The Implication of Gastric Microbiome in the Treatment of Gastric Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Bacteria and Carcinogenesis
3. Gastric Microbiome
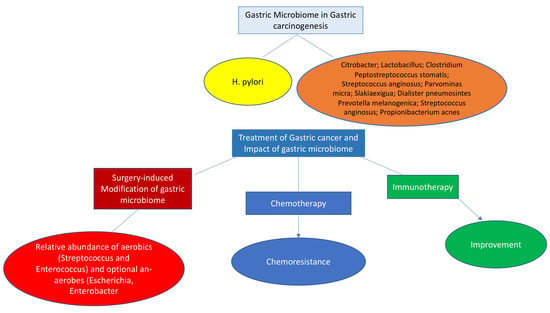
4. Gastric Microbiota in Gastric Carcinogenesis
5. Gut Microbiota in Gastric Cancer
6. Gut Microbiota in Gastric Carcinogenesis
7. Surgery for Gastric Cancer and Microbiome
8. Microbiome and Gastric Cancer Treatment
9. Chemotherapy
10. Microbiome and Cancer Immunotherapy
11. Relation of Gut Microbiome and Immune Response in Gastric Cancer
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carneiro, F. Gastric Cancer. Pathobiology of Human Disease. A Dynamic Encyclopedia of Disease Mechanism, 1st ed.; McManus, L.M., Mitchell, R.N., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 1305–1318. [Google Scholar]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [Green Version]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Assumpcao, P.P.; Araujo, T.M.T.; de Assumpcao, P.B.; Barra, W.F.; Khayat, A.S.; Assumpcao, C.B.; Ishak, G.; Noronha Nunes, D.; Dias-Neto, E.; Gonzaga Vaz Coelho, L. Suicide journey of H. pylori through gastric carcinogenesis: The role of non-H. pylori microbiome and potential consequences for clinical practice. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1591–1597. [Google Scholar] [CrossRef]
- Engstrand, L.; Graham, D.Y. Microbiome and gastric cancer. Dig. Dis. Sci. 2020, 65, 865–873. [Google Scholar] [CrossRef] [Green Version]
- Schulz, C.; Schütte, K.; Mayerle, J.; Malfertheiner, P. The role of the gastric bacterial microbiome in gastric cancer: Helicobacter pylori and beyond. Therap. Adv. Gastroenterol. 2019, 12, 1756284819894062. [Google Scholar] [CrossRef] [Green Version]
- Cavaleiro-Pinto, M.; Peleteiro, B.; Lunet, N.; Barros, H. Helicobacter pylori infection and gastric cardia cancer: Systematic review and meta-analysis. Cancer Causes Contr. 2011, 22, 375–387. [Google Scholar] [CrossRef]
- Blanca Piazuelo, M.; Correa, P. Gastric cancer: Overview. Colomb. Med. 2013, 44, 192–201. [Google Scholar]
- Sheh, A.; Fox, J.G. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes 2013, 4, 505–531. [Google Scholar] [CrossRef] [Green Version]
- Mattarelli, P.; Brandi, G.; Calabrese, C.; Fornari, F.; Prati, G.M.; Biavati, B.; Sgorbati, B. Occurrence of Bifidobacteriaceae in human hypochlorhydria stomach. Microb. Ecol. Health Dis. 2014, 25, 21379. [Google Scholar]
- Monstein, H.J.; Tiveljung, A.; Kraft, C.H.; Borch, K.; Jonasson, J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J. Med. Microbiol. 2000, 49, 817–822. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Cummins, J.; Tangney, M. Bacteria and tumours: Causative agents or opportunistic inhabitants? Infect. Agents Cancer 2013, 8, 11. [Google Scholar] [CrossRef] [Green Version]
- Polk, D.B.; Peek, R.M. Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef] [Green Version]
- Robinson, K.M.; Sieber, K.B.; Hotopp, J.C.D. A review of bacteria-animal lateral gene transfer may inform our understanding of diseases like cancer. PLoS Genet. 2013, 9, e1003877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshiol, J.; Wozniak, A.; Cook, P.; Adaniel, C.; Acevedo, J.; Azócar, L.; Hsing, A.W.; Roa, J.C.; Pasetti, M.F.; Miquel, J.F.; et al. Salmonella enterica serovar Typhi and gallbladder cancer: A case–control study and meta-analysis. Cancer Med. 2016, 5, 3235–3310. [Google Scholar] [CrossRef] [PubMed]
- Eyvazi, S.; Vostakolaei, M.A.; Dilmaghani, A.; Borumandi, O.; Hejazi, M.S.; Kahroba, H.; Tarhriz, V. The oncogenic rolesof bacterial infections in development of cancer. Microb. Pathog. 2020, 141, 104019. [Google Scholar] [CrossRef] [PubMed]
- Luu, T.H.; Michel, C.; Bard, J.-M.; Dravet, F.; Nazih, H.; Bobin-Dubigeon, C. Intestinal proportion of Blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr. Cancer 2017, 69, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Hartung, M.L.; Gruber, D.C.; Koch, K.N.; Grüter, L.; Rehrauer, H.; Tegtmeyer, N.; Backert, S.; Müller, A.H. pylori-induced DNA strand breaks are introduced by nucleotide excision repair endonucleases and promote NF-κB target gene expression. Cell Rep. 2015, 13, 70–79. [Google Scholar] [CrossRef] [Green Version]
- Maekita, T.; Nakazawa, K.; Mihara, M.; Nakajima, T.; Yanaoka, K.; Iguchi, M.; Arii, K.; Kaneda, A.; Tsukamoto, T.; Tatematsu, M.; et al. High levels of aberrant DNA methylation in Helicobacter pylori–infected gastric mucosae and its possible association with gastric cancer risk. Clin. Cancer Res. 2006, 12, 989–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, S.; Yamaoka, Y. Helicobacter pylori virulence factor cytotoxin-associated Gene A (CagA)-mediated gastric pathogenicity. Int. J. Mol. Sci. 2020, 21, 7430. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Yamaoka, Y. Role of vacuolating cytotoxin A in Helicobacter pylori infection and its impact on gastric pathogenesis. Expert Rev. Anti. Infect. Ther. 2020, 18, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Higashi, H.; Tsutsumi, R.; Muto, S.; Sugiyama, T.; Azuma, T.; Asaka, M.; Hatakeyama, M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 2002, 295, 683–686. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Yin, W.; Zhao, R.; Wang, Y.; Song, C.; Wang, H.; Rong, J.; Wang, F.; Xie, Y. Outer inflammatory protein of Helicobacter pylori impacts IL-8 expression, adherence, cell apoptosis and cell cycle of gastric cells independent of its copy number. Med. Microbiol. Immunol. 2020, 209, 621–630. [Google Scholar] [CrossRef]
- Yong, X.; Tang, B.; Li, B.S.; Xie, R.; Hu, C.J.; Luo, G.; Qin, Y.; Dong, H.; Yang, S.M. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun. Sign. 2015, 13, 30. [Google Scholar] [CrossRef] [Green Version]
- Stein, M.; Rappuoli, R.; Covacci, A. Helicobacter Pylori: Physiology and Genetics; Mobley, H.L.T., Mendz, G.L., Hazell, S.L., Eds.; ASM Press: Washington, DC, USA, 2001; Chapter 31; pp. 345–353. [Google Scholar]
- Selbach, M.; Paul, F.E.; Brandt, S.; Guye, P.; Daumke, O.; Backert, S.; Dehio, C.; Mann, M. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe 2009, 5, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Zeaiter, Z.; Cohen, D.; Müsch, A.; Bagnoli, F.; Covacci, A.; Stein, M. Analysis of detergent-resistant membranes of Helicobacter pylori infected gastric adenocarcinoma cells reveals a role for MARK2/ Par1b in CagA-mediated disruption of cellular polarity. Cell Microbiol. 2008, 10, 781–794. [Google Scholar] [CrossRef]
- Yamahashi, Y.; Hatakeyama, M. PAR1b takes the stage in the morphogenetic and motogenetic activity of Helicobacter pylori CagA oncoprotein. Cell Adh. Migr. 2013, 7, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Mimuro, H.; Kiga, K.; Fukumatsu, M.; Ishijima, N.; Morikawa, H.; Nagai, S.; Koyasu, S.; Gilman, R.H.; Kersulyte, D.; et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 2009, 5, 23–24. [Google Scholar] [CrossRef] [Green Version]
- Schulz, C.; Schutte, K.; Koch, N.; Vilchez-Vargas, R.; Wos-Oxley, M.L.; Oxley, A.P.A.; Vital, M.; Malfertheiner, P.; Pieper, D.H. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 2018, 67, 216–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuik, F.; Dicksved, J.; Lam, S.Y.; Fuhler, G.M.; van der Laan, L.; van de Winkel, A.; Konstantinov, S.R.; Spaander, M.; Peppelenbosch, M.P.; Engstrand, L.; et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United Eur. Gastroenterol. J. 2019, 7, 897–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mailhe, M.; Ricaboni, D.; Vitton, V.; Gonzalez, J.M.; Bachar, D.; Dubourg, G.; Cadoret, F.; Robert, C.; Delerce, J.; Levasseur, A.; et al. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol. 2018, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Vasapolli, R.; Schutte, K.; Schulz, C.; Vital, M.; Schomburg, D.; Pieper, D.H.; Vilchez-Vargas, R.; Malfertheiner, P. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology 2019, 157, 1081–1092.e1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, H.; Satoh-Takayama, N. Stomach microbiota, Helicobacter pylori, and group 2 innate lymphoid cells. Exp. Mol. Med. 2020, 52, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xia, C.; Li, Q.; Jin, L.; Zheng, L.; Wu, Z. Comparisons Between Bacterial Communities in Mucosa in Patients with Gastric Antrum Ulcer and a Duodenal Ulcer. Front. Cell. Infect. Microbiol. 2018, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Hu, Y.L.; Pang, W.; Huang, Y.; Zhang, Y.; Zhang, C.J. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Front. Cell. Infect. Microbiol. 2018, 8, 433. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Marques, J.; Hout, A.; Ferreira, R.M.; Weber, M.; Pinto-Ribeiro, I.; van Doorn, L.J.; Knetsch, C.W.; Figueiredo, C. Impact of Host DNA and Sequencing Depth on the Taxonomic Resolution of Whole Metagenome Sequencing for Microbiome Analysis. Front. Microbiol. 2019, 10, 1277. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Shao, L.; Liu, X.; Ji, F.; Mei, Y.; Cheng, Y.; Liu, F.; Yan, C.; Li, L.; Ling, Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019, 40, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Lofgren, J.L.; Whary, M.T.; Ge, Z.; Muthupalani, S.; Taylor, N.S.; Mobley, M.; Potter, A.; Varro, A.; Eibach, D.; Suerbaum, S.; et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 2011, 140, 210–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, H.; Hioki, K.; Popivanova, B.K.; Oguma, K.; Van Rooijen, N.; Ishikawa, T.O.; Oshima, M. Prostaglandin E (2) signaling and bacterial infection recruit tumor-promoting macrophages to mouse gastric tumors. Gastroenterology 2011, 140, 596–607.e597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lertpiriyapong, K.; Whary, M.T.; Muthupalani, S.; Lofgren, J.L.; Gamazon, E.R.; Feng, Y.; Ge, Z.; Wang, T.C.; Fox, J.G. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014, 63, 54–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Z.; Sheh, A.; Feng, Y.; Muthupalani, S.; Ge, L.; Wang, C.; Kurnick, S.; Mannion, A.; Whary, M.T.; Fox, J.G. Helicobacter pylori-infected C57BL/6 mice with different gastrointestinal microbiota have contrasting gastric pathology, microbial and host immune responses. Sci. Rep. 2018, 8, 8014. [Google Scholar] [CrossRef]
- Velazquez, E.M.; Nguyen, H.; Heasley, K.T.; Saechao, C.H.; Gil, L.M.; Rogers, A.W.L.; Miller, B.M.; Rolston, M.R.; Lopez, C.A.; Litvak, Y.; et al. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol. 2019, 4, 1057–1064. [Google Scholar] [CrossRef]
- Youssef, O.; Lahti, L.; Kokkola, A.; Karla, T.; Tikkanen, M.; Ehsan, H.; Carpelan-Holmström, M.; Koskensalo, S.; Böhling, T.; Rautelin, H.; et al. Stool microbiota composition differs in patients with stomach, colon, and rectal neoplasms. Dig. Dis. Sci. 2018, 63, 2950–2958. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Yang, Y.; Wang, H.; Wang, H.; Yu, X.; Lu, Y.; Shen, S.; Teng, L. Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine 2019, 98, e16626. [Google Scholar] [CrossRef]
- Gao, J.J.; Zhang, Y.; Gerhard, M.; Mejias-Luque, R.; Zhang, L.; Vieth, M.; Ma, J.-L.; Bajbouj, M.; Suchanek, S.; Liu, W.-D.; et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front. Cell. Infect. Microbiol. 2018, 8, 202. [Google Scholar] [CrossRef] [Green Version]
- Horvath, A.; Bausys, A.; Sabaliauskaite, R.; Stratilatovas, E.; Jarmalaite, S.; Schuetz, B.; Stiegler, P.; Bausys, R.; Stadlbauer, V.; Strupas, K. Distal gastrectomy with Billroth II reconstruction is associated with oralization of gut microbiome and intestinal inflammation: A proof-of-concept study. Ann. Surg. Oncol. 2021, 28, 1198–1208. [Google Scholar] [CrossRef]
- Morales-Marroquin, E.; Hanson, B.; Greathouse, L.; de la Cruz-Munoz, N.; Messiah, S.E. Comparison of methodological approaches to human gut microbiota changes in response to metabolic and bariatric surgery: A systematic review. Obes. Rev. 2020, 21, e13025. [Google Scholar] [CrossRef]
- Farin, W.; Oñate, F.P.; Plassais, J.; Bonny, C.; Beglinger, C.; Woelnerhanssen, B.; Nocca, D.; Magoules, F.; Le Chatelier, E.; Pons, N.; et al. Impact of laparoscopic Roux-en-Y gastric bypass and sleeve gastrectomy on gut microbiota: A metagenomic comparative analysis. Surg. Obes. Relat. Dis. 2020, 16, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y.F.; Sun, J.N.; Ren, L.F.; Cao, X.L.; Dong, J.H.; Tao, K.; Guan, X.M.; Cui, Y.N.; Su, W. Intestinal Microbiota Is Altered in Patients with Gastric Cancer from Shanxi Province, China. Dig. Dis. Sci. 2019, 64, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Yang, J. Correlations of gastrointestinal hormones with inflammation and intestinal flora in patients with gastric cancer. J. BUON 2019, 24, 1595–1600. [Google Scholar] [PubMed]
- Yu, C.; Su, Z.; Li, Y.; Li, Y.; Liu, K.; Chu, F.; Liu, T.; Chen, R.; Ding, X. Dysbiosis of gut microbiota is associated with gastric carcinogenesis in rats. Biomed. Pharmacother. 2020, 126, 110036. [Google Scholar] [CrossRef]
- Khachfe, H.H.; Salhab, H.A.; Fares, M.Y.; Chahrour, M.A.; Jamali, F.R. Landscape of interventional clinical trials involving gastrectomy for gastric cancer. Ecancermedicalscience 2021, 15, 1218. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, T.; Lu, J.; Wei, K.; Tian, H.; Liu, W.; Xu, T.; Wang, X.; Wang, S.; Yang, R.; et al. Adjuvant treatment and molecular mechanism of probiotic compounds in patients with gastric cancer after gastrectomy. Food. Funct. 2021, 12, 6294–6308. [Google Scholar] [CrossRef]
- Penna, M.; Allum, W. New treatments for gastric cancer: Are they changing clinical practice? Clin. Practice 2013, 10, 649–659. [Google Scholar] [CrossRef]
- Tseng, C.H.; Lin, J.T.; Ho, H.J.; Lai, Z.-L.; Wang, C.-B.; Tang, S.-L.; Wu, C.-Y. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci. Rep. 2016, 6, 20701. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.H.; Huang, K.H.; Chuang, W.H.; Luo, J.-C.; Lin, C.-C.; Ting, P.-H.; Young, S.-H.; Fang, W.-L.; Hou, M.-C.; Lee, F.-Y. The long-term effect of metabolic profile and microbiota status in early gastric cancer patients after subtotal gastrectomy. PLoS ONE 2018, 13, e0206930. [Google Scholar] [CrossRef]
- Eom, B.W.; Lee, H.J.; Yoo, M.W.; Cho, J.J.; Kim, W.H.; Yang, H.-K.; Lee, K.U. Synchronous and metachronous cancers in patients with gastric cancer. J. Surg. Oncol. 2008, 98, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Chon, H.J.; Kang, B.; Kim, K.; Jeung, H.C.; Chung, H.C.; Noh, S.H.; Rha, S.Y. Prediction of metachronous multiple primary cancers following the curative resection of gastric cancer. BMC Cancer 2013, 13, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erawijantari, P.P.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Saito, Y.; Fukuda, S.; Yachida, S.; Yamada, T.; et al. Influence of gastrectomy for gastric cancer treatment on faecal microbiome and metabolome profiles. Gut 2020, 69, 1404–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, Y.; Saku, M.; Kawanaka, H.; Nonaka, M.; Yoshida, K. Features of second primary cancer in patients with gastric cancer. Oncology 2003, 65, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Aron-Wisnewsky, J.; Doré, J.; Clement, K. The importance of the gut microbiota after bariatric surgery. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 590–598. [Google Scholar] [CrossRef]
- Quercia, I.; Dutia, R.; Kotler, D.P.; Belsley, S.; Laferrère, B. Gastrointestinal changes after bariatric surgery. Diabetes Metab. 2014, 40, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Palleja, A.; Kashani, A.; Allin, K.H.; Nielsen, T.; Zhang, C.; Li, Y.; Brach, T.; Liang, S.; Feng, Q.; Jørgensen, N.B.; et al. Roux-E n-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med. 2016, 8, 67. [Google Scholar] [CrossRef] [Green Version]
- Ilhan, Z.E.; Di Baise, J.K.; Isern, N.G.; Hoyt, D.W.; Marcus, A.K.; Kang, D.-W.; Crowell, M.D.; Rittmann, B.E.; Krajmalnik-Brown, R. Distinctive microbiomes and metabolites linked with weight loss after gastric bypass, but not gastric banding. Isme J. 2017, 11, 2047–2058. [Google Scholar] [CrossRef]
- Kong, L.C.; Tap, J.; Aron-Wisnewsky, J.; Pelloux, V.; Basdevant, A.; Bouillot, J.-L.; Zucker, J.-D.; Doré, J.; Clément, K. Gut microbiota after gastric bypass in human obesity: Increased richness and associations of bacterial genera with adipose tissue genes. Am. J. Clin. Nutr. 2013, 98, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Celiker, H. A new proposed mechanism of action for gastric bypass surgery: Air hypothesis. Med. Hypotheses 2017, 107, 81–89. [Google Scholar] [CrossRef]
- Zhang, H.; Di Baise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.S.; Badgwell, B.D. Current Treatment and Recent Progress in Gastric Cancer. CA Cancer J. Clin. 2021, 71, 264–279. [Google Scholar] [CrossRef]
- Pevsner-Fischer, M.; Tuganbaev, T.; Meijer, M.; Zhang, S.H.; Zeng, Z.R.; Chen, M.H.; Elinav, E. Role of the microbiome in non-gastrointestinal cancers. World. J. Clin. Oncol. 2016, 7, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viaud, S.; Flament, C.; Zoubir, M.; Pautier, P.; Le Cesne, A.; Ribrag, V.; Soria, J.C.; Marty, V.; Vielh, P.; Robert, C.; et al. Cyclophosphamide induces differentiation of Th17 cells in cancer patients. Cancer Res. 2011, 71, 661–665. [Google Scholar] [CrossRef] [Green Version]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Kim, Y.G.; Udayanga, K.G.; Totsuka, N.; Weinberg, J.B.; Núñez, G.; Shibuya, A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungiinduced PGE2. Cell Host Microbe 2014, 15, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Scheiermann, J.; Klinman, D.M. Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine 2014, 32, 6377–6389. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geller, L.; Barzliy-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Balibrea, E.; Martinez-Cardus, A.; Gines, A.; de Porras, V.R.; Moutinho, C.; Layos, L.; Manzano, J.L.; Bugés, C.; Bystrup, S.; Esteller, M.; et al. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol. Cancer. Ther. 2015, 14, 1767–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, J.L.; Ran, Z.H.; Shen, J.; Zhang, C.X.; Xiao, S.D. Meta-analysis: The effect of supplementation with probiotics on eradication rates and adverse events during Helicobacter pylori eradication therapy. Aliment. Pharmacol. Ther. 2007, 25, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Shadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Allegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Gharaibeh, R.Z.; Jobin, C. Microbiota and cancer immunotherapy: In search of microbial signals. Gut 2019, 68, 385–388. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Man Lei, Y.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [Green Version]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Bachem, A.; Makhlouf, C.; Binger, K.J.; de Souza, D.P.; Tull, D.; Hochheiser, K.; Whitney, P.G.; Fernandez-Ruiz, D.; Dähling, S.; Kastemüller, W.; et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 2019, 20, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Dummer, R.; de Pril, V.; Lebbé, C.; Hodi, F.S. Patterns of onset and resolution of immune-related adverse events of special interest with ipilimumab: Detailed safety analysis from a phase 3 trial in patients with advanced melanoma. Cancer 2013, 119, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littman, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, J.D.; Abu Sbeih, H.; Sanchez, C.A.; Chang, C.C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef]
- Das, S.; Suarez, G.; Beswick, E.J.; Sierra, J.C.; Graham, D.Y.; Reyes, V.E. Expression of B7-H1 on Gastric Epithelial Cells: Its Potential Role in Regulating T Cells during Helicobacter pylori Infection. J. Immunol. 2006, 176, 3000–3009. [Google Scholar] [CrossRef] [Green Version]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-Y.; Lin, C.-W.; Cheng, K.-S.; Lin, C.; Wang, Y.-M.; Lin, I.-T.; Chou, Y.-H.; Hsu, P.-N. Increased programmed death-ligand-1 expression in human gastric epithelial cells in Helicobacter pylori infection. Clin. Exp. Immunol. 2010, 161, 551–559. [Google Scholar] [CrossRef]
- Liu, X.; Choi, M.G.; Kim, K.; Kim, K.-M.; Kim, S.T.; Park, S.H.; Cristescu, R.; Peter, S.; Lee, J. High PD-L1 expression in gastric cancer (GC) patients and correlation with molecular features. Pathol. Res. Pract. 2020, 216, 152881. [Google Scholar] [CrossRef]
- Pietrocola, F.; Pol, J.; Vacchelli, E.; Rao, S.; Enot, D.P.; Baracco, E.E.; Levesque, S.; Castoldi, F.; Jacquelot, N.; Yamazaki, T.; et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell 2016, 30, 147–160. [Google Scholar] [CrossRef] [Green Version]
- Joglekar, P.; Segre, J.A. Building a Translational Microbiome Toolbox. Cell 2017, 169, 378–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawant, S.S.; Patil, S.M.; Gupta, V.; Kunda, N.K. Microbes as medicines: Harnessing the power of bacteria in advancing cancer treatment. Int. J. Mol. Sci. 2020, 21, 7575. [Google Scholar] [CrossRef] [PubMed]
- Soleimanpour, S.; Hasanian, S.M.; Avan, A.; Yaghoubi, A.; Khazaei, M. Bacteriotherapy in gastrointestinal cancer. Life Sci. 2020, 254, 117754. [Google Scholar] [CrossRef] [PubMed]
- Yaghoubi, A.; Khazaei, M.; Jalili, S.; Hasanian, S.M.; Avan, A.; Soleimanpour, S. Bacteria as a double-action sword in cancer. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188388. [Google Scholar] [CrossRef]
- Mai, X.T.; Huang, J.; Tan, J.; Huang, Y.; Chen, Y. Effects and mechanisms of the secondary structure on the antimicrobial activity and specificity of antimicrobial peptides. J. Pept. Sci. 2015, 21, 561–568. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, Y.; Liu, D.; Chen, Y. Two hits are better than one: Synergistic anticancer activity of a-helical peptides and doxorubicin/epirubicin. Oncotarget 2015, 6, 1769–1778. [Google Scholar] [CrossRef]
- Hu, C.; Chen, X.; Huang, Y.; Chen, Y. Synergistic effect of the pro-apoptosis peptide kla-TAT and the cationic anticancer peptide HPRP-A1. Apoptosis 2018, 23, 132–142. [Google Scholar] [CrossRef]
- Hao, W.; Hu, C.; Huang, Y.; Chen, Y. Coadministration of kla peptide with HPRP-A1 to enhance anticancer activity. PLoS ONE 2019, 14, e0223738. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Hao, X.; Liu, D.; Huang, Y.; Chen, Y. In vitro characterization of the rapid cytotoxicity of anticancer peptide HPRP-A2 through membrane destruction and intracellular mechanism against gastric cancer cell lines. PLoS ONE 2015, 10, e0139578. [Google Scholar] [CrossRef]
- Tareq, F.S.; Kim, J.H.; Lee, M.A.; Lee, H.S.; Lee, Y.J.; Lee, J.S.; Shin, H.J. Ieodoglucomides A and B from a marine-derived bacterium Bacillus licheniformis. Org. Lett. 2012, 14, 1464–1467. [Google Scholar] [CrossRef]
- Xie, J.J.; Zhou, F.; Li, E.M.; Jiang, H.; Du, Z.P.; Lin, R.; Fang, D.S. FW523-3, a novel lipopeptide compound, induces apoptosis in cancer cells. Mol. Med. Rep. 2011, 4, 759–763. [Google Scholar] [PubMed] [Green Version]
- Sunakawa, Y.; Matoba, R.; Inoue, E.; Sakamoto, Y.; Kawabata, R.; Ishiguro, A.; Akamaru, Y.; Kito, Y.; Takahashi, M.; Matsuyama, J.; et al. Genomic pathway of gut microbiome to predict efficacy of nivolumab in advanced gastric cancer: DELIVER trial (JACCRO GC-08). J. Clin. Oncol. 2021, 39 (Suppl. 3), 161. [Google Scholar] [CrossRef]


| S. No. | Bacteria Situation | References |
|---|---|---|
| 1 | Increased: Citrobacter; Lactobacillus; Clostridium Decreased: Helicobacter; Neisseria | [5] |
| 2 | Increased: Peptostreptococcus stomatis; Streptococcus anginosus; Parvominas micra; Slakiaexigua; Dialister pneumosintes | [37,38] |
| 3 | Increased: Prevotella melanogenica; Streptococcus anginosus; Propionibacterium acnes Decreased: H. pylori | [37,38] |
| 4 | Decreased microbiome | [39,40,41] |
| S. No. | Bacteria Situation | References |
|---|---|---|
| 1 | Increased: Lactobacillus; Escherichia coli; Klebsiella; Tyzzerella_3; Veillonella; Streptococcus; Lachnospira | [55,56] |
| 2 | Increased: Escherichia coli; Staphylococcus; Enterococcus; Peptostreptococcus Decreased: Bifidobacterium; Lactobacillus; Bacilli | [56,57] |
| 3 | Increased: Akkermansia; Escherichia; Shigella; Lactobacillus; Dialister Decreased: Bacteroides; Helicobacter | [49,56,57] |
| 4 | Increased: Bifidobacterium; Lactobacillus; Escherichia; Shigella Decreased: Lachnospira, Ruminococcus | [49,56,57,58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pappas-Gogos, G.; Tepelenis, K.; Fousekis, F.; Katsanos, K.; Pitiakoudis, M.; Vlachos, K. The Implication of Gastric Microbiome in the Treatment of Gastric Cancer. Cancers 2022, 14, 2039. https://doi.org/10.3390/cancers14082039
Pappas-Gogos G, Tepelenis K, Fousekis F, Katsanos K, Pitiakoudis M, Vlachos K. The Implication of Gastric Microbiome in the Treatment of Gastric Cancer. Cancers. 2022; 14(8):2039. https://doi.org/10.3390/cancers14082039
Chicago/Turabian StylePappas-Gogos, George, Kostas Tepelenis, Fotis Fousekis, Konstantinos Katsanos, Michail Pitiakoudis, and Konstantinos Vlachos. 2022. "The Implication of Gastric Microbiome in the Treatment of Gastric Cancer" Cancers 14, no. 8: 2039. https://doi.org/10.3390/cancers14082039
APA StylePappas-Gogos, G., Tepelenis, K., Fousekis, F., Katsanos, K., Pitiakoudis, M., & Vlachos, K. (2022). The Implication of Gastric Microbiome in the Treatment of Gastric Cancer. Cancers, 14(8), 2039. https://doi.org/10.3390/cancers14082039