Unravelling Prostate Cancer Heterogeneity Using Spatial Approaches to Lipidomics and Transcriptomics
Abstract
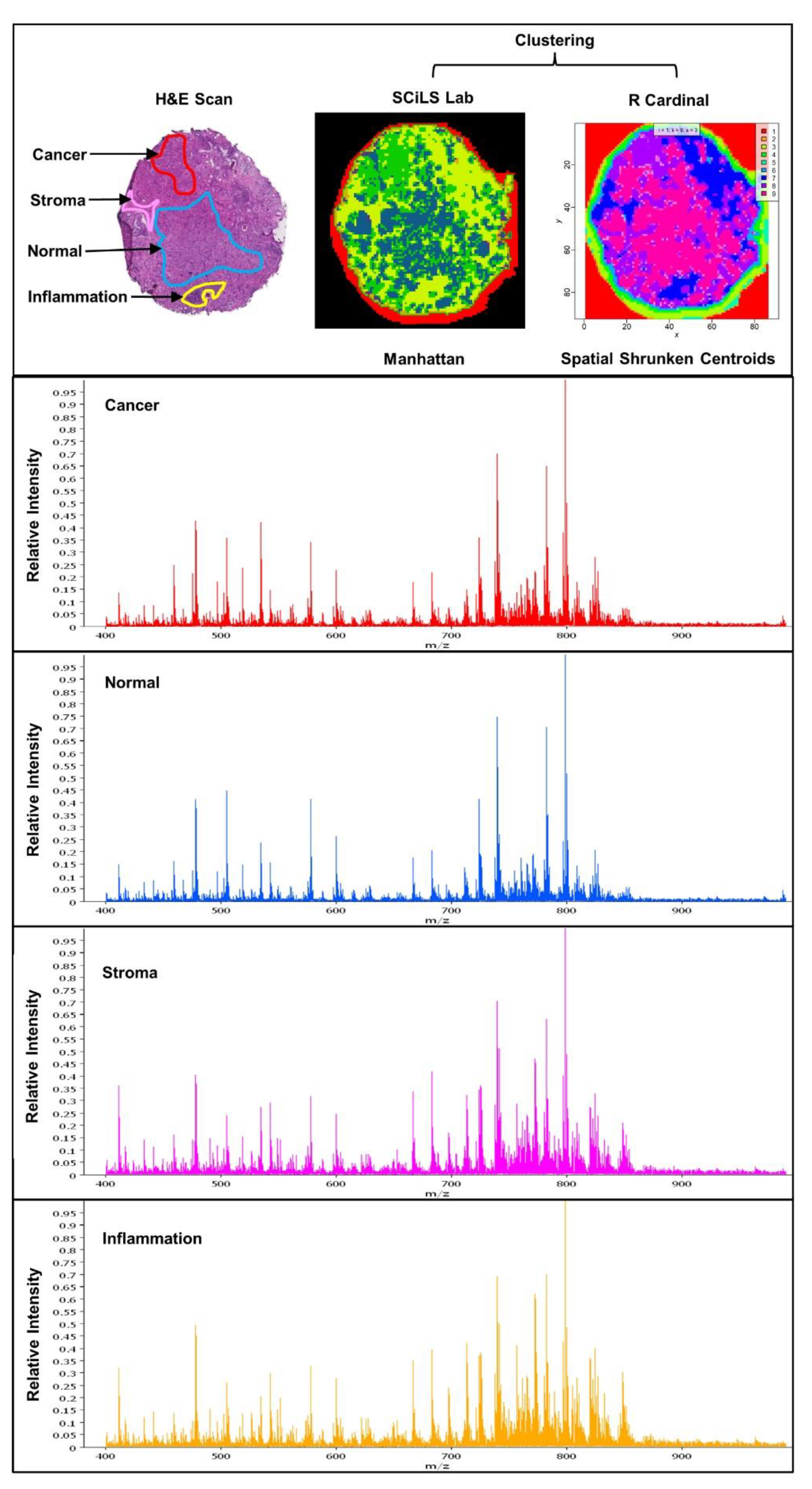
Simple Summary
Abstract
1. Prostate Cancer Heterogeneity
2. Rewired Lipid Metabolism as a Source of Biomarkers and Therapeutic Targets
3. The Need for Spatial Omics Approaches
4. Experimental Approaches in Spatial Lipidomics
5. Spatial Lipidomics Applied to Prostate Cancer
6. Complementary Spatial Transcriptomics to Map Alterations in Lipid Metabolism in PCa
7. Challenges and New Developments in Spatial Lipidomics and Combined Spatial Omics Approaches
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baade, P.D.; Youlden, D.R.; Krnjacki, L.J. International epidemiology of prostate cancer: Geographical distribution and secular trends. Mol. Nutr. Food Res. 2009, 53, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics, 2010. CA A Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef]
- Tonry, C.; Finn, S.; Armstrong, J.; Pennington, S.R. Clinical proteomics for prostate cancer: Understanding prostate cancer pathology and protein biomarkers for improved disease management. Clin. Proteom. 2020, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- McCullough, A.R. Sexual dysfunction after radical prostatectomy. Rev. Urol. 2005, 7 (Suppl. 2), S3–S10. [Google Scholar] [PubMed]
- Zhang, E.; Zhang, M.; Shi, C.; Sun, L.; Shan, L.; Zhang, H.; Song, Y. An overview of advances in multi-omics analysis in prostate cancer. Life Sci. 2020, 260, 118376. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A.; Grading, C. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- Bostwick, D.G.; Qian, J. High-grade prostatic intraepithelial neoplasia. Mod. Pathol. 2004, 17, 360–379. [Google Scholar] [CrossRef]
- Buyyounouski, M.K.; Choyke, P.L.; McKenney, J.K.; Sartor, O.; Sandler, H.M.; Amin, M.B.; Kattan, M.W.; Lin, D.W. Prostate cancer—Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA A Cancer J. Clin. 2017, 67, 245–253. [Google Scholar] [CrossRef]
- Rycaj, K.; Cho, E.J.; Liu, X.; Chao, H.P.; Liu, B.; Li, Q.; Devkota, A.K.; Zhang, D.; Chen, X.; Moore, J.; et al. Longitudinal tracking of subpopulation dynamics and molecular changes during LNCaP cell castration and identification of inhibitors that could target the PSA-/lo castration-resistant cells. Oncotarget 2016, 7, 14220–14240. [Google Scholar] [CrossRef]
- Berglund, E.; Maaskola, J.; Schultz, N.; Friedrich, S.; Marklund, M.; Bergenstrahle, J.; Tarish, F.; Tanoglidi, A.; Vickovic, S.; Larsson, L.; et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018, 9, 2419. [Google Scholar] [CrossRef] [PubMed]
- Yoosuf, N.; Navarro, J.F.; Salmén, F.; Ståhl, P.L.; Daub, C.O. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res. 2020, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Bingham, G.C.; Lee, F.; Naba, A.; Barker, T.H. Spatial-omics: Novel approaches to probe cell heterogeneity and extracellular matrix biology. Matrix Biol. 2020, 91–92, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, S.; Ruzzo, W.L. Spatial modeling of prostate cancer metabolic gene expression reveals extensive heterogeneity and selective vulnerabilities. Sci. Rep. 2020, 10, 3490. [Google Scholar] [CrossRef]
- Drake, R.R.; Angel, P.M.; Wu, J.; Pachynski, R.K.; Ippolito, J.E. How else can we approach prostate cancer biomarker discovery? Expert Rev. Mol. Diagn. 2020, 20, 123–125. [Google Scholar] [CrossRef]
- Gao, B.; Lue, H.W.; Podolak, J.; Fan, S.; Zhang, Y.; Serawat, A.; Alumkal, J.J.; Fiehn, O.; Thomas, G.V. Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites 2019, 9, 82. [Google Scholar] [CrossRef]
- Corona, G.; Baldi, E.; Maggi, M. Androgen regulation of prostate cancer: Where are we now? J. Endocrinol. Investig. 2011, 34, 232–243. [Google Scholar] [CrossRef]
- Swinnen, J.V.; Van Veldhoven, P.P.; Esquenet, M.; Heyns, W.; Verhoeven, G. Androgens markedly stimulate the accumulation of neutral lipids in the human prostatic adenocarcinoma cell line LNCaP. Endocrinology 1996, 137, 4468–4474. [Google Scholar] [CrossRef]
- Swinnen, J.V.; Vanderhoydonc, F.; Elgamal, A.A.; Eelen, M.; Vercaeren, I.; Joniau, S.; Van Poppel, H.; Baert, L.; Goossens, K.; Heyns, W.; et al. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int. J. Cancer 2000, 88, 176–179. [Google Scholar] [CrossRef]
- Heemers, H.; Maes, B.; Foufelle, F.; Heyns, W.; Verhoeven, G.; Swinnen, J.V. Androgens Stimulate Lipogenic Gene Expression in Prostate Cancer Cells by Activation of the Sterol Regulatory Element-Binding Protein Cleavage Activating Protein/Sterol Regulatory Element-Binding Protein Pathway. Mol. Endocrinol. 2001, 15, 1817–1828. [Google Scholar] [CrossRef]
- Swinnen, J.V.; Heemers, H.; van de Sande, T.; de Schrijver, E.; Brusselmans, K.; Heyns, W.; Verhoeven, G. Androgens, lipogenesis and prostate cancer. J. Steroid Biochem. Mol. Biol. 2004, 92, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Bandu, R.; Mok, H.J.; Kim, K.P. Phospholipids as cancer biomarkers: Mass spectrometry-based analysis. Mass Spectrom. Rev. 2018, 37, 107–138. [Google Scholar] [CrossRef] [PubMed]
- Perrotti, F.; Rosa, C.; Cicalini, I.; Sacchetta, P.; Del Boccio, P.; Genovesi, D.; Pieragostino, D. Advances in Lipidomics for Cancer Biomarkers Discovery. Int. J. Mol. Sci. 2016, 17, 1992. [Google Scholar] [CrossRef] [PubMed]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef]
- Han, X.; Gross, R.W. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA 1994, 91, 10635–10639. [Google Scholar] [CrossRef]
- Han, X.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef]
- Sapandowski, A.; Stope, M.; Evert, K.; Evert, M.; Zimmermann, U.; Peter, D.; Page, I.; Burchardt, M.; Schild, L. Cardiolipin composition correlates with prostate cancer cell proliferation. Mol. Cell. Biochem. 2015, 410, 175–185. [Google Scholar] [CrossRef]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z.; et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell. Proteom. 2016, 15, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Mahon, K.L.; Weir, J.M.; Mundra, P.A.; Spielman, C.; Briscoe, K.; Gurney, H.; Mallesara, G.; Marx, G.; Stockler, M.R.; et al. A distinct plasma lipid signature associated with poor prognosis in castration-resistant prostate cancer. Int. J. Cancer 2017, 141, 2112–2120. [Google Scholar] [CrossRef] [PubMed]
- Kiebish, M.A.; Cullen, J.; Mishra, P.; Ali, A.; Milliman, E.; Rodrigues, L.O.; Chen, E.Y.; Tolstikov, V.; Zhang, L.; Panagopoulos, K.; et al. Multi-omic serum biomarkers for prognosis of disease progression in prostate cancer. J. Transl. Med. 2020, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Bjork, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Bostwick, D.G.; Cheng, L. (Eds.) Urologic Surgical Pathology. In Urologic Surgical Pathology; Elsevier Health Sciences: Philadelphia, PA, USA, 2020; pp. 415–525.e42. [Google Scholar]
- Holzlechner, M.; Eugenin, E.; Prideaux, B. Mass spectrometry imaging to detect lipid biomarkers and disease signatures in cancer. Cancer Rep. 2019, 2, e1229. [Google Scholar] [CrossRef]
- Trim, P.J.; Snel, M.F. Small molecule MALDI MS imaging: Current technologies and future challenges. Methods 2016, 104, 127–141. [Google Scholar] [CrossRef]
- McCombie, G.; Staab, D.; Stoeckli, M.; Knochenmuss, R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal. Chem. 2005, 77, 6118–6124. [Google Scholar] [CrossRef]
- Gessel, M.M.; Norris, J.L.; Caprioli, R.M. MALDI imaging mass spectrometry: Spatial molecular analysis to enable a new age of discovery. J. Proteom. 2014, 107, 71–82. [Google Scholar] [CrossRef]
- Karas, M.; Kruger, R. Ion formation in MALDI: The cluster ionization mechanism. Chem. Rev. 2003, 103, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Knochenmuss, R. Ion formation mechanisms in UV-MALDI. Analyst 2006, 131, 966–986. [Google Scholar] [CrossRef] [PubMed]
- Rujoi, M.; Estrada, R.; Yappert, M.C. In Situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Anal. Chem. 2004, 76, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Yang, K.; Zhao, Z.; Guan, S.; Han, X.; Gross, R.W. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte—Matrix interactions. Anal. Chem. 2008, 80, 7576. [Google Scholar] [CrossRef] [PubMed]
- Trim, P.J.; Atkinson, S.J.; Princivalle, A.P.; Marshall, P.S.; West, A.; Clench, M.R. Matrix-assisted laser desorption/ionisation mass spectrometry imaging of lipids in rat brain tissue with integrated unsupervised and supervised multivariant statistical analysis. Rapid Commun. Mass Spectrom. 2008, 22, 1503–1509. [Google Scholar] [CrossRef]
- Ucal, Y.; Durer, Z.A.; Atak, H.; Kadioglu, E.; Sahin, B.; Coskun, A.; Baykal, A.T.; Ozpinar, A. Clinical applications of MALDI imaging technologies in cancer and neurodegenerative diseases. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 795–816. [Google Scholar] [CrossRef]
- Agar, N.Y.; Yang, H.W.; Carroll, R.S.; Black, P.M.; Agar, J.N. Matrix solution fixation: Histology-compatible tissue preparation for MALDI mass spectrometry imaging. Anal. Chem. 2007, 79, 7416–7423. [Google Scholar] [CrossRef]
- Cimino, J.; Calligaris, D.; Far, J.; Debois, D.; Blacher, S.; Sounni, N.E.; Noel, A.; De Pauw, E. Towards lipidomics of low-abundant species for exploring tumor heterogeneity guided by high-resolution mass spectrometry imaging. Int. J. Mol. Sci. 2013, 14, 24560–24580. [Google Scholar] [CrossRef]
- Mutuku, S.M.; Trim, P.J.; Prabhala, B.K.; Irani, S.; Bremert, K.L.; Logan, J.M.; Brooks, D.A.; Stahl, J.; Centenera, M.M.; Snel, M.F.; et al. Evaluation of Small Molecule Drug Uptake in Patient-Derived Prostate Cancer Explants by Mass Spectrometry. Sci. Rep. 2019, 9, 15008. [Google Scholar] [CrossRef]
- Girod, M.; Shi, Y.; Cheng, J.X.; Cooks, R.G. Desorption electrospray ionization imaging mass spectrometry of lipids in rat spinal cord. J. Am. Soc. Mass Spectrom. 2010, 21, 1177–1189. [Google Scholar] [CrossRef]
- Takats, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.D.; Fang, R.; Yuan, J.; Chu, R.K.; Dey, S.K.; Burnum-Johnson, K.E.; Lanekoff, I. Quantitative Mass Spectrometry Imaging of Prostaglandins as Silver Ion Adducts with Nanospray Desorption Electrospray Ionization. Anal. Chem. 2018, 90, 7246–7252. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J. Secondary ion mass spectrometry. Anal. Chem. 2008, 80, 7194–7197. [Google Scholar] [CrossRef] [PubMed]
- Piehowski, P.D.; Davey, A.M.; Kurczy, M.E.; Sheets, E.D.; Winograd, N.; Ewing, A.G.; Heien, M.L. Time-of-flight secondary ion mass spectrometry imaging of subcellular lipid heterogeneity: Poisson counting and spatial resolution. Anal. Chem. 2009, 81, 5593–5602. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Samfors, S.; Fletcher, J.S. Lipid Diversity in Cells and Tissue Using Imaging SIMS. Annu. Rev. Anal. Chem. 2020, 13, 249–271. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Terada, N.; Inoue, T.; Nakayama, K.; Okada, Y.; Yoshikawa, T.; Miyazaki, Y.; Uegaki, M.; Sumiyoshi, S.; Kobayashi, T.; et al. The expression profile of phosphatidylinositol in high spatial resolution imaging mass spectrometry as a potential biomarker for prostate cancer. PLoS ONE 2014, 9, e90242. [Google Scholar] [CrossRef]
- Goto, T.; Terada, N.; Inoue, T.; Kobayashi, T.; Nakayama, K.; Okada, Y.; Yoshikawa, T.; Miyazaki, Y.; Uegaki, M.; Utsunomiya, N.; et al. Decreased expression of lysophosphatidylcholine (16:0/OH) in high resolution imaging mass spectrometry independently predicts biochemical recurrence after surgical treatment for prostate cancer. Prostate 2015, 75, 1821–1830. [Google Scholar] [CrossRef]
- Wang, X.; Han, J.; Hardie, D.B.; Yang, J.; Pan, J.; Borchers, C.H. Metabolomic profiling of prostate cancer by matrix assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry imaging using Matrix Coating Assisted by an Electric Field (MCAEF). Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 755–767. [Google Scholar] [CrossRef]
- Randall, E.C.; Zadra, G.; Chetta, P.; Lopez, B.G.C.; Syamala, S.; Basu, S.S.; Agar, J.N.; Loda, M.; Tempany, C.M.; Fennessy, F.M.; et al. Molecular Characterization of Prostate Cancer with Associated Gleason Score Using Mass Spectrometry Imaging. Mol. Cancer Res. 2019, 17, 1155–1165. [Google Scholar] [CrossRef]
- Iglesias-Gato, D.; Wikstrom, P.; Tyanova, S.; Lavallee, C.; Thysell, E.; Carlsson, J.; Hagglof, C.; Cox, J.; Andren, O.; Stattin, P.; et al. The proteome of primary prostate cancer. Eur. Urol. 2016, 69, 942–952. [Google Scholar] [CrossRef]
- Morse, N.; Jamaspishvili, T.; Simon, D.; Patel, P.G.; Ren, K.Y.M.; Wang, J.; Oleschuk, R.; Kaufmann, M.; Gooding, R.J.; Berman, D.M. Reliable identification of prostate cancer using mass spectrometry metabolomic imaging in needle core biopsies. Lab. Investig. 2019, 99, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Zare, R.N.; Tibshirani, R.J.; Kunder, C.A.; Nolley, R.; Fan, R.; Brooks, J.D.; Sonn, G.A. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. USA 2017, 114, 3334–3339. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Franklin, R.B. Prostatic fluid electrolyte composition for the screening of prostate cancer: A potential solution to a major problem. Prostate Cancer Prostatic Dis. 2009, 12, 17–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bader, D.A.; Hartig, S.M.; Putluri, V.; Foley, C.; Hamilton, M.P.; Smith, E.A.; Saha, P.K.; Panigrahi, A.; Walker, C.; Zong, L.; et al. Mitochondrial pyruvate import is a metabolic vulnerability in androgen receptor-driven prostate cancer. Nat. Metab. 2019, 1, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Balaban, S.; Nassar, Z.D.; Zhang, A.Y.; Hosseini-Beheshti, E.; Centenera, M.M.; Schreuder, M.; Lin, H.M.; Aishah, A.; Varney, B.; Liu-Fu, F.; et al. Extracellular Fatty Acids Are the Major Contributor to Lipid Synthesis in Prostate Cancer. Mol. Cancer Res. 2019, 17, 949–962. [Google Scholar] [CrossRef]
- Kerian, K.S.; Jarmusch, A.K.; Pirro, V.; Koch, M.O.; Masterson, T.A.; Cheng, L.; Cooks, R.G. Differentiation of prostate cancer from normal tissue in radical prostatectomy specimens by desorption electrospray ionization and touch spray ionization mass spectrometry. Analyst 2015, 140, 1090–1098. [Google Scholar] [CrossRef]
- Andersen, M.K.; Høiem, T.S.; Claes, B.S.R.; Balluff, B.; Martin-Lorenzo, M.; Richardsen, E.; Krossa, S.; Bertilsson, H.; Heeren, R.M.A.; Rye, M.B.; et al. Spatial differentiation of metabolism in prostate cancer tissue by MALDI-TOF MSI. Cancer Metab. 2021, 9, 9. [Google Scholar] [CrossRef]
- Butler, L.M.; Mah, C.Y.; Machiels, J.; Vincent, A.D.; Irani, S.; Mutuku, S.M.; Spotbeen, X.; Bagadi, M.; Waltregny, D.; Moldovan, M.; et al. Lipidomic profiling of clinical prostate cancer reveals targetable alterations in membrane lipid composition. Cancer Res. 2021, 81, 4981–4993. [Google Scholar] [CrossRef]
- Young, R.S.E.; Bowman, A.P.; Williams, E.D.; Tousignant, K.D.; Bidgood, C.L.; Narreddula, V.R.; Gupta, R.; Marshall, D.L.; Poad, B.L.J.; Nelson, C.C.; et al. Apocryphal FADS2 activity promotes fatty acid diversification in cancer. Cell Rep. 2021, 34, 108738. [Google Scholar] [CrossRef]
- Salmén, F.; Ståhl, P.L.; Mollbrink, A.; Navarro, J.F.; Vickovic, S.; Frisén, J.; Lundeberg, J. Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat. Protoc. 2018, 13, 2501–2534. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Moncada, R.; Barkley, D.; Wagner, F.; Chiodin, M.; Devlin, J.C.; Baron, M.; Hajdu, C.H.; Simeone, D.M.; Yanai, I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020, 38, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Frisen, J.; Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 2021, 18, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Ke, R.; Mignardi, M.; Pacureanu, A.; Svedlund, J.; Botling, J.; Wahlby, C.; Nilsson, M. In Situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 2013, 10, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, Y.C.; Church, G.M.; Lee, J.H.; Zador, A.M. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 2018, 46, e22. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Wang, X.; Allen, W.E.; Wright, M.A.; Sylwestrak, E.L.; Samusik, N.; Vesuna, S.; Evans, K.; Liu, C.; Ramakrishnan, C.; Liu, J.; et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361. [Google Scholar] [CrossRef]
- Nagarajan, M.B.; Tentori, A.M.; Zhang, W.C.; Slack, F.J.; Doyle, P.S. Spatially resolved and multiplexed MicroRNA quantification from tissue using nanoliter well arrays. Microsyst. Nanoeng. 2020, 6, 51. [Google Scholar] [CrossRef]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef]
- Raj, A.; van den Bogaard, P.; Rifkin, S.A.; van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef]
- Crosetto, N.; Bienko, M.; van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2015, 16, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Teves, J.M.; Won, K.J. Mapping Cellular Coordinates through Advances in Spatial Transcriptomics Technology. Mol. Cells 2020, 43, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.H.; Boettiger, A.N.; Moffitt, J.R.; Wang, S.; Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348, aaa6090. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, J.R.; Hao, J.; Wang, G.; Chen, K.H.; Babcock, H.P.; Zhuang, X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 2016, 113, 11046–11051. [Google Scholar] [CrossRef] [PubMed]
- Lubeck, E.; Coskun, A.F.; Zhiyentayev, T.; Ahmad, M.; Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 2014, 11, 360–361. [Google Scholar] [CrossRef]
- Xia, C.; Fan, J.; Emanuel, G.; Hao, J.; Zhuang, X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. USA 2019, 116, 19490–19499. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, M.; Deng, Y.; Su, G.; Enninful, A.; Guo, C.C.; Tebaldi, T.; Zhang, D.; Kim, D.; Bai, Z.; et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 2020, 183, 1665–1681. [Google Scholar] [CrossRef]
- Dries, R.; Chen, J.; Del Rossi, N.; Khan, M.M.; Sistig, A.; Yuan, G.C. Advances in spatial transcriptomic data analysis. Genome Res. 2021, 31, 1706–1718. [Google Scholar] [CrossRef]
- Jemt, A.; Salmen, F.; Lundmark, A.; Mollbrink, A.; Fernandez Navarro, J.; Stahl, P.L.; Yucel-Lindberg, T.; Lundeberg, J. An automated approach to prepare tissue-derived spatially barcoded RNA-sequencing libraries. Sci. Rep. 2016, 6, 37137. [Google Scholar] [CrossRef]
- Fernández Navarro, J.; Lundeberg, J.; Ståhl, P.L. ST viewer: A tool for analysis and visualization of spatial transcriptomics datasets. Bioinformatics 2019, 35, 1058–1060. [Google Scholar] [CrossRef]
- Andersson, A.; Bergenstråhle, J.; Asp, M.; Bergenstråhle, L.; Jurek, A.; Fernández Navarro, J.; Lundeberg, J. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 2020, 3, 565. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Manzar, N.; Bhatia, V.; Yadav, A.; Nengroo, M.A.; Datta, D.; Carskadon, S.; Gupta, N.; Sigouros, M.; Khani, F.; et al. Androgen deprivation upregulates SPINK1 expression and potentiates cellular plasticity in prostate cancer. Nat. Commun. 2020, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Bowman, A.P.; Bogie, J.F.J.; Hendriks, J.J.A.; Haidar, M.; Belov, M.; Heeren, R.M.A.; Ellis, S.R. Evaluation of lipid coverage and high spatial resolution MALDI-imaging capabilities of oversampling combined with laser post-ionisation. Anal. Bioanal. Chem. 2020, 412, 2277–2289. [Google Scholar] [CrossRef] [PubMed]
- Soltwisch, J.; Kettling, H.; Vens-Cappell, S.; Wiegelmann, M.; Muthing, J.; Dreisewerd, K. Mass spectrometry imaging with laser-induced postionization. Science 2015, 348, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Trim, P.J.; Henson, C.M.; Avery, J.L.; McEwen, A.; Snel, M.F.; Claude, E.; Marshall, P.S.; West, A.; Princivalle, A.P.; Clench, M.R. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem. 2008, 80, 8628–8634. [Google Scholar] [CrossRef]
- Snel, M.F. Ion mobility separation mass spectrometry imaging. In Comprehensive Analytical Chemistry; Elsevier: Oxford, UK, 2019; Volume 83, pp. 237–257. [Google Scholar]
- Spraggins, J.M.; Djambazova, K.V.; Rivera, E.S.; Migas, L.G.; Neumann, E.K.; Fuetterer, A.; Suetering, J.; Goedecke, N.; Ly, A.; Van de Plas, R.; et al. High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (timsTOF) Mass Spectrometry. Anal. Chem. 2019, 91, 14552–14560. [Google Scholar] [CrossRef] [PubMed]
- Ujma, J.; Ropartz, D.; Giles, K.; Richardson, K.; Langridge, D.; Wildgoose, J.; Green, M.; Pringle, S. Cyclic Ion Mobility Mass Spectrometry Distinguishes Anomers and Open-Ring Forms of Pentasaccharides. J. Am. Soc. Mass Spectrom. 2019, 30, 1028–1037. [Google Scholar] [CrossRef]
- Palmer, A.; Phapale, P.; Chernyavsky, I.; Lavigne, R.; Fay, D.; Tarasov, A.; Kovalev, V.; Fuchser, J.; Nikolenko, S.; Pineau, C.; et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods 2017, 14, 57–60. [Google Scholar] [CrossRef]
- Race, A.M.; Bunch, J. Optimisation of colour schemes to accurately display mass spectrometry imaging data based on human colour perception. Anal. Bioanal. Chem. 2015, 407, 2047–2054. [Google Scholar] [CrossRef]
- Tortorella, S.; Tiberi, P.; Bowman, A.P.; Claes, B.S.R.; Scupakova, K.; Heeren, R.M.A.; Ellis, S.R.; Cruciani, G. LipostarMSI: Comprehensive, Vendor-Neutral Software for Visualization, Data Analysis, and Automated Molecular Identification in Mass Spectrometry Imaging. J. Am. Soc. Mass Spectrom. 2020, 31, 155–163. [Google Scholar] [CrossRef]
| Author | Method | Findings |
|---|---|---|
| Butler et al., 2021 | ESI-MS/MS MALDI-MSI | Association of lipid profiles to malignancy status in clinical biopsies and lipid changes in response to metabolic targeting agents |
| Young et al., 2021 | MALDI-MSI OzID | Isomer-resolved lipidomics detects non-canonical fatty acids (reflecting different desaturase activities) present in different regions of the PCa TME, providing support for discrete localisation of desaturase enzymes |
| Andersen et al., 2021 | MALDI TOF MSI | Lipid and metabolite composition was distinct between stromal, non-cancerous epithelium, and PCa. Lysophospholipids had lower abundance in PCa versus non-cancerous epithelium, while PE and PI lipids were higher in PCa. |
| Randall et al., 2019 | MALDI FT-ICR MSI, MALDI TOF MSI | Prostate tumours can be differentiated using different Gleason grades based on metabolomic differences |
| Morse et al., 2019 | DESI-MSI | Logistic regression and PCA/LDA model of lipid and metabolite classifiers can reliably identify cancer and distinguish Gleason grade groups |
| Banerjee et al., 2017 | DESI-MSI | LASSO model identified glucose and citrate as predictors of PCa and normal tissue |
| Wang et al., 2017 | MALDI FT-ICR | Increased energy charge and low abundance of neutral triglycerides in cancerous tissue |
| Goto et al., 2015 | MALDI-MSI | LPC (16:0) and SM (d18:1/16:0) were lower in tumour compared to benign epithelium. LPA (16:0) was an independent predictor of biochemical recurrence after radical prostatectomy |
| Goto et al., 2014 | MALDI-MSI | PI species were more abundant in cancer compared to benign epithelium:
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutuku, S.M.; Spotbeen, X.; Trim, P.J.; Snel, M.F.; Butler, L.M.; Swinnen, J.V. Unravelling Prostate Cancer Heterogeneity Using Spatial Approaches to Lipidomics and Transcriptomics. Cancers 2022, 14, 1702. https://doi.org/10.3390/cancers14071702
Mutuku SM, Spotbeen X, Trim PJ, Snel MF, Butler LM, Swinnen JV. Unravelling Prostate Cancer Heterogeneity Using Spatial Approaches to Lipidomics and Transcriptomics. Cancers. 2022; 14(7):1702. https://doi.org/10.3390/cancers14071702
Chicago/Turabian StyleMutuku, Shadrack M., Xander Spotbeen, Paul J. Trim, Marten F. Snel, Lisa M. Butler, and Johannes V. Swinnen. 2022. "Unravelling Prostate Cancer Heterogeneity Using Spatial Approaches to Lipidomics and Transcriptomics" Cancers 14, no. 7: 1702. https://doi.org/10.3390/cancers14071702
APA StyleMutuku, S. M., Spotbeen, X., Trim, P. J., Snel, M. F., Butler, L. M., & Swinnen, J. V. (2022). Unravelling Prostate Cancer Heterogeneity Using Spatial Approaches to Lipidomics and Transcriptomics. Cancers, 14(7), 1702. https://doi.org/10.3390/cancers14071702






