Proline Metabolism in Malignant Gliomas: A Systematic Literature Review
Abstract
:Simple Summary
Abstract
1. Introduction
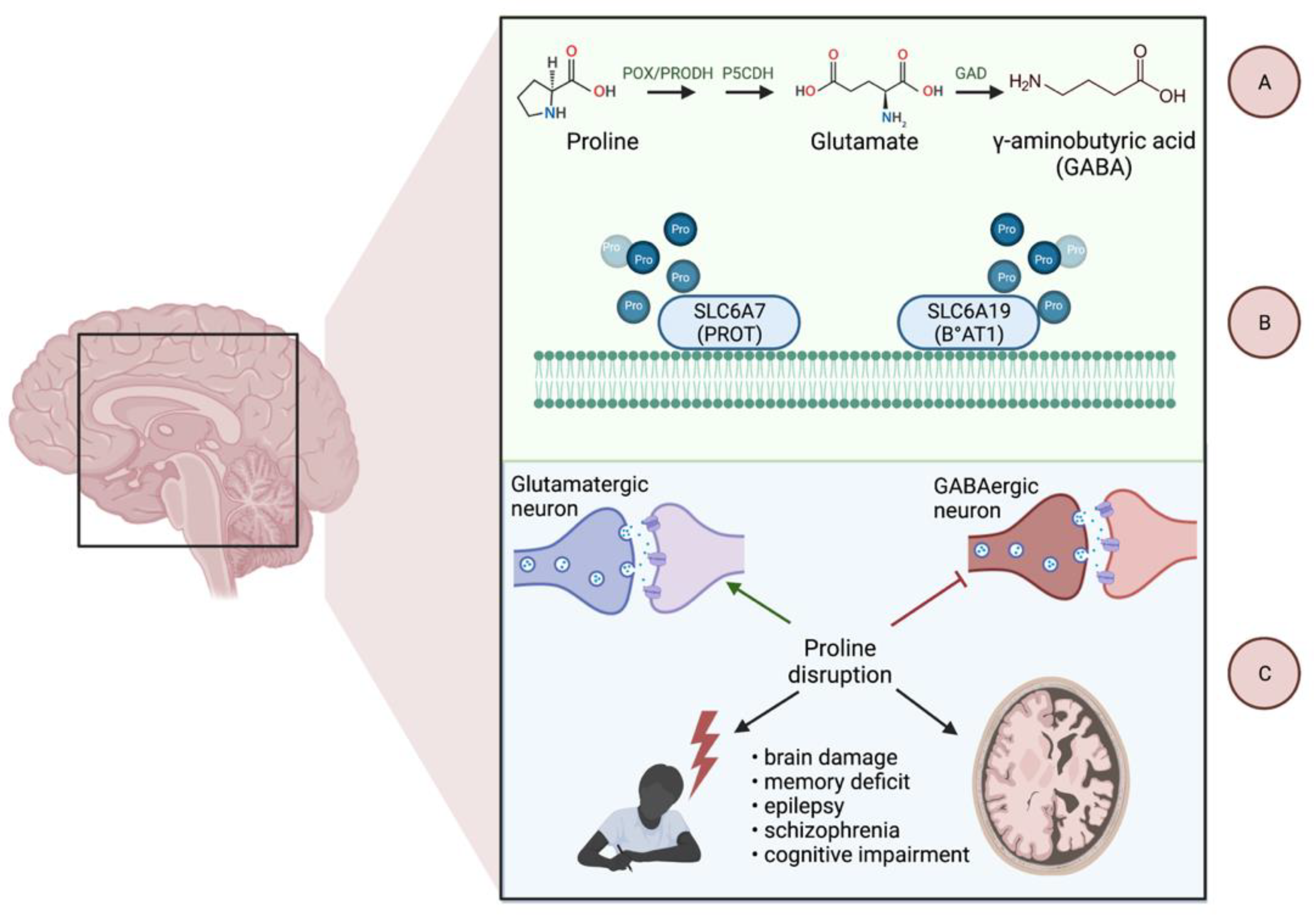
Proline—“An Essential Non-Essential Amino Acid”
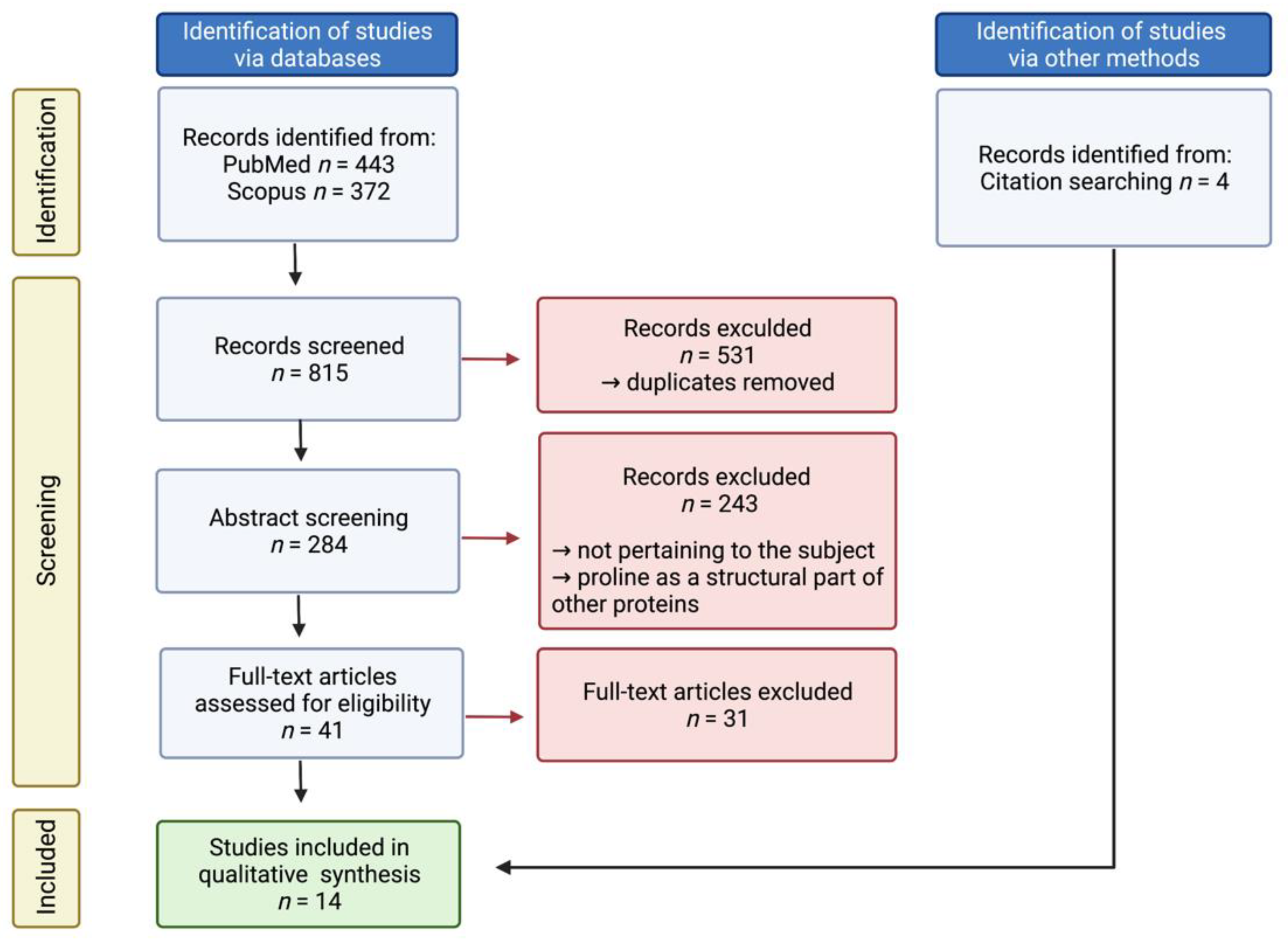
2. Materials and Methods
3. Results
4. Discussion
4.1. Historical Studies on Gliomas
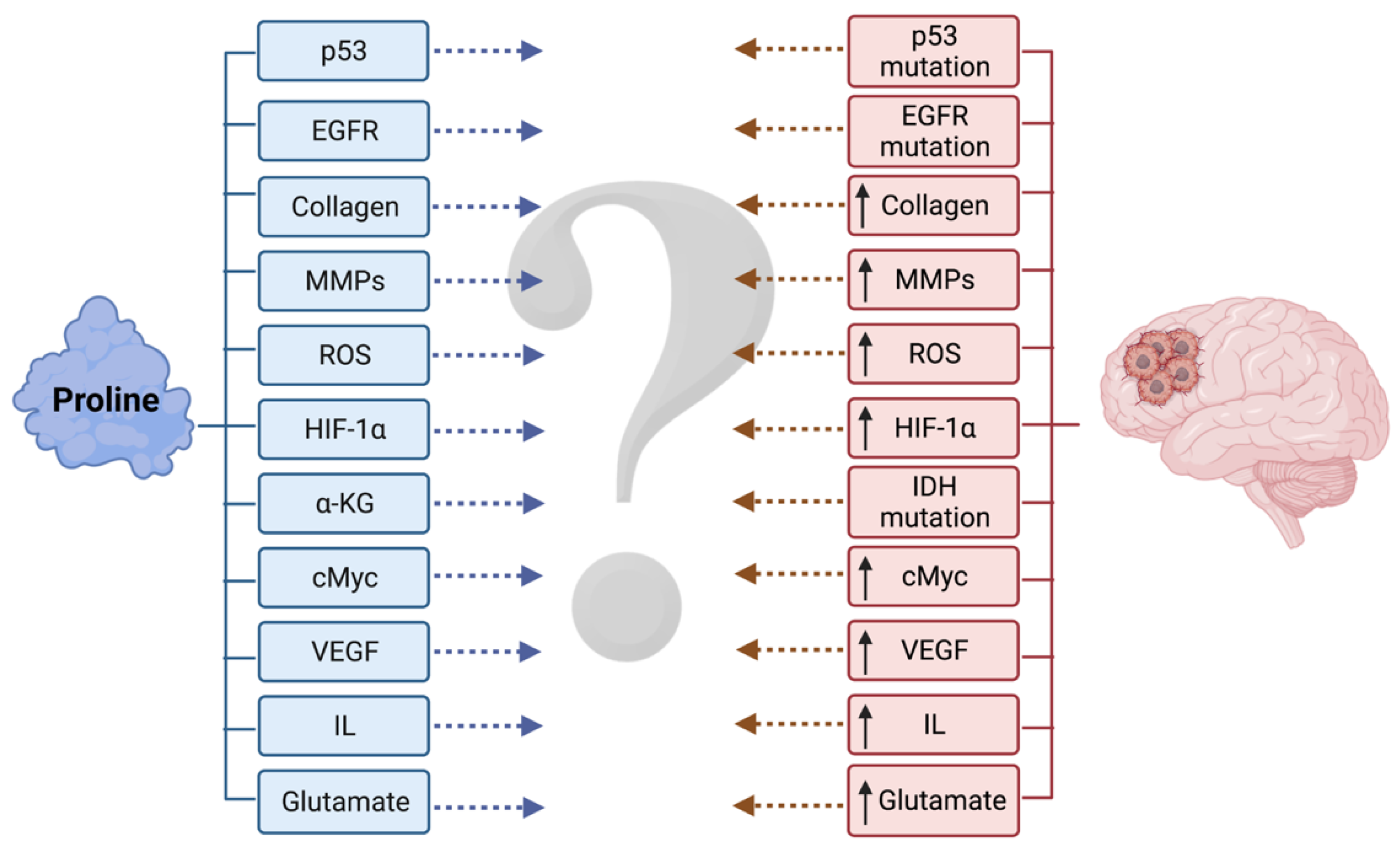
4.2. Role of Proline Metabolism in Gliomas
4.3. Regulatory Role of Proline Metabolism in Gliomas
4.4. Missing Links between Proline Metabolism and Gliomas
4.5. Ambiguous Role of POX/PRODH in General Oncology
4.6. Pro-Neoplastic Role of PYCR in General Oncology
4.7. Prolidase Supports Tumor Metabolism—General Oncology
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Farah, P.; Ondracek, A.; Chen, Y.; Wolinsky, Y.; Stroup, N.E.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013, 15 (Suppl. 2), ii1–ii56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Avgeropoulos, N.G.; Batchelor, T.T. New treatment strategies for malignant gliomas. Oncologist 1999, 4, 209–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Stylli, S.S. Novel Treatment Strategies for Glioblastoma. Cancers 2020, 12, 2883. [Google Scholar] [CrossRef]
- Tanner, J.J.; Fendt, S.M.; Becker, D.F. The Proline Cycle as a Potential Cancer Therapy Target. Biochemistry 2018, 57, 3433–3444. [Google Scholar] [CrossRef]
- Liu, W.; Hancock, C.N.; Fischer, J.W.; Harman, M.; Phang, J.M. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: Involvement of pyridine nucleotides. Sci. Rep. 2015, 5, 17206. [Google Scholar] [CrossRef] [Green Version]
- Burke, L.; Guterman, I.; Palacios Gallego, R.; Britton, R.G.; Burschowsky, D.; Tufarelli, C.; Rufini, A. The Janus-like role of proline metabolism in cancer. Cell Death Discov. 2020, 6, 104. [Google Scholar] [CrossRef]
- Grassian, A.R.; Parker, S.J.; Davidson, S.M.; Divakaruni, A.S.; Green, C.R.; Zhang, X.; Slocum, K.L.; Pu, M.; Lin, F.; Vickers, C.; et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014, 74, 3317–3331. [Google Scholar] [CrossRef] [Green Version]
- Bhavya, B.; Anand, C.R.; Madhusoodanan, U.K.; Rajalakshmi, P.; Krishnakumar, K.; Easwer, H.V.; Deepti, A.N.; Gopala, S. To be Wild or Mutant: Role of Isocitrate Dehydrogenase 1 (IDH1) and 2-Hydroxy Glutarate (2-HG) in Gliomagenesis and Treatment Outcome in Glioma. Cell. Mol. Neurobiol. 2020, 40, 53–63. [Google Scholar] [CrossRef]
- Maus, A.; Peters, G.J. Glutamate and α-ketoglutarate: Key players in glioma metabolism. Amino Acids 2017, 49, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapiero, H.; Mathé, G.; Couvreur, P.; Tew, K.D. II. Glutamine and glutamate. Biomed. Pharmacother. 2002, 56, 446–457. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W. Proline metabolism and cancer. Front. Biosci. 2012, 17, 1835–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phang, J.M.; Liu, W.; Hancock, C.; Christian, K.J. The proline regulatory axis and cancer. Front. Oncol. 2012, 2, 60. [Google Scholar] [CrossRef] [Green Version]
- Phang, J.M. Proline Metabolism in Cell Regulation and Cancer Biology: Recent Advances and Hypotheses. Antioxid. Redox Signal. 2019, 30, 635–649. [Google Scholar] [CrossRef] [Green Version]
- Misiura, M.; Miltyk, W. Current Understanding of the Emerging Role of Prolidase in Cellular Metabolism. Int. J. Mol. Sci. 2020, 21, 5906. [Google Scholar] [CrossRef]
- Lupi, A.; Tenni, R.; Rossi, A.; Cetta, G.; Forlino, A. Human prolidase and prolidase deficiency: An overview on the characterization of the enzyme involved in proline recycling and on the effects of its mutations. Amino Acids 2008, 35, 739–752. [Google Scholar] [CrossRef]
- Spodenkiewicz, M.; Cleary, M.; Massier, M.; Fitsialos, G.; Cottin, V.; Jouret, G.; Poirsier, C.; Doco-Fenzy, M.; Lèbre, A.S. Clinical Genetics of Prolidase Deficiency: An Updated Review. Biology 2020, 9, 108. [Google Scholar] [CrossRef]
- Eni-Aganga, I.; Lanaghan, Z.M.; Balasubramaniam, M.; Dash, C.; Pandhare, J. PROLIDASE: A Review from Discovery to its Role in Health and Disease. Front. Mol. Biosci. 2021, 8, 723003. [Google Scholar] [CrossRef]
- Phang, J.M.; Donald, S.P.; Pandhare, J.; Liu, Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids 2008, 35, 681–690. [Google Scholar] [CrossRef] [Green Version]
- Phang, J.M.; Liu, W.; Hancock, C.N.; Fischer, J.W. Proline metabolism and cancer: Emerging links to glutamine and collagen. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phang, J.M.; Pandhare, J.; Liu, Y. The metabolism of proline as microenvironmental stress substrate. J. Nutr. 2008, 138, 2008S–2015S. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline mechanisms of stress survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogner, A.N.; Stiers, K.M.; Tanner, J.J. Structure, biochemistry, and gene expression patterns of the proline biosynthetic enzyme pyrroline-5-carboxylate reductase (PYCR), an emerging cancer therapy target. Amino Acids 2021, 53, 1817–1834. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhu, S.; Lim, R.R.; Chao, J.R. Proline metabolism and transport in retinal health and disease. Amino Acids 2021, 53, 1789–1806. [Google Scholar] [CrossRef] [PubMed]
- Obara-Michlewska, M.; Szeliga, M. Targeting Glutamine Addiction in Gliomas. Cancers 2020, 12, 310. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wu, G. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 2018, 50, 29–38. [Google Scholar] [CrossRef]
- Patriarca, E.J.; Cermola, F.; D’Aniello, C.; Fico, A.; Guardiola, O.; De Cesare, D.; Minchiotti, G. The Multifaceted Roles of Proline in Cell Behavior. Front. Cell Dev. Biol. 2021, 9, 728576. [Google Scholar] [CrossRef]
- Yu, X.C.; Zhang, W.; Oldham, A.; Buxton, E.; Patel, S.; Nghi, N.; Tran, D.; Lanthorn, T.H.; Bomont, C.; Shi, Z.C.; et al. Discovery and characterization of potent small molecule inhibitors of the high affinity proline transporter. Neurosci. Lett. 2009, 451, 212–216. [Google Scholar] [CrossRef]
- Wyse, A.T.; Netto, C.A. Behavioral and neurochemical effects of proline. Metab. Brain Dis. 2011, 26, 159–172. [Google Scholar] [CrossRef]
- Orešič, M.; Tang, J.; Seppänen-Laakso, T.; Mattila, I.; Saarni, S.E.; Saarni, S.I.; Lönnqvist, J.; Sysi-Aho, M.; Hyötyläinen, T.; Perälä, J.; et al. Metabolome in schizophrenia and other psychotic disorders: A general population-based study. Genome Med. 2011, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafer, I.A.; Scriver, C.R.; Efron, M.L. Familial hyperprolinemia, cerebral dysfunction and renal anomalies occurring in a family with hereditary nephropathy and deafness. N. Engl. J. Med. 1962, 267, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Mitsubuchi, H.; Nakamura, K.; Matsumoto, S.; Endo, F. Inborn errors of proline metabolism. J. Nutr. 2008, 138, 2016S–2020S. [Google Scholar] [CrossRef] [PubMed]
- Hollinshead, K.E.R.; Munford, H.; Eales, K.L.; Bardella, C.; Li, C.; Escribano-Gonzalez, C.; Thakker, A.; Nonnenmacher, Y.; Kluckova, K.; Jeeves, M.; et al. Oncogenic IDH1 Mutations Promote Enhanced Proline Synthesis through PYCR1 to Support the Maintenance of Mitochondrial Redox Homeostasis. Cell Rep. 2018, 22, 3107–3114. [Google Scholar] [CrossRef] [Green Version]
- Cappelletti, P.; Tallarita, E.; Rabattoni, V.; Campomenosi, P.; Sacchi, S.; Pollegioni, L. Proline oxidase controls proline, glutamate, and glutamine cellular concentrations in a U87 glioblastoma cell line. PLoS ONE 2018, 13, e0196283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, A.G.K.; Biasibetti-Brendler, H.; Sidegum, D.S.V.; Loureiro, S.O.; Figueiró, F.; Wyse, A.T.S. Effect of Proline on Cell Death, Cell Cycle, and Oxidative Stress in C6 Glioma Cell Line. Neurotox. Res. 2021, 39, 327–334. [Google Scholar] [CrossRef]
- Gonullu, E.; Silav, G.; Kaya, M.; Arslan, M.; Gonullu, H.; Arslan, H.; Cebi, A.; Demir, H. Paraoxonase and Prolidase Activity in Patietns With Malignant Gliomas. J. Neurol. Sci.-Turk. 2012, 29, 778–782. [Google Scholar]
- Verma, A.; Singh, K.; Pandey, S.; Srivastava, R. Prolidase Activity and Oxidative Stress in Patients with Glioma. J. Clin. Diagn. Res. 2018, 12, BC07–BC10. [Google Scholar] [CrossRef]
- Shao, W.; Hao, Z.Y.; Chen, Y.F.; Du, J.; He, Q.; Ren, L.L.; Gao, Y.; Song, N.; Song, Y.; He, H.; et al. OIP5-AS1 specifies p53-driven POX transcription regulated by TRPC6 in glioma. J. Mol. Cell Biol. 2021, 13, 409–421. [Google Scholar] [CrossRef]
- Zhao, H.; Heimberger, A.B.; Lu, Z.; Wu, X.; Hodges, T.R.; Song, R.; Shen, J. Metabolomics profiling in plasma samples from glioma patients correlates with tumor phenotypes. Oncotarget 2016, 7, 20486–20495. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Weinstein, S.J.; Kitahara, C.M.; Karoly, E.D.; Sampson, J.N.; Albanes, D. A prospective study of serum metabolites and glioma risk. Oncotarget 2017, 8, 70366–70377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björkblom, B.; Jonsson, P.; Tabatabaei, P.; Bergström, P.; Johansson, M.; Asklund, T.; Bergenheim, A.T.; Antti, H. Metabolic response patterns in brain microdialysis fluids and serum during interstitial cisplatin treatment of high-grade glioma. Br. J. Cancer 2020, 122, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, P.; Antti, H.; Späth, F.; Melin, B.; Björkblom, B. Identification of Pre-Diagnostic Metabolic Patterns for Glioma Using Subset Analysis of Matched Repeated Time Points. Cancers 2020, 12, 3349. [Google Scholar] [CrossRef] [PubMed]
- Panosyan, E.H.; Lin, H.J.; Koster, J.; Lasky, J.L. In search of druggable targets for GBM amino acid metabolism. BMC Cancer 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhu, A.H.; Kant, S.; Kesarwani, P.; Ahmed, K.; Forsyth, P.; Nakano, I.; Chinnaiyan, P. Integrative cross-platform analyses identify enhanced heterotrophy as a metabolic hallmark in glioblastoma. Neuro Oncol. 2019, 21, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Loreck, D.J.; Galarraga, J.; Van der Feen, J.; Phang, J.M.; Smith, B.H.; Cummins, C.J. Regulation of the pentose phosphate pathway in human astrocytes and gliomas. Metab. Brain Dis. 1987, 2, 31–46. [Google Scholar] [CrossRef]
- Lefauconnier, J.M.; Portemer, C.; Chatagner, F. Free amino acids and related substances in human glial tumours and in fetal brain: Comparison with normal adult brain. Brain Res. 1976, 117, 105–113. [Google Scholar] [CrossRef]
- Hagedorn, C.H.; Phang, J.M. Transfer of reducing equivalents into mitochondria by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch. Biochem. Biophys. 1983, 225, 95–101. [Google Scholar] [CrossRef]
- Hagedorn, C.H.; Phang, J.M. Catalytic transfer of hydride ions from NADPH to oxygen by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch. Biochem. Biophys. 1986, 248, 166–174. [Google Scholar] [CrossRef]
- Surazynski, A.; Miltyk, W.; Palka, J.; Phang, J.M. Prolidase-dependent regulation of collagen biosynthesis. Amino Acids 2008, 35, 731–738. [Google Scholar] [CrossRef]
- Insolia, V.; Priori, E.C.; Gasperini, C.; Coppa, F.; Cocchia, M.; Iervasi, E.; Ferrari, B.; Besio, R.; Maruelli, S.; Bernocchi, G.; et al. Prolidase enzyme is required for extracellular matrix integrity and impacts on postnatal cerebellar cortex development. J. Comp. Neurol. 2020, 528, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Insolia, V.; Piccolini, V.M. Brain morphological defects in prolidase deficient mice: First report. Eur. J. Histochem. 2014, 58, 2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, L.S.; Huang, P.H. The pathobiology of collagens in glioma. Mol. Cancer Res. 2013, 11, 1129–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mughal, A.A.; Zhang, L.; Fayzullin, A.; Server, A.; Li, Y.; Wu, Y.; Glass, R.; Meling, T.; Langmoen, I.A.; Leergaard, T.B.; et al. Patterns of Invasive Growth in Malignant Gliomas-The Hippocampus Emerges as an Invasion-Spared Brain Region. Neoplasia 2018, 20, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Inda, M.M.; Bonavia, R.; Seoane, J. Glioblastoma multiforme: A look inside its heterogeneous nature. Cancers 2014, 6, 226–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Nunno, V.; Franceschi, E.; Tosoni, A.; Gatto, L.; Lodi, R.; Bartolini, S.; Brandes, A.A. Glioblastoma: Emerging Treatments and Novel Trial Designs. Cancers 2021, 13, 3750. [Google Scholar] [CrossRef]
- Saadeh, F.S.; Mahfouz, R.; Assi, H.I. EGFR as a clinical marker in glioblastomas and other gliomas. Int. J. Biol. Markers 2018, 33, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Vallentgoed, W.R.; French, P.J. Finding the Right Way to Target EGFR in Glioblastomas; Lessons from Lung Adenocarcinomas. Cancers 2018, 10, 489. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.R.D.; Regad, T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct. Target. Ther. 2017, 2, 17040. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Li, Y.; Bhattacharya, A.; Zhang, Y. PEPD is a pivotal regulator of p53 tumor suppressor. Nat. Commun. 2017, 8, 2052. [Google Scholar] [CrossRef] [Green Version]
- Surazynski, A.; Donald, S.P.; Cooper, S.K.; Whiteside, M.A.; Salnikow, K.; Liu, Y.; Phang, J.M. Extracellular matrix and HIF-1 signaling: The role of prolidase. Int. J. Cancer 2008, 122, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Zareba, I.; Palka, J. Prolidase-proline dehydrogenase/proline oxidase-collagen biosynthesis axis as a potential interface of apoptosis/autophagy. Biofactors 2016, 42, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, J.J.; Fu, X.L.; Guang, R.; To, S.T. Advances in the targeting of HIF-1α and future therapeutic strategies for glioblastoma multiforme (Review). Oncol. Rep. 2017, 37, 657–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, B.; Khwaja, F.W.; Severson, E.A.; Matheny, S.L.; Brat, D.J.; Van Meir, E.G. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 2005, 7, 134–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Fabiani, M.B.; Haase, S.; Comba, A.; Carney, S.; McClellan, B.; Banerjee, K.; Alghamri, M.S.; Syed, F.; Kadiyala, P.; Nunez, F.J.; et al. Genetic Alterations in Gliomas Remodel the Tumor Immune Microenvironment and Impact Immune-Mediated Therapies. Front. Oncol. 2021, 11, 631037. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Holsinger, R.M.; Kruse, C.A.; Flügel, A.; Graeber, M.B. The potential for genetically altered microglia to influence glioma treatment. CNS Neurol. Disord. Drug Targets 2013, 12, 750–762. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Jin, X.; Sohn, Y.W.; Jeon, H.Y.; Kim, E.J.; Ham, S.W.; Jeon, H.M.; Chang, S.Y.; Oh, S.Y.; Yin, J.; et al. Tumoral RANKL activates astrocytes that promote glioma cell invasion through cytokine signaling. Cancer Lett. 2014, 353, 194–200. [Google Scholar] [CrossRef]
- Han, J.; Alvarez-Breckenridge, C.A.; Wang, Q.E.; Yu, J. TGF-β signaling and its targeting for glioma treatment. Am. J. Cancer Res. 2015, 5, 945–955. [Google Scholar]
- D’Aniello, C.; Patriarca, E.J.; Phang, J.M.; Minchiotti, G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front. Oncol. 2020, 10, 776. [Google Scholar] [CrossRef]
- Liu, W.; Glunde, K.; Bhujwalla, Z.M.; Raman, V.; Sharma, A.; Phang, J.M. Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer Res. 2012, 72, 3677–3686. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Mao, C.; Liu, S.; Xiao, D.; Shi, Y.; Tao, Y. Proline dehydrogenase in cancer: Apoptosis, autophagy, nutrient dependency and cancer therapy. Amino Acids 2021, 53, 1891–1902. [Google Scholar] [CrossRef] [PubMed]
- Zareba, I.; Celinska-Janowicz, K.; Surazynski, A.; Miltyk, W.; Palka, J. Proline oxidase silencing induces proline-dependent pro-survival pathways in MCF-7 cells. Oncotarget 2018, 9, 13748–13757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinaldi, M.; Caffo, M.; Minutoli, L.; Marini, H.; Abbritti, R.V.; Squadrito, F.; Trichilo, V.; Valenti, A.; Barresi, V.; Altavilla, D.; et al. ROS and Brain Gliomas: An Overview of Potential and Innovative Therapeutic Strategies. Int. J. Mol. Sci. 2016, 17, 984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Phang, J.M. Proline dehydrogenase (oxidase) in cancer. Biofactors 2012, 38, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Annibali, D.; Whitfield, J.R.; Favuzzi, E.; Jauset, T.; Serrano, E.; Cuartas, I.; Redondo-Campos, S.; Folch, G.; Gonzàlez-Juncà, A.; Sodir, N.M.; et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat. Commun. 2014, 5, 4632. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Iafrate, A.J.; Ho, Q.; Curry, W.T.; Batchelor, T.T.; Flaherty, K.T.; Onozato, M.L.; Lelic, N.; Sundaram, S.; Cahill, D.P.; et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin. Cancer Res. 2016, 22, 4452–4465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Borchert, G.L.; Donald, S.P.; Diwan, B.A.; Anver, M.; Phang, J.M. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res. 2009, 69, 6414–6422. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Chiao, C.C.; Phan, N.N.; Li, C.Y.; Sun, Z.D.; Jiang, J.Z.; Hung, J.H.; Chen, Y.L.; Yen, M.C.; Weng, T.Y.; et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am. J. Cancer Res. 2020, 10, 95–113. [Google Scholar]
- Zareba, I.; Surazynski, A.; Chrusciel, M.; Miltyk, W.; Doroszko, M.; Rahman, N.; Palka, J. Functional Consequences of Intracellular Proline Levels Manipulation Affecting PRODH/POX-Dependent Pro-Apoptotic Pathways in a Novel in Vitro Cell Culture Model. Cell Physiol. Biochem. 2017, 43, 670–684. [Google Scholar] [CrossRef]
- Liu, Y.; Borchert, G.L.; Surazynski, A.; Phang, J.M. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene 2008, 27, 6729–6737. [Google Scholar] [CrossRef] [Green Version]
- Tołoczko-Iwaniuk, N.; Dziemiańczyk-Pakieła, D.; Celińska-Janowicz, K.; Zaręba, I.; Klupczyńska, A.; Kokot, Z.J.; Nowaszewska, B.K.; Reszeć, J.; Borys, J.; Miltyk, W. Proline-Dependent Induction of Apoptosis in Oral Squamous Cell Carcinoma (OSCC)-The Effect of Celecoxib. Cancers 2020, 12, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celińska-Janowicz, K.; Zaręba, I.; Lazarek, U.; Teul, J.; Tomczyk, M.; Pałka, J.; Miltyk, W. Constituents of propolis: Chrysin, caffeic acid, p-coumaric acid, and ferulic acid induce PRODH/POX-dependent apoptosis in human tongue squamous cell carcinoma cell (CAL-27). Front. Pharmacol. 2018, 9, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivares, O.; Mayers, J.R.; Gouirand, V.; Torrence, M.E.; Gicquel, T.; Borge, L.; Lac, S.; Roques, J.; Lavaut, M.N.; Berthezène, P.; et al. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat. Commun. 2017, 8, 16031. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mao, C.; Wang, M.; Liu, N.; Ouyang, L.; Liu, S.; Tang, H.; Cao, Y.; Wang, X.; Xiao, D.; et al. Cancer progression is mediated by proline catabolism in non-small cell lung cancer. Oncogene 2020, 39, 2358–2376. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Chang, L.; Tian, H.; Wang, L.; Zhang, Y.; Yang, T.; Li, G.; Hu, W.; Shah, K.; Chen, G.; et al. 1-Pyrroline-5-carboxylate released by prostate Cancer cell inhibit T cell proliferation and function by targeting SHP1/cytochrome c oxidoreductase/ROS Axis. J. Immunother. Cancer 2018, 6, 148. [Google Scholar] [CrossRef]
- Elia, I.; Broekaert, D.; Christen, S.; Boon, R.; Radaelli, E.; Orth, M.F.; Verfaillie, C.; Grünewald, T.G.P.; Fendt, S.M. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 2017, 8, 15267. [Google Scholar] [CrossRef]
- Fang, H.; Du, G.; Wu, Q.; Liu, R.; Chen, C.; Feng, J. HDAC inhibitors induce proline dehydrogenase (POX) transcription and anti-apoptotic autophagy in triple negative breast cancer. Acta Biochim. Biophys. Sin. 2019, 51, 1064–1070. [Google Scholar] [CrossRef]
- Maxwell, S.A.; Rivera, A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. J. Biol. Chem. 2003, 278, 9784–9789. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.; Jain, M.; Madhusudhan, N.; Sheppard, N.G.; Strittmatter, L.; Kampf, C.; Huang, J.; Asplund, A.; Mootha, V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014, 5, 3128. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Wang, L.; Zhang, Y.; Yan, Z.; Liu, L.; Chen, G. PYCR1 promotes the progression of non-small-cell lung cancer under the negative regulation of miR-488. Biomed. Pharmacother. 2019, 111, 588–595. [Google Scholar] [CrossRef]
- Cai, F.; Miao, Y.; Liu, C.; Wu, T.; Shen, S.; Su, X.; Shi, Y. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol. Lett. 2018, 15, 731–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, S.; Zhang, C.; Shan, J. Pyrroline-5-Carboxylate Reductase 1 Accelerates the Migration and Invasion of Nonsmall Cell Lung Cancer. Cancer Biother. Radiopharm. 2019, 34, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Luo, L.; Xie, Y.; Zhao, Y.; Yao, J.; Liu, X. PYCR1 knockdown inhibits the proliferation, migration, and invasion by affecting JAK/STAT signaling pathway in lung adenocarcinoma. Mol. Carcinog. 2020, 59, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Kuo, M.L.; Su, L.; Xue, L.; Luh, F.; Zhang, H.; Wang, J.; Lin, T.G.; Zhang, K.; Chu, P.; et al. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 2017, 38, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Song, Y.; Ye, Y.; He, S.; Ma, X.; Zhang, M.; Ni, J.; Wang, J.; Xia, W. PYCR1 interference inhibits cell growth and survival via c-Jun N-terminal kinase/insulin receptor substrate 1 (JNK/IRS1) pathway in hepatocellular cancer. J. Transl. Med. 2019, 17, 343. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Ericksen, R.E.; Escande-Beillard, N.; Lee, Q.Y.; Loh, A.; Denil, S.; Steckel, M.; Haegebarth, A.; Wai Ho, T.S.; Chow, P.; et al. Metabolic pathway analyses identify proline biosynthesis pathway as a promoter of liver tumorigenesis. J. Hepatol. 2020, 72, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Zhu, L.; Liao, M.; Zhuo, W.; Yang, S.; Wu, W.; Wang, D. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med. Oncol. 2017, 34, 27. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Li, S.; Yuan, Z.; Zhou, L. Pyrroline-5-carboxylate reductase 1 (PYCR1) upregulation contributes to gastric cancer progression and indicates poor survival outcome. Ann. Transl. Med. 2020, 8, 937. [Google Scholar] [CrossRef]
- Du, S.; Sui, Y.; Ren, W.; Zhou, J.; Du, C. PYCR1 promotes bladder cancer by affecting the Akt/Wnt/β-catenin signaling. J. Bioenerg. Biomembr. 2021, 53, 247–258. [Google Scholar] [CrossRef]
- Ye, Y.; Wu, Y.; Wang, J. Pyrroline-5-carboxylate reductase 1 promotes cell proliferation via inhibiting apoptosis in human malignant melanoma. Cancer Manag. Res. 2018, 10, 6399–6407. [Google Scholar] [CrossRef] [Green Version]
- Wilk, P.; Wątor, E.; Weiss, M.S. Prolidase—A protein with many faces. Biochimie 2021, 183, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Celik, S.; Kızıltan, R.; Yılmaz, E.M.; Yılmaz, Ö.; Demir, H. Potential diagnostic and prognostic significance of plasma prolidase activity in gastric cancer. Biomark. Med. 2017, 11, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Cechowska-Pasko, M.; Pałka, J.; Wojtukiewicz, M.Z. Enhanced prolidase activity and decreased collagen content in breast cancer tissue. Int. J. Exp. Pathol. 2006, 87, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Arioz, D.T.; Camuzcuoglu, H.; Toy, H.; Kurt, S.; Celik, H.; Aksoy, N. Serum prolidase activity and oxidative status in patients with stage I endometrial cancer. Int. J. Gynecol. Cancer 2009, 19, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Gecit, I.; Aslan, M.; Gunes, M.; Pirincci, N.; Esen, R.; Demir, H.; Ceylan, K. Serum prolidase activity, oxidative stress, and nitric oxide levels in patients with bladder cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 739–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guszczyn, T.; Sobolewski, K. Deregulation of collagen metabolism in human stomach cancer. Pathobiology 2004, 71, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Sayır, F.; Şehitoğulları, A.; Demir, H.; Aslan, M.; Çobanoğlu, U.; Bilgin, C. Serum prolidase activity, total oxidant/antioxidant, and nitric oxide levels in patients with esophageal squamous cell carcinoma. Turk. J. Thorac. Cardiovasc. Surg. 2019, 27, 206–211. [Google Scholar] [CrossRef]
- Palka, J.; Surazynski, A.; Karna, E.; Orlowski, K.; Puchalski, Z.; Pruszynski, K.; Laszkiewicz, J.; Dzienis, H. Prolidase activity disregulation in chronic pancreatitis and pancreatic cancer. Hepatogastroenterology 2002, 49, 1699–1703. [Google Scholar]

| Author | Year | Title | Materials/Methods | Highlights |
|---|---|---|---|---|
| Hollinshead et al. [34] | 2018 | Oncogenic IDH1 Mutations Promote Enhanced Proline Synthesis through PYCR1 to Support the Maintenance of Mitochondrial Redox Homeostasis | Cell cultures: human anaplastic oligodendroglioma (grade 3) wild-type and IDH-mutant | IDH-mutant high-grade oligodendroglioma utilizes proline cycle as a redox shuttle to maintain redox balance. PYCR1 expression is increased in IDH1-mutated gliomas. Glutamine-derived proline synthesis is increased. An adaptation for hypoxic conditions: sparing oxygen allows for anabolism. |
| Cappelletti et al. [35] | 2018 | Proline oxidase controls proline, glutamate, and glutamine cellular concentrations in a U87 glioblastoma cell line | Cell cultures: human U87 GBM wild-type and POX-mutant | U87 human GBM cells with wild-type and impaired-activity POX/PRODH were studied to assess connections between proline and glutamate metabolism. A decrease in POX/PRODH activity caused a transient drop in proline, glutamate, and glutamine concentration. |
| Ferreira et al. [36] | 2020 | Effect of Proline on Cell Death, Cell Cycle, and Oxidative Stress in C6 Glioma Cell Line | Cell cultures: rat C6 GBM | The influence of proline extracellular concentration on GBM cultures was assessed. With increasing concentration: no cytotoxicity, no apoptosis, more ROS, and an increase in NF- κB—in general, a pro-proliferative influence. |
| Gönullu [37] | 2012 | Paraoxonase and Prolidase Activity in Patients with Malignant Gliomas | Serum of GBM patients | In patients diagnosed with GBM and anaplastic astrocytoma (grade 3), serum prolidase activity was lower than that in the healthy controls. |
| Verma et al. [38] | 2018 | Prolidase Activity and Oxidative Stress in Patients with Glioma | Serum and excised GBM tissue | In GBM patients, prolidase activity was elevated both in the serum and in tumor tissue. |
| Shao et al. [39] | 2021 | OIP5-AS1 specifies p53-driven POX transcription regulated by TRPC6 in glioma | Cell cultures, animal model | Molecular mechanism of how POX/PRODH is regulated by p53 through TRPC6. In general, POX/PRODH suppressed glioma. POX/PRODH in glioma was sixfold lower than that in the brain. |
| Zhao et al. [40] | 2016 | Metabolomics profiling in plasma samples from glioma patients correlates with tumor phenotypes | Database of plasma metabolomics | Metabolic pathways and metabolites of proline and arginine had the highest impact in differentiating high vs. low grade, IDH mutation vs. wild type. |
| Huang et al. [41] | 2017 | A prospective study of serum metabolites and glioma risk | Plasma of GBM patients and matched controls | Proline/arginine metabolism related to malignant glioma: lower metabolite concentration in the plasma of GBM patients. |
| Björkblom et al. [42] | 2020 | Metabolic response patterns in brain microdialysis fluids and serum during interstitial cisplatin treatment of high-grade glioma | Brain and tumor microdialysis fluid | Compared with the unaffected brain, glioma microdialysis fluid had a 4.4-fold higher proline concentration. |
| Jonsson et al. [43] | 2020 | Identification of Pre-Diagnostic Metabolic Patterns for Glioma Using Subset Analysis of Matched Repeated Time Points | Plasma sample metabolomic profile | A study in a large biobank of plasma samples; selected glioma cases prior to diagnosis were matched with controls. Along with a number of other amino acids, plasma proline concentration in glioma patients was significantly higher even prior to diagnosis, with potential to consider proline as a pre-diagnostic glioma biomarker. |
| Panosyan et al. [44] | 2017 | In search of druggable targets for GBM amino acid metabolism | Database of gene expression | In large datasets of gene expression, the authors found that POX/PRODH gene expression was lower in GBM than in non-GBM. Among GBM samples, there was a 50/50 equilibrium in high vs. low POX/PRODH expression, and overall survival (OS) was shorter in high-POX/PRODH patients, with a high level of significance. Although significant, the authors reflected upon POX/PRODH somewhat superficially. Additionally, the authors emphasized the role of glutamate. |
| Prabhu et al. [45] | 2019 | Integrative cross-platform analyses identify enhanced heterotrophy as a metabolic hallmark in glioblastoma | GBM and low-grade glioma tissues metabolomic and genomic profiles | In a search for a glioma metabolic reprogramming program, the authors found that proline metabolism was significantly altered. |
| Loreck et al. [46] | 1987 | Regulation of the pentose phosphate pathway in human astrocytes and gliomas | Cells cultures | A historical study. Proline cycle in gliomas was absent; no effect was observed on glucose metabolism due to the addition of proline to the medium: “POX is low to absent”. |
| Lefauconnier et al. [47] | 1976 | Free Amino Acids and Related Substances in Human Glial Tumours and in Fetal Brain–Comparison with Normal Adult Brain | Tissues: excised GBM, fetal and adult brain | A historical study. Proline concentration in GBM was 10x higher than that in the brain. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawicka, M.M.; Sawicki, K.; Łysoń, T.; Polityńska, B.; Miltyk, W. Proline Metabolism in Malignant Gliomas: A Systematic Literature Review. Cancers 2022, 14, 2030. https://doi.org/10.3390/cancers14082030
Sawicka MM, Sawicki K, Łysoń T, Polityńska B, Miltyk W. Proline Metabolism in Malignant Gliomas: A Systematic Literature Review. Cancers. 2022; 14(8):2030. https://doi.org/10.3390/cancers14082030
Chicago/Turabian StyleSawicka, Magdalena M., Karol Sawicki, Tomasz Łysoń, Barbara Polityńska, and Wojciech Miltyk. 2022. "Proline Metabolism in Malignant Gliomas: A Systematic Literature Review" Cancers 14, no. 8: 2030. https://doi.org/10.3390/cancers14082030
APA StyleSawicka, M. M., Sawicki, K., Łysoń, T., Polityńska, B., & Miltyk, W. (2022). Proline Metabolism in Malignant Gliomas: A Systematic Literature Review. Cancers, 14(8), 2030. https://doi.org/10.3390/cancers14082030






