Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression Analysis
2.2. Cells and Culture Conditions
2.3. Cell Viability
2.4. SiRNA Transfection
2.5. Western Blot
2.6. Antibodies
2.7. Glutamine and Glutamate Determination
2.8. Glycolytic Enzyme Assays
2.9. Bioluminescent Luciferase ATP Assay
2.10. Evaluation of ATP and AMP Levels
2.11. Evaluation of Cytosolic Reactive Oxygen Species Production
2.12. Evaluation of Proliferation and Apoptosis
2.13. Evaluation of Autophagic Vacuoles with Monodansylcadaverine
2.14. Analysis of Actin Cytoskeleton and Evaluation of Cell Morphology
2.15. In Vivo Studies
2.16. Histology and Immunohistochemistry
2.17. Statistical Analysis
3. Results
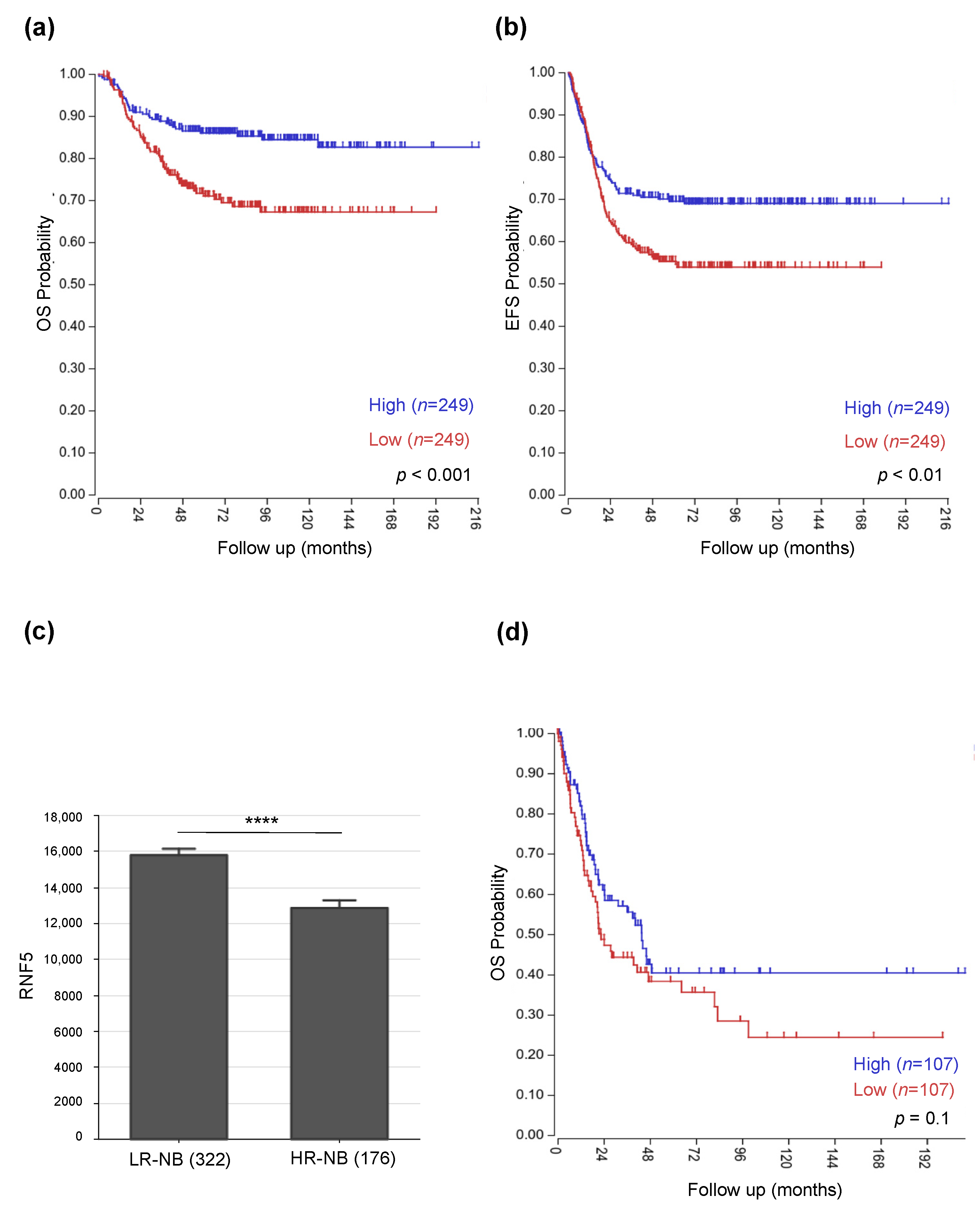
3.1. Low RNF5 Expression Determines a Poor Prognostic Outcome in Neuroblastoma and Melanoma Patients
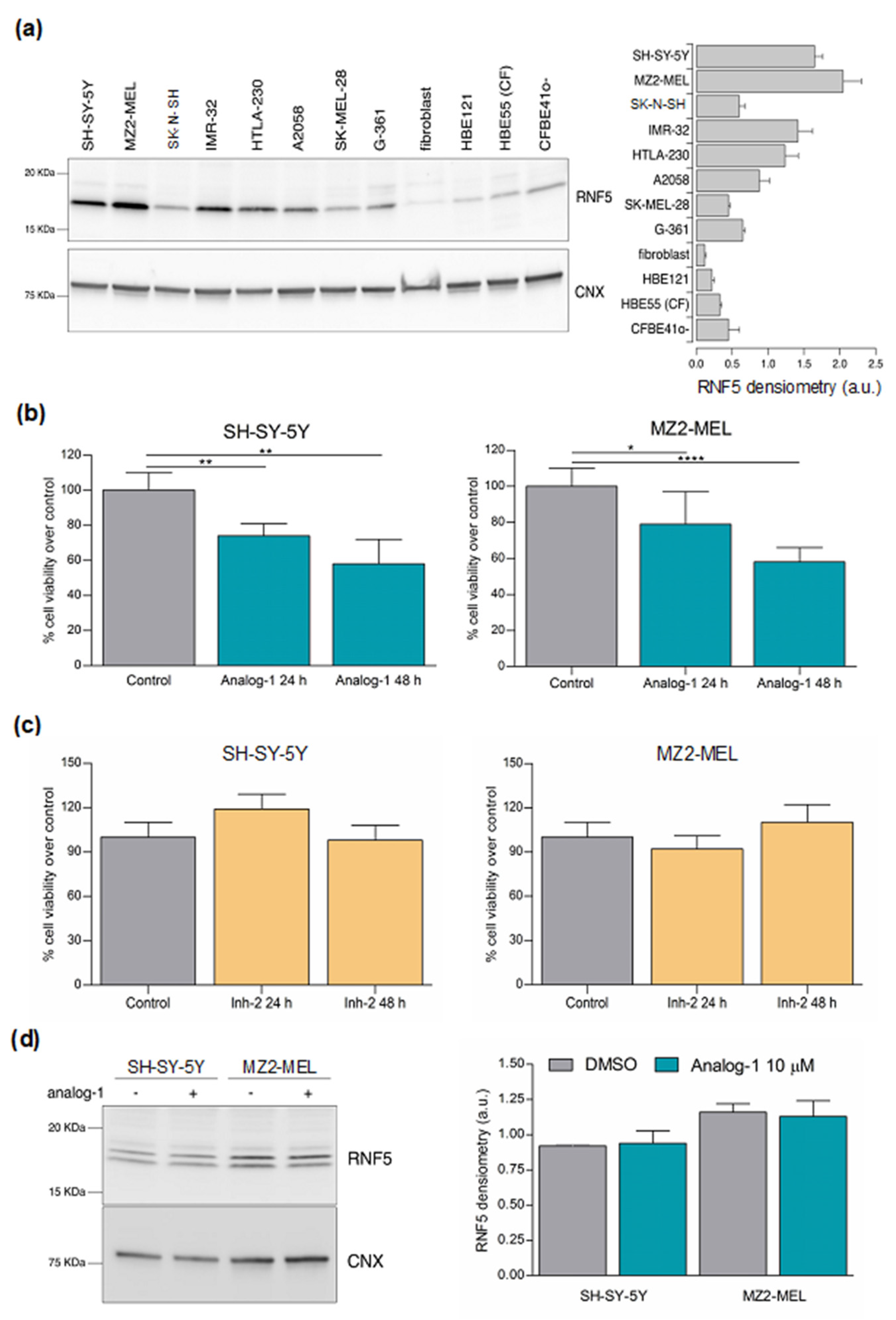
3.2. RNF5 Is Expressed by Neuroblastoma and Melanoma Cell Lines and Its Activation by Analog-1 Reduces Tumor Cell Viability
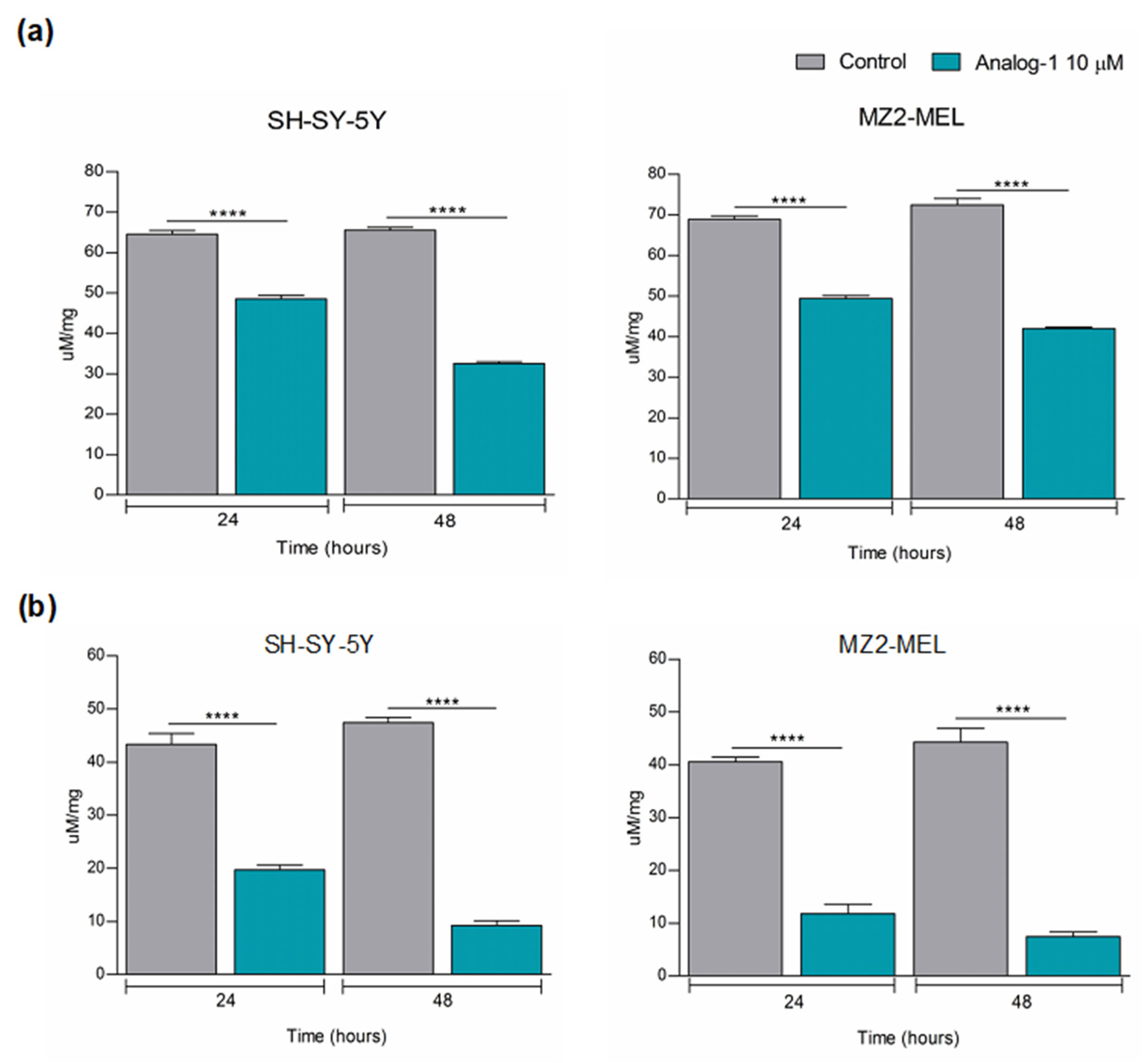
3.3. Analog-1 Modulates the Metabolism of Neuroblastoma and Melanoma Cell Lines
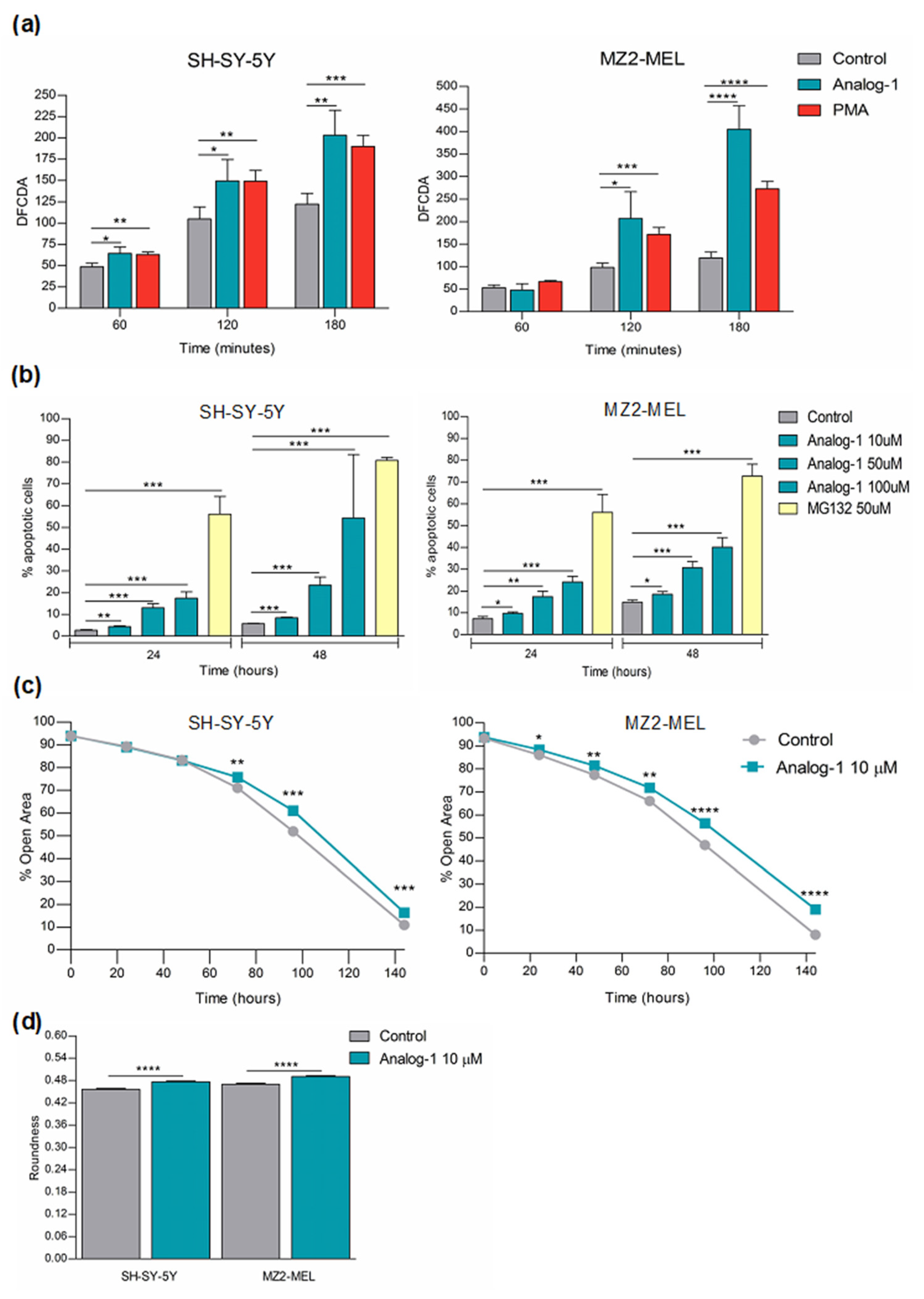
3.4. Analog-1 Induces Apoptosis by Promoting Oxidative Stress and Decreases the Proliferation of Neuroblastoma and Melanoma Cells
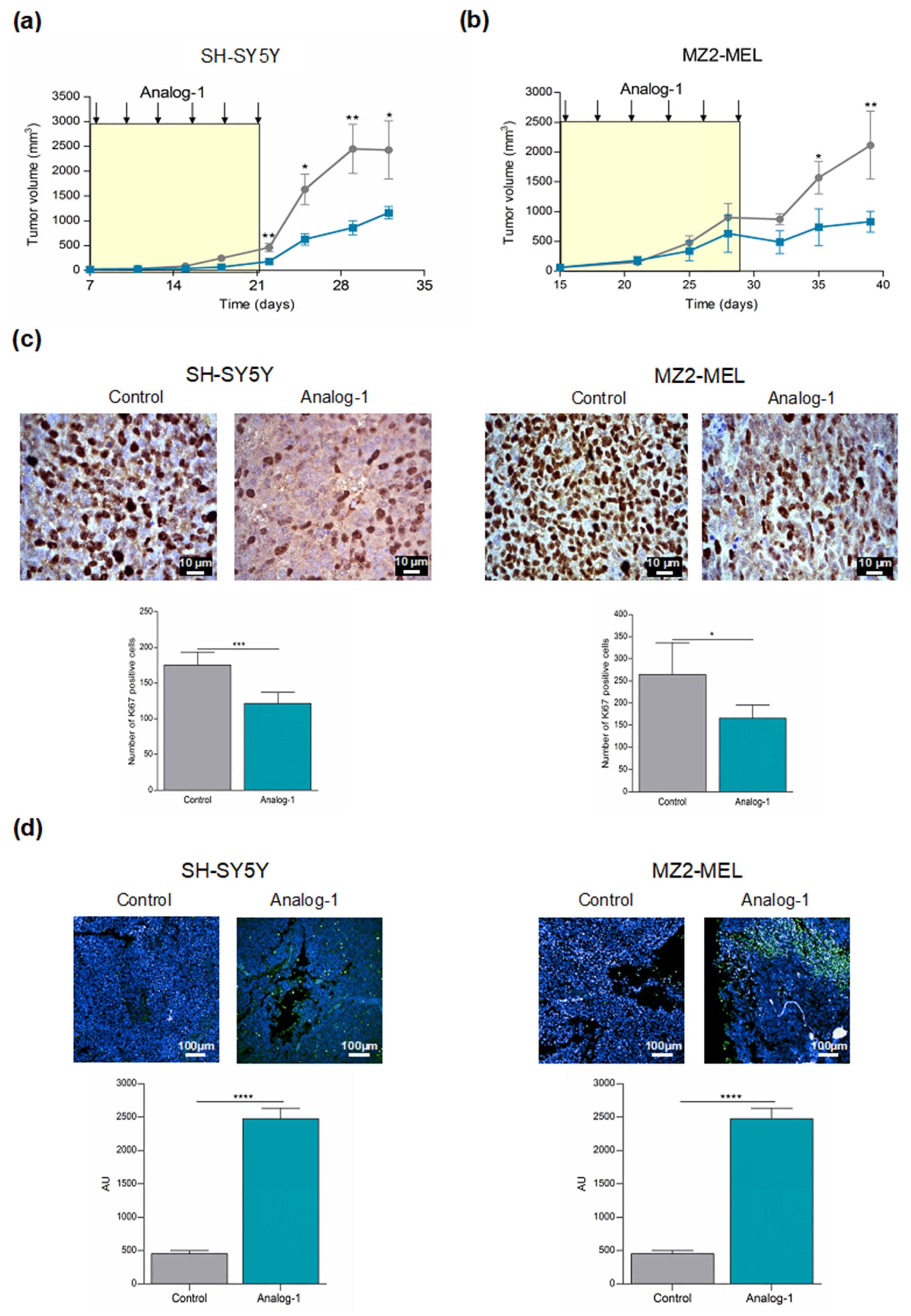
3.5. Analog-1 Delays Tumor Growth in Human Neuroblastoma and Melanoma Animal Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, S.; Lorick, K.L.; Jensen, J.P.; Weissman, A.M. RING finger ubiquitin protein ligases: Implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 2003, 13, 5–14. [Google Scholar] [CrossRef]
- Ciechanover, A.A.H. No Title THE UBIQUITIN SYSTEM. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar]
- Tsai, Y.C.; Weissman, A.M. The unfolded protein response, degradation from the endoplasmic reticulum, and cancer. Genes Cancer 2010, 1, 764–778. [Google Scholar] [PubMed]
- Kuang, E.; Qi, J.; Ronai, Z. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem. Sci. 2013, 38, 453–460. [Google Scholar]
- Didier, C.; Broday, L.; Bhoumik, A.; Israeli, S.; Takahashi, S.; Nakayama, K.; Thomas, S.M.; Turner, C.E.; Henderson, S.; Sabe, H.; et al. RNF5, a RING Finger Protein That Regulates Cell Motility by Targeting Paxillin Ubiquitination and Altered Localization. Mol. Cell. Biol. 2003, 23, 5331–5345. [Google Scholar] [CrossRef]
- Younger, J.M.; Chen, L.; Ren, H.Y.; Rosser, M.F.N.; Turnbull, E.L.; Fan, C.Y.; Patterson, C.; Cyr, D.M. Sequential Quality-Control Checkpoints Triage Misfolded Cystic Fibrosis Transmembrane Conductance Regulator. Cell 2006, 126, 571–582. [Google Scholar] [CrossRef]
- Grove, D.E.; Fan, C.Y.; Ren, H.Y.; Cyr, D.M. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRΔF508. Mol. Biol. Cell 2011, 22, 301–314. [Google Scholar] [CrossRef]
- Sondo, E.; Falchi, F.; Caci, E.; Ferrera, L.; Giacomini, E.; Pesce, E.; Tomati, V.; Mandrup Bertozzi, S.; Goldoni, L.; Armirotti, A.; et al. Pharmacological Inhibition of the Ubiquitin Ligase RNF5 Rescues F508del-CFTR in Cystic Fibrosis Airway Epithelia. Cell Chem. Biol. 2018, 25, 891–905. [Google Scholar]
- Bromberg, K.D.; Kluger, H.M.; Delaunay, A.; Abbas, S.; Di Vito, K.A.; Krajewski, S.; Ronai, Z. Increased expression of the E3 ubiquitin ligase RNF5 is associated with decreased survival in breast cancer. Cancer Res. 2007, 67, 8172–8179. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Liu, H.; Li, B. Six genes as potential diagnosis and prognosis biomarkers for hepatocellular carcinoma through data mining. J. Cell. Physiol. 2019, 234, 9787–9792. [Google Scholar]
- Khateb, A.; Deshpande, A.; Feng, Y.; Finlay, D.; Lee, J.S.; Lazar, I.; Fabre, B.; Li, Y.; Fujita, Y.; Zhang, T.; et al. The ubiquitin ligase RNF5 determines acute myeloid leukemia growth and susceptibility to histone deacetylase inhibitors. Nat. Commun. 2021, 12, 5397. [Google Scholar] [CrossRef]
- Li, Y.; Tinoco, R.; Elmén, L.; Segota, I.; Xian, Y.; Fujita, Y.; Sahu, A.; Zarecki, R.; Marie, K.; Feng, Y.; et al. Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5 −/− mice. Nat. Commun. 2019, 10, 1492. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Khelifa, S.; Ratnikov, B.; Scott, D.A.; Feng, Y.; Parisi, F.; Ruller, C.; Lau, E.; Kim, H.; Brill, L.M.; et al. Regulation of Glutamine Carrier Proteins by RNF5 Determines Breast Cancer Response to ER Stress-Inducing Chemotherapies. Cancer Cell 2015, 27, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wan, X.; Yu, T.; Huang, Z.; Shen, C.; Qi, Q.; Xiang, S.; Chen, X.; Arbely, E.; Ling, Z.Q.; et al. Acetylation Stabilizes Phosphoglycerate Dehydrogenase by Disrupting the Interaction of E3 Ligase RNF5 to Promote Breast Tumorigenesis. Cell Rep. 2020, 32, 108021. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xuan, C.; Jin, M.; An, Q.; Zhuo, B.; Chen, X.; Wang, L.; Wang, Y.; Sun, Q.; Shi, Y. Ubiquitin ligase rnf5 serves an important role in the development of human glioma. Oncol. Lett. 2019, 18, 4659–4666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Spector, T.D.; Deloukas, P.; Bell, J.T.; Engelhardt, B.E. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol. 2015, 16, 14. [Google Scholar] [CrossRef] [PubMed]
- Cirenajwis, H.; Ekedahl, H.; Lauss, M.; Harbst, K.; Carneiro, A.; Enoksson, J.; Rosengren, F.; Werner-Hartman, L.; Törngren, T.; Kvist, A.; et al. Molecular stratification of metastatic melanoma using gene expression profiling-Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget 2015, 6, 12297–12309. [Google Scholar] [CrossRef]
- Pastorino, F.; Brignole, C.; Marimpietri, D.; Pagnan, G.; Morando, A.; Ribatti, D.; Semple, S.C.; Gambini, C.; Allen, T.M.; Ponzoni, M. Targeted liposomal c-myc antisense oligodeoxynucleotides induce apoptosis and inhibit tumor growth and metastases in human melanoma models. Clin. Cancer Res. 2003, 9, 4595–4605. [Google Scholar]
- Brignole, C.; Bensa, V.; Fonseca, N.A.; Del Zotto, G.; Bruno, S.; Cruz, A.F.; Malaguti, F.; Carlini, B.; Morandi, F.; Calarco, E.; et al. Cell surface Nucleolin represents a novel cellular target for neuroblastoma therapy. J. Exp. Clin. Cancer Res. 2021, 40, 180. [Google Scholar] [CrossRef]
- Pagnan, G.; Stuart, D.D.; Pastorino, F.; Raffaghello, L.; Montaldo, P.G.; Allen, T.M.; Calabretta, B.; Ponzoni, M. Delivery of c-myb antisense oligodeoxynucleotides to human neuroblastoma cells via disialoganglioside GD2- targeted immunoliposomes: Antitumor effects. J. Natl. Cancer Inst. 2000, 92, 253–261. [Google Scholar] [CrossRef]
- Sondo, E.; Tomati, V.; Caci, E.; Esposito, A.I.; Pfeffer, U.; Pedemonte, N.; Galietta, L.J.V. Rescue of the mutant CFTR chloride channel by pharmacological correctors and low temperature analyzed by gene expression profiling. Am. J. Physiol.-Cell Physiol. 2011, 301, C872–C885. [Google Scholar] [CrossRef] [PubMed]
- Scudieri, P.; Caci, E.; Bruno, S.; Ferrera, L.; Schiavon, M.; Sondo, E.; Tomati, V.; Gianotti, A.; Zegarra-Moran, O.; Pedemonte, N.; et al. Association of TMEM16A chloride channel overexpression with airway goblet cell metaplasia. J. Physiol. 2012, 590, 6141–6155. [Google Scholar] [CrossRef] [PubMed]
- Capurro, V.; Tomati, V.; Sondo, E.; Renda, M.; Borrelli, A.; Pastorino, C.; Guidone, D.; Venturini, A.; Giraudo, A.; Mandrup Bertozzi, S.; et al. Partial rescue of f508del-cftr stability and trafficking defects by double corrector treatment. Int. J. Mol. Sci. 2021, 22, 5262. [Google Scholar] [CrossRef] [PubMed]
- Ravera, S.; Vaccaro, D.; Cuccarolo, P.; Columbaro, M.; Capanni, C.; Bartolucci, M.; Panfoli, I.; Morelli, A.; Dufour, C.; Cappelli, E.; et al. Mitochondrial respiratory chain Complex i defects in Fanconi anemia complementation group A. Biochimie 2013, 95, 1828–1837. [Google Scholar] [CrossRef]
- Cappelli, E.; Cuccarolo, P.; Stroppiana, G.; Miano, M.; Bottega, R.; Cossu, V.; Degan, P.; Ravera, S. Defects in mitochondrial energetic function compels Fanconi Anaemia cells to glycolytic metabolism. Biochim. Biophys. Acta-Mol. Basis Dis. 2017, 1863, 1214–1221. [Google Scholar] [CrossRef]
- Ravera, S.; Aluigi, M.G.; Calzia, D.; Ramoino, P.; Morelli, A.; Panfoli, I. Evidence for ectopic aerobic ATP production on C6 glioma cell plasma membrane. Cell. Mol. Neurobiol. 2011, 31, 313–321. [Google Scholar] [CrossRef]
- Ravera, S.; Panfoli, I.; Aluigi, M.G.; Calzia, D.; Morelli, A. Characterization of Myelin Sheath FoF1-ATP Synthase and its Regulation by IF1. Cell Biochem. Biophys. 2011, 59, 63–70. [Google Scholar] [CrossRef]
- Marini, C.; Bianchi, G.; Buschiazzo, A.; Ravera, S.; Martella, R.; Bottoni, G.; Petretto, A.; Emionite, L.; Monteverde, E.; Capitanio, S.; et al. Divergent targets of glycolysis and oxidative phosphorylation result in additive effects of metformin and starvation in colon and breast cancer. Sci. Rep. 2016, 6, 19569. [Google Scholar] [CrossRef]
- Biederbick, H.P.E.A.; Kern, H.F. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 1995, 66, 3–14. [Google Scholar]
- Kuang, E.; Okumura, C.Y.M.; Sheffy-Levin, S.; Varsano, T.; Shu, V.C.; Qi, J.; Niesman, I.R.; Yang, H.; López-Otín, C.; Yang, W.Y.; et al. Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection. PLoS Genet. 2012, 8, e1003007. [Google Scholar] [CrossRef]
- Tomati, V.; Sondo, E.; Armirotti, A.; Caci, E.; Pesce, E.; Marini, M.; Gianotti, A.; Ju Jeon, Y.; Cilli, M.; Pistorio, A.; et al. Genetic Inhibition of the Ubiquitin Ligase Rnf5 Attenuates Phenotypes Associated to F508del Cystic Fibrosis Mutation. Sci. Rep. 2015, 5, 12138. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, F.; Qian, H. Defueling the cancer: ATP synthase as an emerging target in cancer therapy. Mol. Ther.-Oncolytics 2021, 23, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Mowery, Y.M.; Pizzo, S.V. Targeting cell surface F1F0 ATP synthase in cancer therapy. Cancer Biol. Ther. 2008, 7, 1836–1838. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B.; Mammalian, P.; Exhibit, C.; Metabolism, A. Understanding the Warburg Effect: Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Cuezva, J.M. The H+-ATP synthase: A gate to ROS-mediated cell death or cell survival. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 1099–1112. [Google Scholar] [CrossRef]
- Kim, M.S.; Gernapudi, R.; Cedeño, Y.C.; Polster, B.M.; Martinez, R.; Shapiro, P.; Kesari, S.; Nurmemmedov, E.; Passaniti, A. Targeting breast cancer metabolism with a novel inhibitor of mitochondrial ATP synthesis. Oncotarget 2020, 11, 3863–3885. [Google Scholar] [CrossRef]
- Fan, W.H.; Hou, Y.; Meng, F.K.; Wang, X.F.; Luo, Y.N.; Ge, P.F. Proteasome inhibitor MG-132 induces C6 glioma cell apoptosis via oxidative stress. Acta Pharmacol. Sin. 2011, 32, 619–625. [Google Scholar] [CrossRef]
- John, M.D.; Maris, M. Recent Advances in Neuroblastomae. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Prim. 2016, 2, 16078. [Google Scholar] [CrossRef]
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Chen, H.; Zhang, C.; You, S.; Zhao, Y.; Lin, X.; Yu, Y.; Fang, F.; Fang, T.; et al. RING-finger protein 5 promotes hepatocellular carcinoma progression and predicts poor prognosis. Hum. Cell 2021, 34, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Qing, G.; Li, B.; Vu, A.; Skuli, N.; Walton, Z.E.; Liu, X.; Mayes, P.A.; Wise, D.R.; Thompson, C.B.; Maris, J.M.; et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 2012, 22, 631–644. [Google Scholar] [CrossRef]
- Ratnikov, B.; Aza-Blanc, P.; Ronai, Z.A.; Smith, J.W.; Osterman, A.L.; Scott, D.A. Glutamate and asparagine cataplerosis underlie glutamine addiction in melanoma. Oncotarget 2015, 6, 7379–7389. [Google Scholar] [CrossRef]
- Ye, P.; Chi, X.; Cha, J.-H.; Luo, S.; Yang, G.; Yan, X.; Yang, W.-H. Potential of E3 Ubiquitin Ligases in Cancer Immunity: Opportunities and Challenges. Cells 2021, 10, 3309. [Google Scholar] [CrossRef]
- Bae, E.; Hope, J.L.; Otero, D.C.; Faso, H.A.; Palete, A.B.; Ronai, Z.A.; Bradley, L.M. RNF5-deficiency inhibits CD8+ T cell exhaustion in chronic antigen stimulation. J. Immunol. 2021, 206 (Suppl. S1), 12–14. [Google Scholar]
- Ren, P.; Yue, M.; Xiao, D.; Xiu, R.; Gan, L.; Liu, H.; Qing, G. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J. Pathol. 2015, 235, 90–100. [Google Scholar] [CrossRef]
- Baenke, F.; Chaneton, B.; Smith, M.; Van Den Broek, N.; Hogan, K.; Tang, H.; Viros, A.; Martin, M.; Galbraith, L.; Girotti, M.R.; et al. Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells. Mol. Oncol. 2016, 10, 73–84. [Google Scholar] [CrossRef]
- Feng, Y. Identification and Characterization of IMD-0354 as a Glutamine Carrier Protein Inhibitor in Melanoma. Mol. Cancer Ther. 2021, 20, 816–832. [Google Scholar] [CrossRef]
- Wang, Q.; Beaumont, K.A.; Otte, N.J.; Font, J.; Bailey, C.G.; Van Geldermalsen, M.; Sharp, D.M.; Tiffen, J.C.; Ryan, R.M.; Jormakka, M.; et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer 2014, 135, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Li, J.Y. ATP5A1 and ATP5B are highly expressed in glioblastoma tumor cells and endothelial cells of microvascular proliferation. J. Neurooncol. 2016, 126, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Muys, B.R.; Sousa, J.F.; Plaça, J.R.; de Araújo, L.F.; Sarshad, A.A.; Anastasakis, D.G.; Wang, X.; Li, X.L.; de Molfetta, G.A.; Ramão, A.; et al. miR-450a acts as a tumor suppressor in ovarian cancer by regulating energy metabolism. Cancer Res. 2019, 79, 3294–3305. [Google Scholar] [CrossRef] [PubMed]
- Speransky, S.; Serafini, P.; Caroli, J.; Bicciato, S.; Lippman, M.E.; Bishopric, N.H. A novel RNA aptamer identifies plasma membrane ATP synthase beta subunit as an early marker and therapeutic target in aggressive cancer. Breast Cancer Res. Treat. 2019, 176, 271–289. [Google Scholar] [CrossRef]
- Yuan, L.; Chen, L.; Qian, K.; Wang, G.; Lu, M.; Qian, G.; Cao, X.; Jiang, W.; Xiao, Y.; Wang, X. A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by co-expression analysis. Oncol. Rep. 2018, 39, 525–536. [Google Scholar] [CrossRef]
- Hong, S.; Pedersen, P.L. ATP Synthase and the Actions of Inhibitors Utilized To Study Its Roles in Human Health, Disease, and Other Scientific Areas. Microbiol. Mol. Biol. Rev. 2008, 72, 590–641. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Kadenbach, B.; Ramzan, R.; Wen, L.; Vogt, S. New extension of the Mitchell Theory for oxidative phosphorylation in mitochondria of living organisms. Biochim. Biophys. Acta-Gen. Subj. 2009, 1800, 205–212. [Google Scholar] [CrossRef]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef]
- Azad, M.B.; Chen, Y.; Gibson, S.B. Regulation of autophagy by reactive oxygen species (ROS): Implications for cancer progression and treatment. Antioxid. Redox Signal. 2008, 11, 777–790. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. Regulation of autophagy by ROS: Physiology and pathology. Trends Biochem. Sci. 2011, 36, 30–38. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Principi, E.; Sondo, E.; Bianchi, G.; Ravera, S.; Morini, M.; Tomati, V.; Pastorino, C.; Zara, F.; Bruno, C.; Eva, A.; et al. Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors. Cancers 2022, 14, 1802. https://doi.org/10.3390/cancers14071802
Principi E, Sondo E, Bianchi G, Ravera S, Morini M, Tomati V, Pastorino C, Zara F, Bruno C, Eva A, et al. Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors. Cancers. 2022; 14(7):1802. https://doi.org/10.3390/cancers14071802
Chicago/Turabian StylePrincipi, Elisa, Elvira Sondo, Giovanna Bianchi, Silvia Ravera, Martina Morini, Valeria Tomati, Cristina Pastorino, Federico Zara, Claudio Bruno, Alessandra Eva, and et al. 2022. "Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors" Cancers 14, no. 7: 1802. https://doi.org/10.3390/cancers14071802
APA StylePrincipi, E., Sondo, E., Bianchi, G., Ravera, S., Morini, M., Tomati, V., Pastorino, C., Zara, F., Bruno, C., Eva, A., Pedemonte, N., & Raffaghello, L. (2022). Targeting of Ubiquitin E3 Ligase RNF5 as a Novel Therapeutic Strategy in Neuroectodermal Tumors. Cancers, 14(7), 1802. https://doi.org/10.3390/cancers14071802







