The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies
Abstract
:Simple Summary
Abstract
1. Introduction
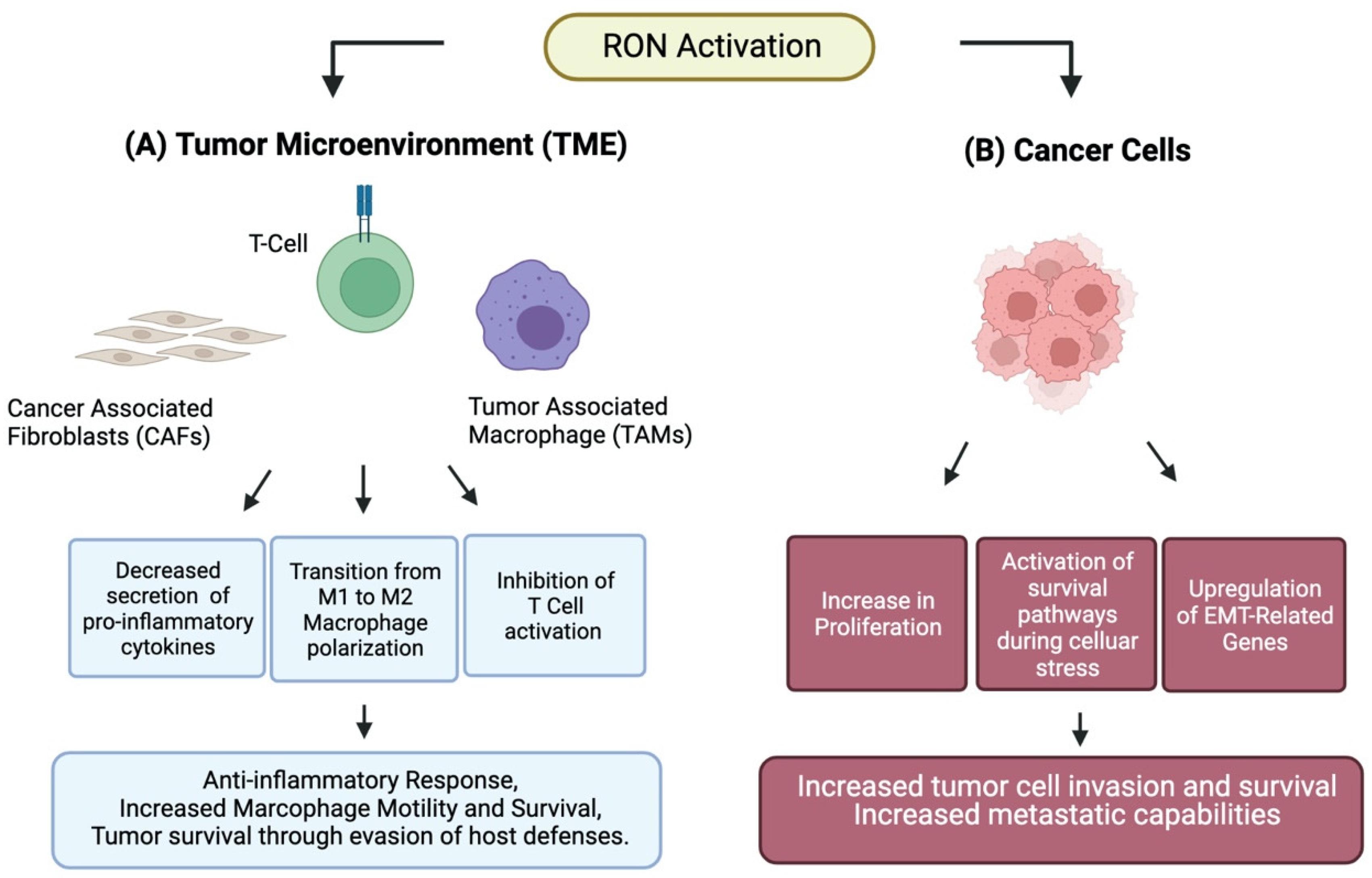
2. RON Attenuates Inflammation
3. RON Isoforms
4. RON Alters Macrophage Polarization
5. RON in Cancer Cells
6. RON Crosstalk and Other RTKs
6.1. RON and EGFR Crosstalk in Cancer
6.2. RON and MET Crosstalk in Cancer
6.3. RON Crosstalk with Other RTKs
7. RON in Metastasis
8. RON and Adaptation to Cellular Stress
9. Clinical Trials
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ronsin, C.; Muscatelli, F.; Mattei, M.G.; Breathnach, R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene 1993, 8, 1195–1202. [Google Scholar]
- Wang, M.-H.; Ronsin, C.; Gesnel, M.-C.; Coupey, L.; Skeel, A.; Leonard, E.J.; Breathnach, R. Identification of the ron Gene Product as the Receptor for the Human Macrophage Stimulating Protein. Science 1994, 266, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, G.; Follenzi, A.; Naldini, L.; Collesi, C.; Santoro, M.M.; Gallo, K.; Godowski, P.; Comoglio, P. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. EMBO J. 1994, 13, 3524–3532. [Google Scholar] [CrossRef]
- Yoshimura, T.; Yuhki, N.; Wang, M.H.; Skeel, A.; Leonard, E. Cloning, sequencing, and expression of human macrophage stimulating protein (MSP, MST1) confirms MSP as a member of the family of kringle proteins and locates the MSP gene on chromosome 3. J. Biol. Chem. 1993, 268, 15461–15468. [Google Scholar] [CrossRef]
- Wang, M.-H.; Wang, D.; Chen, Y.-Q. Oncogenic and invasive potentials of human macrophage-stimulating protein receptor, the RON receptor tyrosine kinase. Carcinogenesis 2003, 24, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Danilkovitch-Miagkova, A.; Miagkov, A.; Skeel, A. Oncogenic Mutants of RON and MET Receptor Tyrosine Kinases Cause Activation of the ß-Catenin Pathway. Mol. Cell. Biol. 2001, 21, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-Q.; Zhou, Y.-Q.; Angeloni, D.; Kurtz, A.L.; Qiang, X.-Z.; Wang, M.-H. Overexpression and Activation of the RON Receptor Tyrosine Kinase in a Panel of Human Colorectal Carcinoma Cell Lines. Exp. Cell Res. 2000, 261, 229–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.-Q.; Wang, M.-H.; Kung, H.-F. Macrophage Stimulating Protein Activates Ras by both Activation and Translocation of SOS Nucleotide Exchange Factor. Biochem. Biophys. Res. Commun. 1995, 216, 110–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudino, G.; Avantaggiato, V.; Follenzi, A. The proto-oncogene RON is involved in development of epithelial, bone and neuro-endocrine tissues. Oncogene 1995, 11, 2627–2637. [Google Scholar] [PubMed]
- Quantin, B.; Schuhbaur, B.; Gesnel, M.-C.; Dollé, P.; Breathnach, R. Restricted expression of the ron gene encoding the macrophage stimulating protein receptor during mouse development. Dev. Dyn. 1995, 204, 383–390. [Google Scholar] [CrossRef] [PubMed]
- van den Akker, E.; van Dijk, T.; Parren-van Amelsvoort, M.; Grossmann, K.S.; Schaeper, U.; Toney-Earley, K.; Waltz, S.E.; Löwenberg, B.; von Lindern, M. Tyrosine kinase receptor RON functions downstream of the erythropoietin receptor to induce expansion of erythroid progenitors. Blood 2004, 103, 4457–4465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunelleschi, S.; Penengo, L.; Lavagno, L. Macrophage Stimulating Protein (MSP) evokes superoxide anion production by human macrophages of different origin. Br. J. Pharmacol. 2001, 134, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggiora, P.; Marchio, S.; Stella, M.C.; Giai, M.; Belfiore, A.; De Bortoli, M.; Di Renzo, M.F.; Costantino, A.; Sismondi, P.; Comoglio, P.M. Overexpression of the RON gene in human breast carcinoma. Oncogene 1998, 16, 2927–2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.-Q.; He, C.; Chen, Y.-Q.; Wang, D.; Wang, M.-H. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: Generation of different splicing RON variants and their oncogenic potential. Oncogene 2003, 22, 186–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thobe, M.N.; Gray, J.K.; Gurusamy, D.; Paluch, A.M.; Wagh, P.K.; Pathrose, P.; Lentsch, A.B.; Waltz, S.E. The Ron receptor promotes prostate tumor growth in the TRAMP mouse model. Oncogene 2011, 30, 4990–4998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Greninger, P.; Rhodes, D.; Koopman, L.; Violette, S.; Bardeesy, N.; Settleman, J. A Gene Expression Signature Associated with “K-Ras Addiction” Reveals Regulators of EMT and Tumor Cell Survival. Cancer Cell 2009, 15, 489–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, R.M.; Toney, K.; Fenoglio-Preiser, C.; Revelo-Penafiel, M.P.; Hingorani, S.; Tuveson, D.A.; Waltz, S.; Lowy, A.M. The RON Receptor Tyrosine Kinase Mediates Oncogenic Phenotypes in Pancreatic Cancer Cells and Is Increasingly Expressed during Pancreatic Cancer Progression. Cancer Res. 2007, 67, 6075–6082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan-Collins, J.; Thomas, R.M.; Yu, P.; Jaquish, D.; Mose, E.; French, R.; Stuart, W.; McClaine, R.; Aronow, B.; Hoffman, R.M.; et al. Silencing of RON Receptor Signaling Promotes Apoptosis and Gemcitabine Sensitivity in Pancreatic Cancers. Cancer Res. 2010, 70, 1130–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fialin, C.; Larrue, C.; Vergez, F.; Sarry, J.E.; Bertoli, S.; Mas, V.M.-D.; Demur, C.; Delabesse, E.; Payrastre, B.; Manenti, S.; et al. The short form of RON is expressed in acute myeloid leukemia and sensitizes leukemic cells to cMET inhibitors. Leukemia 2013, 27, 325–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, X.; Zhang, X.; Fan, J.; Tong, Y.; Li, S.; Jin, J.; Yao, H. The RON receptor tyrosine kinase is a potential therapeutic target in Burkitt lymphoma. Cancer Biol. Ther. 2013, 14, 370–377. [Google Scholar] [CrossRef] [Green Version]
- Maggiora, P.; Lorenzato, A.; Fracchioli, S.; Costa, B.; Castagnaro, M.; Arisio, R.; Katsaros, D.; Massobrio, M.; Comoglio, P.; Di Renzo, M.F. The RON and MET oncogenes are co-expressed in human ovarian carcinomas and cooperate in activating invasiveness. Exp. Cell Res. 2003, 288, 382–389. [Google Scholar] [CrossRef]
- Chen, Q.; Seol, D.W.; Carr, B.; Zarnegar, R. Co-expression and regulation ofMet andRon proto-oncogenes in human hepatocellular carcinoma tissues and cell lines. Hepatology 1997, 26, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Q.; Zhou, Y.-Q.; Fisher, J.H.; Wang, M.-H. Targeted expression of the receptor tyrosine kinase RON in distal lung epithelial cells results in multiple tumor formation: Oncogenic potential of RON in vivo. Oncogene 2002, 21, 6382–6386. [Google Scholar] [CrossRef] [Green Version]
- Willett, C.G.; Wang, M.-H.; Emanuel, R.L.; Graham, S.A.; Smith, D.I.; Shridhar, V.; Sugarbaker, D.J.; Sunday, M.E. Macrophage-stimulating Protein and Its Receptor in Non-small-cell Lung Tumors: Induction of Receptor Tyrosine Phosphorylation and Cell Migration. Am. J. Respir. Cell Mol. Biol. 1998, 18, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.-Y.; Liu, H.-S.; Cheng, H.-L.; Tzai, T.-S.; Guo, H.-R.; Ho, C.-L.; Chow, N.-H. Collaboration of RON and Epidermal Growth Factor Receptor in Human Bladder Carcinogenesis. J. Urol. 2006, 176, 2262–2267. [Google Scholar] [CrossRef] [PubMed]
- Gurusamy, D.; Gray, J.K.; Pathrose, P.; Kulkarni, R.M.; Finkleman, F.D.; Waltz, S.E. Myeloid-Specific Expression of Ron Receptor Kinase Promotes Prostate Tumor Growth. Cancer Res. 2013, 73, 1752–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallakin, A.; Kutcher, L.W.; McDowell, S.A.; Kong, S.; Schuster, R.; Lentsch, A.B.; Aronow, B.J.; Leikauf, G.D.; Waltz, S.E. Gene Expression Profiles of Mst1r-Deficient Mice during Nickel-Induced Acute Lung Injury. Am. J. Respir. Cell Mol. Biol. 2006, 34, 15–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lentsch, A.B.; Pathrose, P.; Kader, S.; Kuboki, S.; Collins, M.H.; Waltz, S.E. The Ron Receptor Tyrosine Kinase Regulates Acute Lung Injury and Suppresses Nuclear Factor κB Activation. Shock 2007, 27, 274–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.-Q.; Chen, Y.-Q.; Fisher, J.H.; Wang, M.-H. Activation of the RON Receptor Tyrosine Kinase by Macrophage-stimulating Protein Inhibits Inducible Cyclooxygenase-2 Expression in Murine Macrophages. J. Biol. Chem. 2002, 277, 38104–38110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaidis, N.M.; Gray, J.K.; Gurusamy, D.; Fox, W.; Stuart, W.D.; Huber, N.; Waltz, S.E. Ron Receptor Tyrosine Kinase Negatively Regulates Tnfα Production in Alveolar Macrophages by Inhibiting Nf-κB Activity and Adam17 Production. Shock 2010, 33, 197–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, D.C.; Clements, J.E.; Henderson, A.J. RON Receptor Tyrosine Kinase, a Negative Regulator of Inflammation, Is Decreased during Simian Immunodeficiency Virus–Associated Central Nervous System Disease. J. Immunol. 2013, 191, 4280–4287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, H.; Cho, S.; Loh, T.J.; Zhou, J.; Ghigna, C.; Biamonti, G.; Green, M.R.; Zheng, X.; Shen, H. A 2-nt RNA enhancer on exon 11 promotes exon 11 inclusion of the Ron proto-oncogene. Oncol. Rep. 2014, 31, 450–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakedis, J.; French, R.; Babicky, M.; Jaquish, D.; Mose, E.; Cheng, P.; Holman, P.; Howard, H.; Miyamoto, J.; Porras, P.; et al. Characterization of RON protein isoforms in pancreatic cancer: Implications for biology and therapeutics. Oncotarget 2016, 7, 45959–45975. [Google Scholar] [CrossRef] [PubMed]
- Chakedis, J.; French, R.; Babicky, M.L.; Jaquish, D.; Howard, H.; Mose, E.; Lam, R.; Holman, P.; Miyamoto, J.; Walterscheid, Z.; et al. A novel protein isoform of the RON tyrosine kinase receptor transforms human pancreatic duct epithelial cells. Oncogene 2015, 35, 3249–3259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, S.-C.A.; Gundlapalli, H.; Ekiz, H.A.; Jiang, A.; Fernandez, E.; Welm, A.L. Blocking Short-Form Ron Eliminates Breast Cancer Metastases through Accumulation of Stem-Like CD4+ T Cells That Subvert Immunosuppression. Cancer Discov. 2021, 11, 3178–3197. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; Hu, S.; He, J.; Wu, Z.; Qi, Z.; Huang, M.; Liu, R.; Lin, Y.; Tan, C.; et al. Short-form RON (sf-RON) enhances glucose metabolism to promote cell proliferation via activating β-catenin/SIX1 signaling pathway in gastric cancer. Cell Biol. Toxicol. 2021, 37, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, A.; Rajput, A.; Wan, G. RON kinase isoforms demonstrate variable cell motility in normal cells. Heliyon 2016, 2, e00153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welm, A.L.; Sneddon, J.B.; Taylor, C.; Nuyten, D.S.A.; van de Vijver, M.J.; Hasegawa, B.H.; Bishop, J.M. The macrophage-stimulating protein pathway promotes metastasis in a mouse model for breast cancer and predicts poor prognosis in humans. Proc. Natl. Acad. Sci. USA 2007, 104, 7570–7575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eyob, H.; Ekiz, H.A.; Derose, Y.S.; Waltz, S.; Williams, M.; Welm, A.L. Inhibition of Ron Kinase Blocks Conversion of Micrometastases to Overt Metastases by Boosting Antitumor Immunity. Cancer Discov. 2013, 3, 751–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babicky, M.L.; Harper, M.M.; Chakedis, J.; Cazes, A.; Mose, E.S.; Jaquish, D.V.; French, R.P.; Childers, B.; Alakus, H.; Schmid, M.C.; et al. MST1R kinase accelerates pancreatic cancer progression via effects on both epithelial cells and macrophages. Oncogene 2019, 38, 5599–5611. [Google Scholar] [CrossRef] [PubMed]
- Krishnaswamy, S.; Mohammed, A.K.; Amer, O.E.; Tripathi, G.; Alokail, M.S.; Al-Daghri, N.M. Recepteur d’Origine nantais (RON) tyrosine kinase splicing variants lacking exons 18 and 19 occur ubiquitously in lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 20778–20786. [Google Scholar] [PubMed]
- Graves-Deal, R.; Bogatcheva, G.; Rehman, S.; Lu, Y.; Higginbotham, J.N.; Singh, B. Broad-spectrum receptor tyrosine kinase inhibitors overcome de novo and acquired modes of resistance to EGFR-targeted therapies in colorectal cancer. Oncotarget 2019, 10, 1320–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilangumaran, S.; Villalobos-Hernandez, A.; Bobbala, D.; Ramanathan, S. The hepatocyte growth factor (HGF)–MET receptor tyrosine kinase signaling pathway: Diverse roles in modulating immune cell functions. Cytokine 2016, 82, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Khoi, P.N.; Li, S.; Thuan, U.T.; Sah, D.K.; Kang, T.W.; Nguyen, T.T.; Lian, S.; Xia, Y.; Jung, Y.D. Lysophosphatidic Acid Upregulates Recepteur D’origine Nantais Expression and Cell Invasion via Egr-1, AP-1, and NF-κB Signaling in Bladder Carcinoma Cells. Int. J. Mol. Sci. 2020, 21, 304. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-S.; Hsu, P.Y.; Lai, M.-D.; Chang, H.-Y.; Ho, C.-L.; Cheng, H.-L.; Chen, H.-T.; Lin, Y.-J.; Wu, T.-J.; Tzai, T.-S.; et al. An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis 2010, 31, 1456–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, J.; Nimnual, A.S.; Shroyer, K.R.; Joy, C.; Ischenko, I.; Chandler, C.S.; Dong, L.M.; Hayman, M.J.; Chan, E.L. Ron tyrosine kinase receptor synergises with EGFR to confer adverse features in head and neck squamous cell carcinoma. Br. J. Cancer 2013, 109, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, D.M.; Short, K.; Stueven, N. MST1R/RON and EGFR in a complex with syndecans sustain carcinoma S-phase progression by preventing p38MAPK activation. Cancer Biol. 2018; preprint. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, C.; Brown, N.E.; Vasiliauskas, J.; Pathrose, P.; Starnes, S.L.; Waltz, S.E. Prostate Epithelial RON Signaling Promotes M2 Macrophage Activation to Drive Prostate Tumor Growth and Progression. Mol. Cancer Res. 2020, 18, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.; Lin, Y.H.; Goossen, E.A.; DeVette, C.I.; Albertella, M.R.; Thomson, S.; Mulvihill, M.J.; Welm, A.L. The RON Receptor Tyrosine Kinase Promotes Metastasis by Triggering MBD4-Dependent DNA Methylation Reprogramming. Cell Rep. 2014, 6, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Benight, N.M.; Wagh, P.K.; Zinser, G.M.; Peace, B.E.; Stuart, W.D.; Vasiliauskas, J.; Pathrose, P.; Starnes, S.L.; Waltz, S.E. HGFL supports mammary tumorigenesis by enhancing tumor cell intrinsic survival and influencing macrophage and T-cell responses. Oncotarget 2015, 6, 17445–17461. [Google Scholar] [CrossRef] [Green Version]
- Faham, N.; Zhao, L.; Welm, A.L.; Chiavi, G.L.; Gentile, A.; Comoglio, P. mTORC1 is a key mediator of RON-dependent breast cancer metastasis with therapeutic potential. NPJ Breast Cancer 2018, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Benvenuti, S.; Lazzari, L.; Arnesano, A. Ron Kinase Transphosphorylation Sustains MET Oncogene Addiction. Cancer Res. 2011, 71, 1945–1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, K.; Karnad, A.; Zhao, S.; Freeman, J.W. Roles of c-Met and RON kinases in tumor progression and their potential as therapeutic targets. Oncotarget 2015, 6, 3507–3518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Follenzi, A.; Bakovic, S.; Gual, P.; Stella, M.C.; Longati, P.; Comoglio, P. Cross-talk between the proto-oncogenes Met and Ron. Oncogene 2000, 19, 3041–3049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batth, I.; Yun, H.; Hussain, S.; Meng, P.; Osmulski, P.; Huang, T.H.-M.; Bedolla, R.; Profit, A.; Reddick, R.; Kumar, A. Crosstalk between RON and androgen receptor signaling in the development of castration resistant prostate cancer. Oncotarget 2016, 7, 14048–14063. [Google Scholar] [CrossRef] [PubMed]
- Weng, T.-H.; Yao, M.Y.; Xu, X.-M.; Hu, C.-Y.; Yao, S.-H.; Liu, Y.-Z.; Wu, Z.-G.; Tang, T.-M.; Fu, P.-F.; Wang, M.-H.; et al. RON and MET Co-overexpression Are Significant Pathological Characteristics of Poor Survival and Therapeutic Targets of Tyrosine Kinase Inhibitors in Triple-Negative Breast Cancer. Cancer Res. Treat. 2020, 52, 973–986. [Google Scholar] [CrossRef]
- Hu, C.-Y.; Xu, X.M.; Hong, B.; Wu, Z.-G.; Qian, Y.; Weng, T.-H.; Liu, Y.-Z.; Tang, T.-M.; Wang, M.-H.; Yao, H.-P. Aberrant RON and MET Co-overexpression as Novel Prognostic Biomarkers of Shortened Patient Survival and Therapeutic Targets of Tyrosine Kinase Inhibitors in Pancreatic Cancer. Front. Oncol. 2019, 9, 1377. [Google Scholar] [CrossRef]
- Yin, B.; Liu, Z.; Wang, Y.; Wang, X.; Liu, W.; Yu, P.; Duan, X.; Liu, C.; Chen, Y.; Zhang, Y.; et al. RON and c-Met facilitate metastasis through the ERK signaling pathway in prostate cancer cells. Oncol. Rep. 2017, 37, 3209–3218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraoka, R.S.; Sun, W.Y.; Colbert, M.C.; Waltz, S.E.; Witte, D.P.; Degen, J.L.; Degen, S.J.F. The Ron/STK receptor tyrosine kinase is essential for peri-implantation development in the mouse. J. Clin. Investig. 1999, 103, 1277–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderwerff, B.R.; Church, K.J.; Kawas, L.H.; Harding, J.W. Comparative characterization of the HGF/Met and MSP/Ron systems in primary pancreatic adenocarcinoma. Cytokine 2019, 123, 154762. [Google Scholar] [CrossRef] [PubMed]
- Collesi, C.; Santoro, M.M.; Gaudino, G.; Comoglio, P.M. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol. Cell. Biol. 1996, 16, 5518–5526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potratz, J.C.; Saunders, D.N.; Wai, D.H.; Ng, T.L.; McKinney, S.E.; Carboni, J.M.; Gottardis, M.M.; Triche, T.J.; Jürgens, H.; Pollak, M.N.; et al. Synthetic Lethality Screens Reveal RPS6 and MST1R as Modifiers of Insulin-like Growth Factor-1 Receptor Inhibitor Activity in Childhood Sarcomas. Cancer Res. 2010, 70, 8770–8781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrotto, P.; Corso, S.; Gamberini, S.; Comoglio, P.M.; Giordano, S. Interplay between scatter factor receptors and B plexins controls invasive growth. Oncogene 2004, 23, 5131–5137. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.T.; Babicky, M.; Jaquish, D.; French, R.; Marayuma, K.; Mose, E.; Niessen, S.; Hoover, H.; Shields, D.; Cheresh, D.; et al. The RON-receptor regulates pancreatic cancer cell migration through phosphorylation-dependent breakdown of the hemidesmosome. Int. J. Cancer 2012, 131, 1744–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate Partners for Tumor Cell Migration, Invasion, and Metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagh, P.K.; Gray, J.K.; Zinser, G.M.; Vasiliauskas, J.; James, L.; Monga, S.P.; E Waltz, S. β-Catenin is required for Ron receptor-induced mammary tumorigenesis. Oncogene 2011, 30, 3694–3704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhao, L.; DeRose, Y.S.; Lin, Y.-C.; Bieniasz, M.; Eyob, H.; Buys, S.S.; Neumayer, L.; Welm, A.L. Short-Form Ron Promotes Spontaneous Breast Cancer Metastasis through Interaction with Phosphoinositide 3-Kinase. Genes Cancer 2011, 2, 753–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, B.G.; Wicker, C.A.; Bourn, J.R.; Lower, E.E.; Takiar, V.; Waltz, S.E. MST1R (RON) expression is a novel prognostic biomarker for metastatic progression in breast cancer patients. Breast Cancer Res. Treat. 2020, 181, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Yao, H.-P.; Zhou, Y.-Q.; Zhou, J.; Zhang, R.; Wang, M.-H. Prevention of BMS-777607-induced polyploidy/senescence by mTOR inhibitor AZD8055 sensitizes breast cancer cells to cytotoxic chemotherapeutics. Mol. Oncol. 2014, 8, 469–482. [Google Scholar] [CrossRef] [Green Version]
- Park, J.S.; Park, J.H.; Khoi, P.N.; Joo, Y.E.; Jung, Y.D. MSP-induced RON activation upregulates uPAR expression and cell invasiveness via MAPK, AP-1 and NF- B signals in gastric cancer cells. Carcinogenesis 2011, 32, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Milan, M.; Benvenuti, S.; Balderacchi, A.M.; Virzì, A.R.; Gentile, A.; Senetta, R.; Cassoni, P.; Comoglio, P.M.; Stella, G.M. RON tyrosine kinase mutations in brain metastases from lung cancer. ERJ Open Res. 2018, 4, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stella, G.M.; Senetta, R.; Inghilleri, S.; Di Cantogno, L.V.; Mantovani, C.; Piloni, D.; Scudeller, L.; Meloni, F.; Papotti, M.; Ricardi, U.; et al. MET mutations are associated with aggressive and radioresistant brain metastatic non-small-cell lung cancer. Neuro-Oncology 2016, 18, 598–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berning, P.; Schaefer, C.; Clemens, D.; Korsching, E.; Dirksen, U.; Potratz, J. The CXCR4 antagonist plerixafor (AMD3100) promotes proliferation of Ewing sarcoma cell lines in vitro and activates receptor tyrosine kinase signaling. Cell Commun. Signal. 2018, 16, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesi, F.; Monti, P.; Leone, B.E.; Zerbi, A.; Vecchi, A.; Piemonti, L.; Mantovani, A.; Allavena, P. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res. 2004, 64, 8420–8427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, G.; Liao, H.-J. Receptor Tyrosine Kinases in the Nucleus. Cold Spring Harb. Perspect. Biol. 2013, 5, a008979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.-Y.; Chang, T.C.; Huang, W.Y.; Lee, C.-T.; Yen, C.-J.; Tsai, Y.-S.; Tzai, T.-S.; Chen, S.-H.; Chow, N.-H. RON Nuclear Translocation under Hypoxia Potentiates Chemoresistance to DNA Double-Strand Break-Inducing Anticancer Drugs. Mol. Cancer Ther. 2016, 15, 276–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.-Y.; Liu, H.-S.; Lai, M.-D.; Tsai, Y.-S.; Tzai, T.-S.; Cheng, H.-L.; Chow, N.-H. Hypoxia Promotes Nuclear Translocation and Transcriptional Function in the Oncogenic Tyrosine Kinase RON. Cancer Res. 2014, 74, 4549–4562. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Huang, L.; Zhou, Y.T.; Wei, T.; Yang, L.; Li, C. RON and RONΔ160 promote gastric cancer cell proliferation, migration, and adaption to hypoxia via interaction with β-catenin. Aging 2019, 11, 2735–2748. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Torres, S.J.; Benight, N.M.; Karns, R.A.; Lower, E.E.; Guan, J.-L.; Waltz, S.E. HGFL-mediated RON signaling supports breast cancer stem cell phenotypes via activation of non-canonical β-catenin signaling. Oncotarget 2017, 8, 58918–58933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, J.-Y.; Sharma, S.; Zhou, Y.-Q.; Yao, H.-P.; Hu, X.; Zhang, R.; Wang, M.-H. Synergistic Activities of MET/RON Inhibitor BMS-777607 and mTOR Inhibitor AZD8055 to Polyploid Cells Derived from Pancreatic Cancer and Cancer Stem Cells. Mol. Cancer Ther. 2014, 13, 37–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LoRusso, P.M.; Gounder, M.; Jalal, S.I.; André, V.; Kambhampati, S.R.P.; Loizos, N.; Hall, J.; Holzer, T.; Nasir, A.; Cosaert, J.; et al. Phase 1 study of narnatumab, an anti-RON receptor monoclonal antibody, in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Howell, G.M.; Humphrey, L.E.; Wang, J.; Brattain, M.G. Ron knockdown and Ron monoclonal antibody IMC-RON8 sensitize pancreatic cancer to histone deacetylase inhibitors (HDACi). PLoS ONE 2013, 8, e69992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Phase 1 Study of Crizotinib in Combination with Enzalutamide in Metastatic Castration-Resistant Prostate Cancer before or after Progression on Docetaxel. Available online: https://clinicaltrials.gov/ct2/show/NCT02207504 (accessed on 21 February 2022).
- Tripathi, A.; Supko, J.G.; Gray, K.P.; Melnick, Z.J.; Regan, M.M.; Taplin, M.-E.; Choudhury, A.D.; Pomerantz, M.M.; Bellmunt, J.; Yu, C.; et al. Dual Blockade of c-MET and the Androgen Receptor in Metastatic Castration-resistant Prostate Cancer: A Phase I Study of Concurrent Enzalutamide and Crizotinib. Clin. Cancer Res. 2020, 26, 6122–6131. [Google Scholar] [CrossRef] [PubMed]
- A Phase II Study of Crizotinib in Patients with c-MET or RON-Positive Metastatic Urothelial Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT02612194 (accessed on 23 February 2022).
- Saleh, M.; Cassier, P.A.; Eberst, L.; Naik, G.; Morris, V.K.; Pant, S.; Terret, C.; Gao, L.; Long, A.; Mao, H.; et al. Phase I Study of Ramucirumab Plus Merestinib in Previously Treated Metastatic Colorectal Cancer: Safety, Preliminary Efficacy, and Pharmacokinetic Findings. Oncologist 2020, 25, e1628–e1639. [Google Scholar] [CrossRef] [PubMed]
- He, A.R.; Cohen, R.B.; Denlinger, C.S.; Sama, A.; Birnbaum, A.; Hwang, J.; Sato, T.; Lewis, N.; Mynderse, M.; Niland, M.; et al. First-in-Human Phase I Study of Merestinib, an Oral Multikinase Inhibitor, in Patients with Advanced Cancer. Oncologist 2019, 24, e930–e942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randomized, Double-Blind, Phase 2 Study of Ramucirumab or Merestinib or Placebo Plus Cisplatin and Gemcitabine as First-Line Treatment in Patients with Advanced or Metastatic Biliary Tract Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT02711553 (accessed on 22 February 2022).


| Title | Identifier | Sponsor | Phase/End Date | N | Tumor Type | Treatment | Drug Type | Conclusion |
|---|---|---|---|---|---|---|---|---|
| A Study of IMC-RON8 in Advanced Solid Tumors | NCT01119456 | Eli Lilly and Company | Phase I/Nov 2013 | 39 | Advanced Solid Tumors | IMC-RON8 (Other names) LY3012219 Narnatumab Dose escalation: 5, 10, 15, 20, or 25 mg/kg IV weekly for a 4 week cycle 15, 20, 25, 30, 35, or 40 mg/kg IV every two weeks for a 4 week cycle | Monoclonal Antibody | No complete or partial responses. However, only 1 patient achieved therapeutic drug concentration values >140 μL/mL. |
| A Phase I Study of LY2801653 in Patients with Advanced Cancer | NCT01285037 | Eli Lilly and Company | Phase I/II/Sept 2017 | 186 | Adenocarcinoma of colon or rectum (CRC) HNSCC Uveal Melanoma with liver metastasis (UM) Cholangiocarcinoma (CCA) | Adenocarcinoma: Merestinib HNSCC:Merestinib 120 mg PO × 28 days + Cetuximab UM: Merestinib 120 mg PO × 28 days CCA: Merestinib 120 mg PO qd × 28 days + Cisplatin +/− Gemcitabine Gastric: Merestinib 120 mg PO qd × 28 days + Ramucirumab | LY2801653:small molecule inhibitor c-MET/RON, multi-kinase inhibitor Cetuximab:EGFR inhibitor Cisplatin: alkylating agent Gemcitabine: antimetabolite Ramucirumab: VEGF inhibitor | LY2801653 120 mg qd identified as treatment dose. Three PR and one CR within the triple therapy CCA cohort and dual therapy cohort respectively. Overall, 32% achieved a best response of SD, 48% had PD, and one death occurred due to AE. mPFS 1.7 months HNSCC, 1.8 months CRC, 1.8 months UM, 1.9 CCA, gastric not reported |
| A Study of Ramucirumab (LY3009806) or Merestinib (LY2801653) in Advanced or Metastatic Biliary Tract Cancer | NCT02711553 | Eli Lilly and Company | Phase II/Feb 2018 Modified to end Dec 2022 | 306 | Biliary Tract Cancer Advanced Cancer Metastatic Cancer | A1: Ramucirumab + cisplatin + gemcitabine intravenously (IV) on Days 1 and 8, every 21 days. A2: Placebo + cisplatin + gemcitabine IV on days 1 and 8, every 21 days. B1: Merestinib PO daily + cisplatin + gemcitabine IV on days 1 and 8, every 21 days. B2: Placebo PO daily + cisplatin + gemcitabine IV on days 1 and 8, every 21 days. | Ramucirumab:VEGF inhibitor Merestinib:small molecule inhibitor c-MET/RON, multi-kinase inhibitor | No significant difference in progression-free survival (PFS), overall survival (OS), disease control rate (DCR) between trial drugs and placebo. Secondary endpoint for overall response rate (ORR) noted significant in Merestinib vs. Placebo cohort with two-sided p-value 0.015 with Odds ratio 0.4. |
| LGI-GU-URO-CRI-001: A Phase II Study of Crizotinib in Patients with c-MET or RON Positive Metastatic Urothelial Cancer | NCT02612194 | Earle Burgess | Phase II/Nov 2019 | 46 | Stage IV Urinary Bladder Neoplasms Stage IV Ureteral Neoplasms Stage IV Urethral Neoplasms | Cohort 1: Crizotinib c-MET high (>50%) RON null (0–9%) Cohort 2: Crizotinib c-MET (10–100%), RON (10–100%) Cohort 3: Crizotinib c-MET null (0–10%), RON (10–100%) | Crizotinib: c-MET inhibitor | Study terminated due to low accrual. |
| An Open-Label, Phase Ia/Ib Study of Ramucirumab in Combination with Other Targeted Agents in Advanced Cancers | NCT02745769 | Eli Lilly and Company | Phase Ia/Ib/Jan 2019 | 23 | Stage IV Colon Cancer Mantle Cell Lymphoma | Arm 1: Ramucirumab 8 mg/kg IV day 1 and day 15 + Merestinib 80 mg PO qd × 28 days until disease progression. Arm 2: Ramucirumab 8 mg/kg IV day 1 and day 15 + Abemaciclib PO bid × 28 days until disease progression. (Cancelled without enrollement) | Ramucirumab:VEGF inhibitor Merestinib:small molecule inhibitor c-MET/RON, multi-kinase inhibitor Abemaciclib: CDK4/6 inhibitor | Therapeutic doses achieved in combination. In mCRC 43% of patients with >Grade 3 adverse effects with dual treatment. Stable disease was noted in (52%), partial response (0%), complete response (0%). mPFS was 3.3 months. mOS was 8.0 months. |
| A Phase I Study of Crizotinib in Combination with Enzalutamide in Metastatic Castration-resistant Prostate Cancer Before or After Progression of Docetaxel | NCT02207504 | Dana-Farber Cancer Institute Pfizer Astellas Pharma Inc | Phase I/Jan 2022 | 24 | Castration Resistant Prostate Cancer | Crizotinib PO qd 250 mg qd + Enzalutamide 160 mg qd Crizotinib PO 200 mg bid+ Enzalutamide 160 mg qd Crizotinib 250 mg bid + Enzalutamide 150 mg qd | Crizotinib: small molecule inhibitor of c-MET/RON, ROS, ALK Enzalutamide: androgen receptor inhibitor | Crizotinib 250 mg bid was identified as the maximum tolerated dose Concurrent treatment with Crizotinib and Enzalutamide resulted in a significant 74% reduction in systemic Crizotinib exposure likely attributed to enzalutamide inducing CYP3A4. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cazes, A.; Childers, B.G.; Esparza, E.; Lowy, A.M. The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies. Cancers 2022, 14, 2037. https://doi.org/10.3390/cancers14082037
Cazes A, Childers BG, Esparza E, Lowy AM. The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies. Cancers. 2022; 14(8):2037. https://doi.org/10.3390/cancers14082037
Chicago/Turabian StyleCazes, Alex, Betzaira G. Childers, Edgar Esparza, and Andrew M. Lowy. 2022. "The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies" Cancers 14, no. 8: 2037. https://doi.org/10.3390/cancers14082037
APA StyleCazes, A., Childers, B. G., Esparza, E., & Lowy, A. M. (2022). The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies. Cancers, 14(8), 2037. https://doi.org/10.3390/cancers14082037





