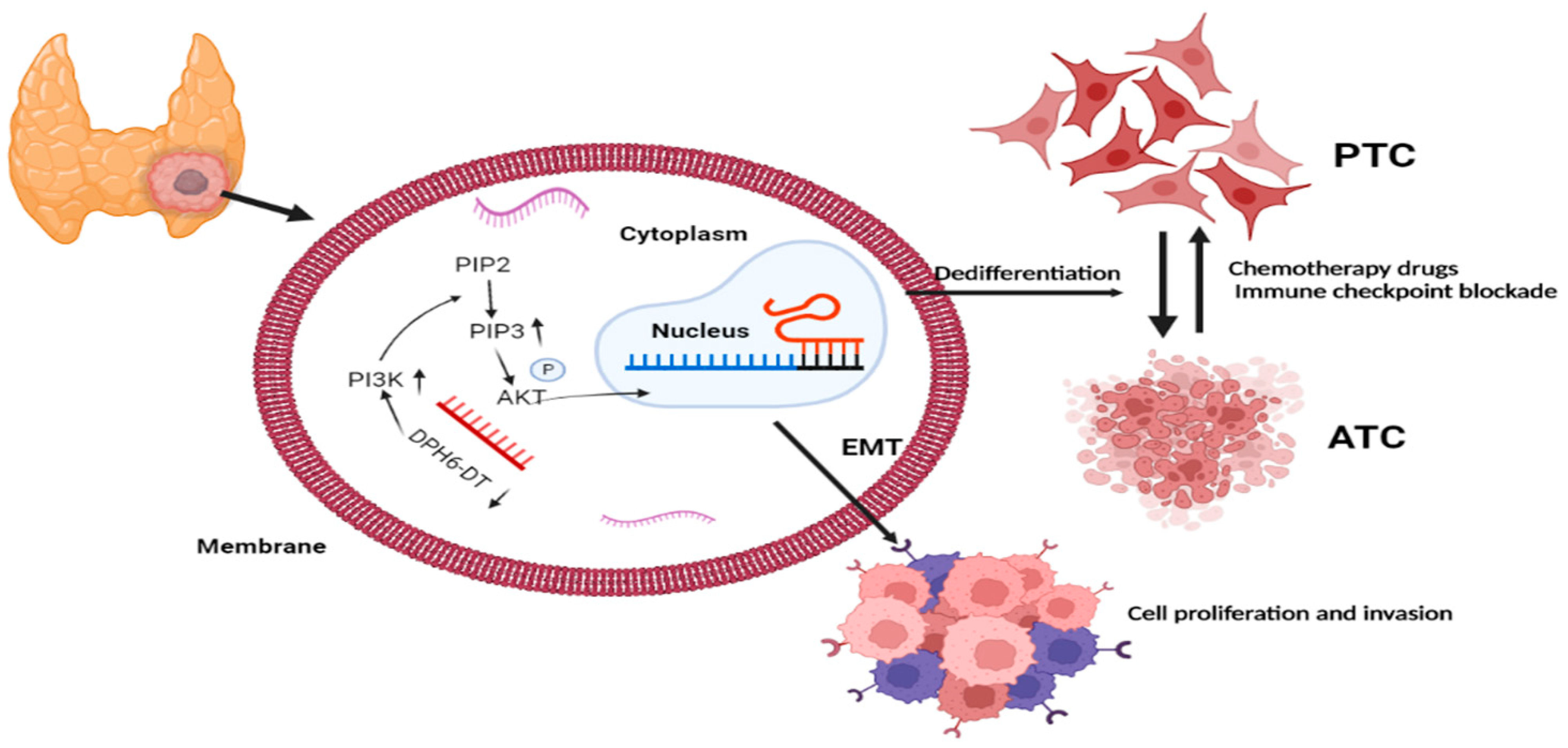
3.7. Downregulation of DPH6-DT Promote Proliferation and Metastasis by Activating the PI3K-AKT Signaling Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.; Sun, K.; Zheng, R.; Zeng, H.; Wang, S.; Chen, R.; Wei, W.; He, J. Cancer incidence and mortality in China, 2015. J. Natl. Cancer Cent. 2021, 1, 2–11. [Google Scholar] [CrossRef]
- Wang, W.; Shen, C.; Zhao, Y.; Sun, B.; Bai, N.; Li, X. Identification and validation of potential novel biomarkers to predict distant metastasis in differentiated thyroid cancer. Ann. Transl. Med. 2021, 9, 1053. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.W.; Kim, H.J.; Kim, Y.H.; Park, S.H.; Chwae, Y.J.; Lee, J.; Soh, E.Y.; Kim, J.H.; Park, T.J. B-RafV600E inhibits sodium iodide symporter expression via regulation of DNA methyltransferase 1. Exp. Mol. Med. 2014, 46, e120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.; Liu, R.; Zhu, G.; Wang, H.; Xing, M. Robust Thyroid Gene Expression and Radioiodine Uptake Induced by Simultaneous Suppression of BRAF V600E and Histone Deacetylase in Thyroid Cancer Cells. J. Clin. Endocrinol. Metab. 2016, 101, 962–971. [Google Scholar] [CrossRef] [Green Version]
- Hardin, H.; Montemayor-Garcia, C.; Lloyd, R.V. Thyroid cancer stem-like cells and epithelial-mesenchymal transition in thyroid cancers. Hum. Pathol. 2013, 44, 1707–1713. [Google Scholar] [CrossRef]
- Riesco-Eizaguirre, G.; Wert-Lamas, L.; Perales-Paton, J.; Sastre-Perona, A.; Fernandez, L.P.; Santisteban, P. The miR-146b-3p/PAX8/NIS Regulatory Circuit Modulates the Differentiation Phenotype and Function of Thyroid Cells during Carcinogenesis. Cancer Res. 2015, 75, 4119–4130. [Google Scholar] [CrossRef] [Green Version]
- Galdiero, M.R.; Varricchi, G.; Marone, G. The immune network in thyroid cancer. Oncoimmunology 2016, 5, e1168556. [Google Scholar] [CrossRef] [Green Version]
- Gewirtz, D.A. An autophagic switch in the response of tumor cells to radiation and chemotherapy. Biochem. Pharmacol. 2014, 90, 208–211. [Google Scholar] [CrossRef]
- Jin, S.M.; Jang, H.W.; Sohn, S.Y.; Kim, N.K.; Joung, J.Y.; Cho, Y.Y.; Kim, S.W.; Chung, J.H. Role of autophagy in the resistance to tumour necrosis factor-related apoptosis-inducing ligand-induced apoptosis in papillary and anaplastic thyroid cancer cells. Endocrine 2014, 45, 256–262. [Google Scholar] [CrossRef]
- Schaukowitch, K.; Kim, T.K. Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience 2014, 264, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Kraus, W.L. From discovery to function: The expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Seyed Saleh, S.H.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-kappaB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, S.; Chen, D.; Yuwen, D.; Zhang, J.; Wei, X.; Han, X.; Guan, X. SOX2-OT induced by PAI-1 promotes triple-negative breast cancer cells metastasis by sponging miR-942-5p and activating PI3K/Akt signaling. Cell. Mol. Life Sci. 2022, 79, 59. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Duan, Y.; Sang, Y.; Li, Y.; Zhang, H.; Liang, Y.; Liu, Y.; Zhang, N.; Yang, Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J. Cell. Physiol. 2019, 234, 9105–9117. [Google Scholar] [CrossRef]
- Ashrafizaveh, S.; Ashrafizadeh, M.; Zarrabi, A.; Husmandi, K.; Zabolian, A.; Shahinozzaman, M.; Aref, A.R.; Hamblin, M.R.; Nabavi, N.; Crea, F.; et al. Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett. 2021, 508, 104–114. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Yu, J.; Yao, X.; Yang, S.; Li, W.; Xu, L.; Zhao, L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol. Cancer 2021, 20, 6. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, Z.; Li, S.; Wang, X.; Zhou, X. LncRNA SPRY4-IT was concerned with the poor prognosis and contributed to the progression of thyroid cancer. Cancer Gene Ther. 2018, 25, 39–46. [Google Scholar] [CrossRef]
- Li, H.M.; Yang, H.; Wen, D.Y.; Luo, Y.H.; Liang, C.Y.; Pan, D.H.; Ma, W.; Chen, G.; He, Y.; Chen, J.Q. Overexpression of LncRNA HOTAIR is Associated with Poor Prognosis in Thyroid Carcinoma: A Study Based on TCGA and GEO Data. Horm. Metab. Res. 2017, 49, 388–399. [Google Scholar] [CrossRef]
- Fan, M.; Li, X.; Jiang, W.; Huang, Y.; Li, J.; Wang, Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp. Ther. Med. 2013, 5, 1143–1146. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudian-Sani, M.R.; Jalali, A.; Jamshidi, M.; Moridi, H.; Alghasi, A.; Shojaeian, A.; Mobini, G.R. Long Non-Coding RNAs in Thyroid Cancer: Implications for Pathogenesis, Diagnosis, and Therapy. Oncol. Res. Treat. 2019, 42, 136–142. [Google Scholar] [CrossRef]
- Jing, W.; Li, X.; Peng, R.; Lv, S.; Zhang, Y.; Cao, Z.; Tu, J.; Ming, L. The diagnostic and prognostic significance of long noncoding RNAs expression in thyroid cancer: A systematic review and meta-analysis. Pathol. Res. Pract. 2018, 214, 327–334. [Google Scholar] [CrossRef] [PubMed]
- French, J.D.; Bible, K.; Spitzweg, C.; Haugen, B.R.; Ryder, M. Leveraging the immune system to treat advanced thyroid cancers. Lancet Diabetes Endocrinol. 2017, 5, 469–481. [Google Scholar] [CrossRef]
- Wang, X.; Peng, W.; Li, C.; Qin, R.; Zhong, Z.; Sun, C. Identification of an immune-related signature indicating the dedifferentiation of thyroid cells. Cancer Cell Int. 2021, 21, 231. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Li, X.; He, Y.; Wu, S.; Wang, S.; Sun, J.; He, Y.; Lun, Y.; Zhang, J. Immune Cell Confrontation in the Papillary Thyroid Carcinoma Microenvironment. Front. Endocrinol. 2020, 11, 570604. [Google Scholar] [CrossRef]
- Kim, B.H. The expression of tumor-associated macrophages in papillary thyroid carcinoma. Endocrinol. Metab. 2013, 28, 178–179. [Google Scholar] [CrossRef] [Green Version]
- Cameselle-Garcia, S.; Abdulkader-Sande, S.; Sanchez-Ares, M.; Rodriguez-Carnero, G.; Garcia-Gomez, J.; Gude-Sampedro, F.; Abdulkader-Nallib, I.; Cameselle-Teijeiro, J.M. PD-L1 expression and immune cells in anaplastic carcinoma and poorly differentiated carcinoma of the human thyroid gland: A retrospective study. Oncol. Lett. 2021, 22, 553. [Google Scholar] [CrossRef]
- Tesselaar, M.H.; Smith, J.W.; Nagarajah, J.; Netea-Maier, R.T.; Plantinga, T.S. Pathological processes and therapeutic advances in radioiodide refractory thyroid cancer. J. Mol. Endocrinol. 2017, 59, R141–R154. [Google Scholar] [CrossRef] [Green Version]
- Lei, Q.; Wang, D.; Sun, K.; Wang, L.; Zhang, Y. Resistance Mechanisms of Anti-PD1/PDL1 Therapy in Solid Tumors. Front. Cell Dev. Biol. 2020, 8, 672. [Google Scholar] [CrossRef]
- Gunda, V.; Gigliotti, B.; Ndishabandi, D.; Ashry, T.; McCarthy, M.; Zhou, Z.; Amin, S.; Freeman, G.J.; Alessandrini, A.; Parangi, S. Combinations of BRAF inhibitor and anti-PD-1/PD-L1 antibody improve survival and tumour immunity in an immunocompetent model of orthotopic murine anaplastic thyroid cancer. Br. J. Cancer 2018, 119, 1223–1232. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 2020, 26, 903–909. [Google Scholar] [CrossRef] [Green Version]
- Nishant, A.; Rehan, A.; Arman, A.B.; Adrian, A.; Harindra, A.; Sylvia, L.A.; Todd, A.J.; Miruna, B.; Saianand, B.; Stephen, B.B.; et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar]
- Dom, G.; Tarabichi, M.; Unger, K.; Thomas, G.; Oczko-Wojciechowska, M.; Bogdanova, T.; Jarzab, B.; Dumont, J.E.; Detours, V.; Maenhaut, C. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br. J. Cancer 2012, 107, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Yi, M.; Nissley, D.V.; McCormick, F.; Stephens, R.M. ssGSEA score-based Ras dependency indexes derived from gene expression data reveal potential Ras addiction mechanisms with possible clinical implications. Sci. Rep. 2020, 10, 10258. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Varma, S.; Doroshow, J.H.; Pommier, Y. Using CellMiner 1.6 for Systems Pharmacology and Genomic Analysis of the NCI-60. Clin. Cancer Res. 2015, 21, 3841–3852. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.X.; Wu, J.; Guo, M.L.; Zhang, Y.; Ma, S.G. Suppression of long non-coding RNA TNRC6C-AS1 protects against thyroid carcinoma through DNA demethylation of STK4 via the Hippo signalling pathway. Cell Prolif. 2019, 52, e12564. [Google Scholar] [CrossRef]
- Bai, D.; Guo, C.; Wang, A.; Pang, G.; Gao, J.; Wang, C.; Zhao, D.; Yang, J.; Ren, J. LncRNA CASC15 promotes the proliferation of papillary thyroid carcinoma cells by regulating the miR-7151-5p/WNT7A axis. Pathol. Res. Pract. 2021, 225, 153561. [Google Scholar] [CrossRef]
- Aashiq, M.; Silverman, D.A.; Na’ara, S.; Takahashi, H.; Amit, M. Radioiodine-Refractory Thyroid Cancer: Molecular Basis of Redifferentiation Therapies, Management, and Novel Therapies. Cancers 2019, 11, 1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, J.M.; Ahn, B.C. Molecular mechanisms of radioactive iodine refractoriness in differentiated thyroid cancer: Impaired sodium iodide symporter (NIS) expression owing to altered signaling pathway activity and intracellular localization of NIS. Theranostics 2021, 11, 6251–6277. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, L.; Jenkins, J.; Purvis, G.; Lee, J.; Franco, A.T. The Thyroid Tumor Microenvironment: Potential Targets for Therapeutic Intervention and Prognostication. Horm. Cancer 2020, 11, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Naoum, G.E.; Morkos, M.; Kim, B.; Arafat, W. Novel targeted therapies and immunotherapy for advanced thyroid cancers. Mol. Cancer 2018, 17, 51. [Google Scholar] [CrossRef]
- Varricchi, G.; Loffredo, S.; Marone, G.; Modestino, L.; Fallahi, P.; Ferrari, S.M.; de Paulis, A.; Antonelli, A.; Galdiero, M.R. The Immune Landscape of Thyroid Cancer in the Context of Immune Checkpoint Inhibition. Int. J. Mol. Sci. 2019, 20, 3934. [Google Scholar] [CrossRef] [Green Version]
- Elia, G.; Ferrari, S.M.; Galdiero, M.R.; Ragusa, F.; Paparo, S.R.; Ruffilli, I.; Varricchi, G.; Fallahi, P.; Antonelli, A. New insight in endocrine-related adverse events associated to immune checkpoint blockade. Best Pract. Res. Clin. Endocrinol. Metab. 2020, 34, 101370. [Google Scholar] [CrossRef]
- Chowdhury, S.; Veyhl, J.; Jessa, F.; Polyakova, O.; Alenzi, A.; MacMillan, C.; Ralhan, R.; Walfish, P.G. Programmed death-ligand 1 overexpression is a prognostic marker for aggressive papillary thyroid cancer and its variants. Oncotarget 2016, 7, 32318–32328. [Google Scholar] [CrossRef]
- Antonelli, A.; Ferrari, S.M.; Fallahi, P.; Frascerra, S.; Piaggi, S.; Gelmini, S.; Lupi, C.; Minuto, M.; Berti, P.; Benvenga, S.; et al. Dysregulation of secretion of CXC alpha-chemokine CXCL10 in papillary thyroid cancer: Modulation by peroxisome proliferator-activated receptor-gamma agonists. Endocr. Relat. Cancer 2009, 16, 1299–1311. [Google Scholar] [CrossRef] [Green Version]
- Spartalis, E.; Athanasiadis, D.I.; Chrysikos, D.; Spartalis, M.; Boutzios, G.; Schizas, D.; Garmpis, N.; Damaskos, C.; Paschou, S.A.; Ioannidis, A.; et al. Histone Deacetylase Inhibitors and Anaplastic Thyroid Carcinoma. Anticancer Res. 2019, 39, 1119–1127. [Google Scholar] [CrossRef]
- Renko, K.; Schache, S.; Hoefig, C.S.; Welsink, T.; Schwiebert, C.; Braun, D.; Becker, N.P.; Kohrle, J.; Schomburg, L. An Improved Nonradioactive Screening Method Identifies Genistein and Xanthohumol as Potent Inhibitors of Iodothyronine Deiodinases. Thyroid 2015, 25, 962–968. [Google Scholar] [CrossRef]
- LaCasce, A.S.; Bociek, R.G.; Sawas, A.; Caimi, P.; Agura, E.; Matous, J.; Ansell, S.M.; Crosswell, H.E.; Islas-Ohlmayer, M.; Behler, C.; et al. Brentuximab vedotin plus bendamustine: A highly active first salvage regimen for relapsed or refractory Hodgkin lymphoma. Blood 2018, 132, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Casella, C.; Ministrini, S.; Galani, A.; Mastriale, F.; Cappelli, C.; Portolani, N. The New TNM Staging System for Thyroid Cancer and the Risk of Disease Downstaging. Front. Endocrinol. 2018, 9, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, Y.; Lin, Y.; Liang, J. Radioactive Iodine-Refractory Differentiated Thyroid Cancer and Redifferentiation Therapy. Endocrinol. Metab. 2019, 34, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Jiang, H.; Wen, D.; Hu, J.; Han, L.; Liu, W.; Xu, W.; Shi, X.; Wei, W.; Liao, T.; et al. Transcriptome Analyses Identify a Metabolic Gene Signature Indicative of Dedifferentiation of Papillary Thyroid Cancer. J. Clin. Endocrinol. Metab. 2019, 104, 3713–3725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, H.Y.; Choi, H.; Paeng, J.C.; Cheon, G.J.; Chung, J.K.; Kang, K.W. Comprehensive gene expression analysis for exploring the association between glucose metabolism and differentiation of thyroid cancer. BMC Cancer 2019, 19, 1260. [Google Scholar] [CrossRef] [Green Version]
- Giuliani, C. The Flavonoid Quercetin Induces AP-1 Activation in FRTL-5 Thyroid Cells. Antioxidants 2019, 8, 112. [Google Scholar] [CrossRef] [Green Version]
- Hou, P.; Bojdani, E.; Xing, M. Induction of thyroid gene expression and radioiodine uptake in thyroid cancer cells by targeting major signaling pathways. J. Clin. Endocrinol. Metab. 2010, 95, 820–828. [Google Scholar] [CrossRef] [Green Version]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]



| Variable Total (%) N = 492 | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95%CI) | p | OR (95%CI) | p | |
| Age 47.01 ± 16.03 | 1.16 (1.10–1.22) | <0.001 | 1.15 (1.08–1.23) | <0.001 |
| Gender Female 361 (77.37) Male 131 (26.63) | Ref. 1.92 (0.69–5.29) | 0.21 | - | - |
| TNM Stage I 276 (56.10) II 52 (10.57) III 109 (22.15) IV 55 (11.18) | Ref. 5.67 (0.80–40.27) 10.25 (2.13–49.45) 16.47 (3.18–85.33) | 0.083 0.004 <0.001 | Ref. 0.77 (0.01–63.96) 0.62 (0.01–47.80) 0.10 (0–15.71) | 0.906 0.827 0.375 |
| T Stage T1 135 (27.44) T2 162 (32.93) T3 172 (34.96) T4 23 (4.67) | Ref. 1.10 (0.18–6.59) 1.69 (0.33–8.73) 11.52 (2.31–57.57) | 0.920 0.531 0.003 | Ref. 1.49 (0.02–100.44) 0.59 (0.01–41.61) 13.61 (0.11–1620.48) | 0.852 0.809 0.284 |
| N Stage N0 271 (55.08) N1 221 (44.92) | Ref. 1.14 (0.43–3.05) | 0.078 | - | - |
| M Stage M0 483 (98.17) M1 9 (1.83) | Ref. 5.39 (1.22–23.83) | 0.026 | Ref. 0.51 (0.05–4.68) | 0.55 |
| Multifocality Multifocal 228 (46.34) Unifocal 264 (53.66) | Ref. 3.91 (0.88–17.34) | 0.073 | - | - |
| Bilaterality Bilateral 84 (17.07) Unilateral 386 (78.46) Isthmus 22 (4.47) | Ref. 0.95 (0.21–4.26) 1.05 (0.09–11.79) | 0.942 0.967 | - | - |
| Risk score | 2.99 (2.08–4.31) | <0.001 | 2.38 (1.45–4.25) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Bai, N.; Li, X. Comprehensive Analysis of the Prognosis and Drug Sensitivity of Differentiation-Related lncRNAs in Papillary Thyroid Cancer. Cancers 2022, 14, 1353. https://doi.org/10.3390/cancers14051353
Wang W, Bai N, Li X. Comprehensive Analysis of the Prognosis and Drug Sensitivity of Differentiation-Related lncRNAs in Papillary Thyroid Cancer. Cancers. 2022; 14(5):1353. https://doi.org/10.3390/cancers14051353
Chicago/Turabian StyleWang, Wenlong, Ning Bai, and Xinying Li. 2022. "Comprehensive Analysis of the Prognosis and Drug Sensitivity of Differentiation-Related lncRNAs in Papillary Thyroid Cancer" Cancers 14, no. 5: 1353. https://doi.org/10.3390/cancers14051353
APA StyleWang, W., Bai, N., & Li, X. (2022). Comprehensive Analysis of the Prognosis and Drug Sensitivity of Differentiation-Related lncRNAs in Papillary Thyroid Cancer. Cancers, 14(5), 1353. https://doi.org/10.3390/cancers14051353





