Bone Progenitors Pull the Strings on the Early Metabolic Rewiring Occurring in Prostate Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.1.1. Cell Lines
2.1.2. Co-Culture System
2.2. RNA Isolation and Sequencing
2.3. Bioinformatics Analyses
2.3.1. Differential Expression Analysis and Reactome Pathway Database
2.3.2. Pathway Enrichment Analysis
2.3.3. Metastatic PCa Patient Cohorts
2.3.4. Principal Component Analysis (PCA)
2.3.5. Risk Scoring System Analysis
2.4. Secretome Analysis of Conditioned Media
2.5. PKA Inhibition and ATP Content Measurement
2.6. RT-qPCR
2.7. Animals
2.8. MDA-PCa-183 Patient-Derived Xenograft (PDX) Generation and RNA Sequencing
2.9. Statistical Analysis
3. Results
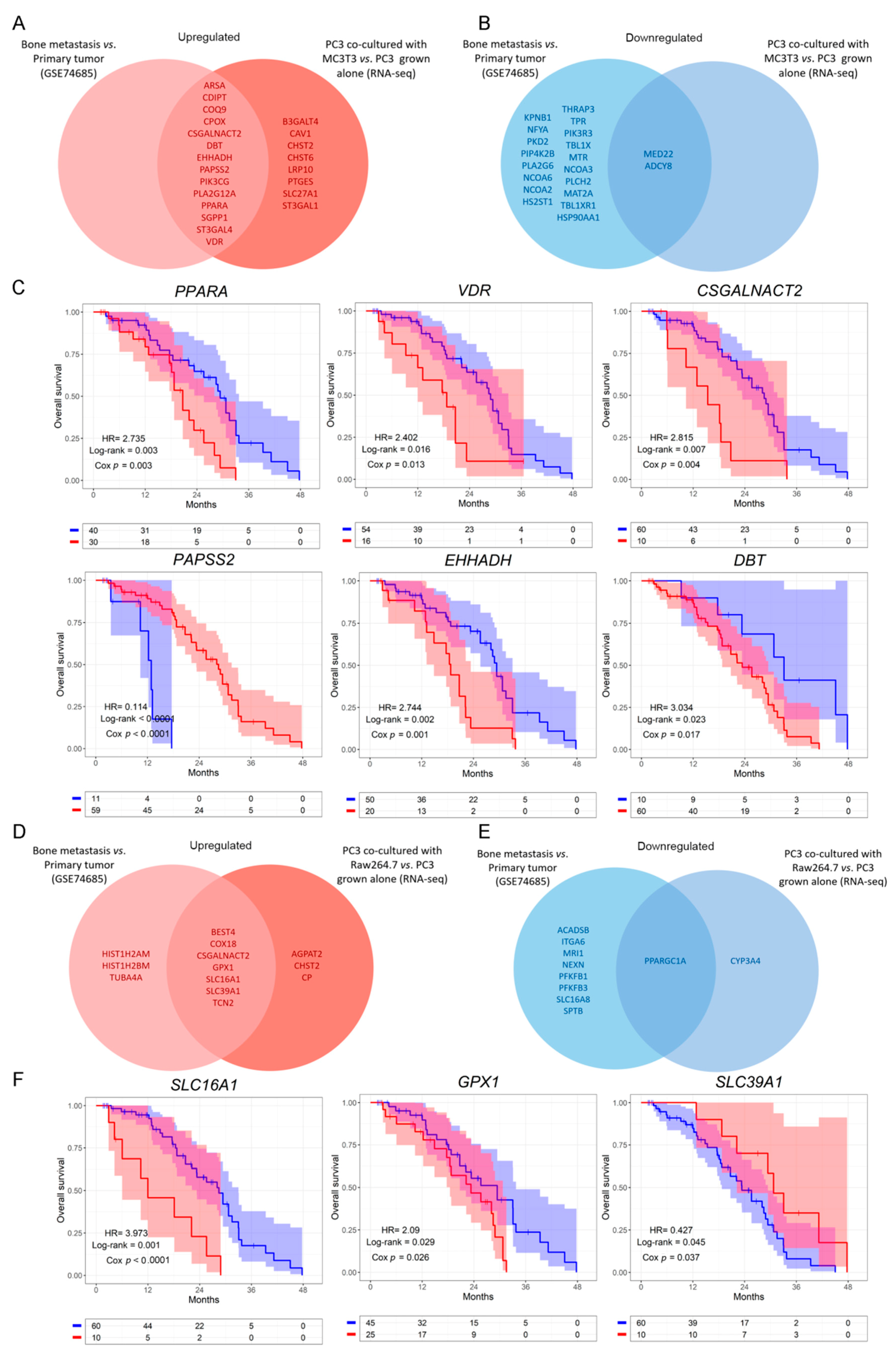
3.1. Differential Transcriptomic Analysis of PC3 and Bone Cells Growing in a Co-Culture Transwell System
3.2. Clinical Correlation of Metabolic Genes Dysregulated in PC3 Co-Cultured with Bone Progenitors and Human PCa Metastatic Samples
3.3. Analysis of the Metabolic-Related Gene Expression Profile Associated with Overall Survival in Human Metastatic PCa
3.4. Defining a Metabolic Gene Signature Associated with Metastatic PCa
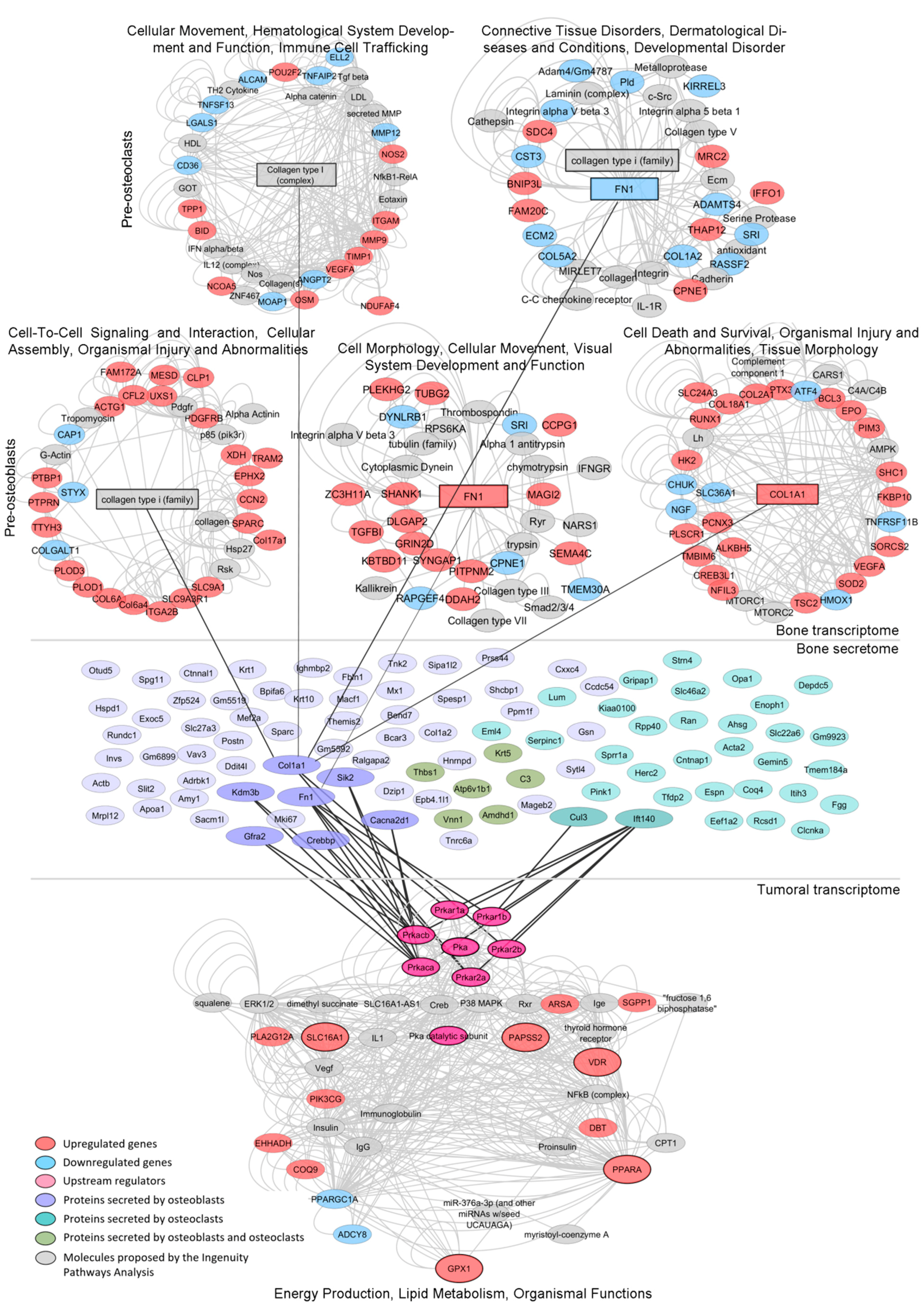
3.5. Mechanism-Centric Approach to Delineate Potential Drivers of the Metabolic Rewiring of PCa Cells
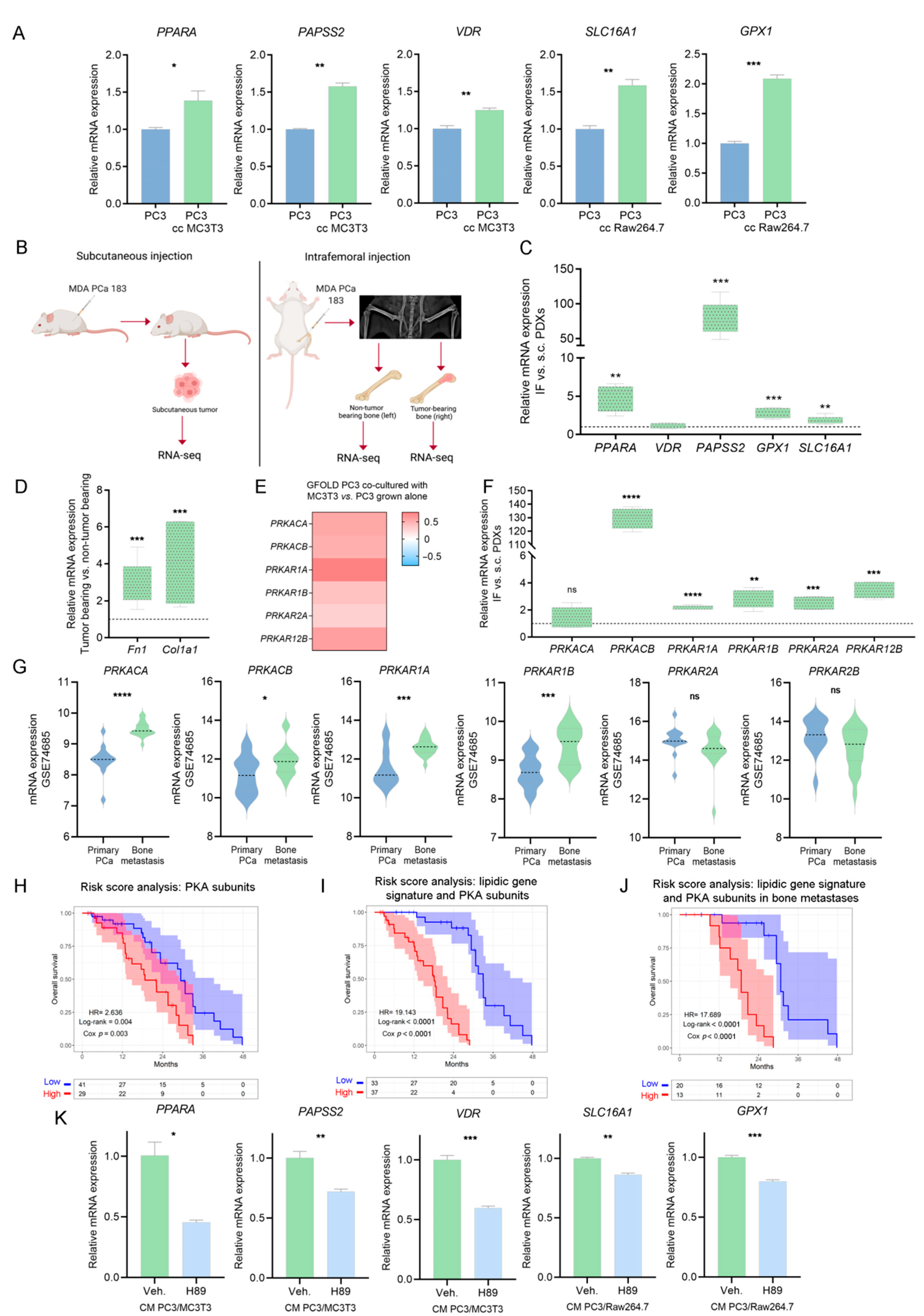
3.6. Integrative Transcriptomics and Secretomics Analyses Pin-Point a Regulatory Axis of Tumoral Metabolism Associated with the Bone Niche
3.7. Validation of PKA as a Driver of the Metabolic Rewiring in PCa Cells Induced by Bone-Secreted Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Cancer Facts and Figures 2022. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2022.html (accessed on 1 November 2021).
- Nevedomskaya, E.; Baumgart, S.J.; Haendler, B. Recent Advances in Prostate Cancer Treatment and Drug Discovery. Int. J. Mol. Sci. 2018, 19, 1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrioli, R.; Francini, E.; Fiaschi, A.I.; Laera, L.; Roviello, G. Targeted Therapies for Prostate Cancer. Cancer Investig. 2015, 33, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-F.; Shen, J.; Li, X.; Rengan, R.; Silvestris, N.; Wang, M.; Derosa, L.; Zheng, X.; Belli, A.; Zhang, X.-L.; et al. Incidence of patients with bone metastases at diagnosis of solid tumors in adults: A large population-based study. Ann. Transl. Med. 2020, 8, 482. [Google Scholar] [CrossRef] [PubMed]
- Bergers, G.; Fendt, S.-M. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer 2021, 21, 162–180. [Google Scholar] [CrossRef]
- Gazi, E.; Gardner, P.; Lockyer, N.P.; Hart, C.A.; Brown, M.D.; Clarke, N.W. Direct evidence of lipid translocation between adipocytes and prostate cancer cells with imaging FTIR microspectroscopy. J. Lipid Res. 2007, 48, 1846–1856. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.D.; Hart, C.; Gazi, E.; Gardner, P.; Lockyer, N.; Clarke, N. Influence of omega-6 PUFA arachidonic acid and bone marrow adipocytes on metastatic spread from prostate cancer. Br. J. Cancer 2010, 102, 403–413. [Google Scholar] [CrossRef]
- Herroon, M.K.; Rajagurubandara, E.; Hardaway, A.L.; Powell, K.; Turchick, A.; Feldmann, D.; Podgorski, I. Bone marrow adipocytes promote tumor growth in bone via FABP4-dependent mechanisms. Oncotarget 2013, 4, 2108–2123. [Google Scholar] [CrossRef] [Green Version]
- Diedrich, J.D.; Rajagurubandara, E.; Herroon, M.K.; Mahapatra, G.; Hüttemann, M.; Podgorski, I. Bone marrow adipocytes promote the Warburg phenotype in metastatic prostate tumors via HIF-1α activation. Oncotarget 2016, 7, 64854–64877. [Google Scholar] [CrossRef] [Green Version]
- Phan, L.M.; Yeung, S.-C.J.; Lee, M.-H. Cancer metabolic reprogramming: Importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014, 11, 1–19. [Google Scholar] [CrossRef]
- Zadra, G.; Photopoulos, C.; Loda, M. The fat side of prostate cancer. Biochim. Biophys. Acta 2013, 1831, 1518–1532. Available online: http://europepmc.org/articles/PMC3766375 (accessed on 30 November 2021). [CrossRef] [Green Version]
- Swinnen, J.V.; Roskams, T.; Joniau, S.; Van Poppel, H.; Oyen, R.; Baert, L.; Heyns, W.; Verhoeven, G. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int. J. cancer 2002, 98, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Graner, E.; Febbo, P.; Weinstein, L.; Bhattacharya, N.; Onody, T.; Bubley, G.; Balk, S.; Loda, M. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol. Cancer Res. 2003, 1, 707–715. [Google Scholar] [PubMed]
- Cao, Z.; Xu, Y.; Guo, F.; Chen, X.; Ji, J.; Xu, H.; He, J.; Yu, Y.; Sun, Y.; Lu, X.; et al. FASN Protein Overexpression Indicates Poor Biochemical Recurrence-Free Survival in Prostate Cancer. Dis. Markers 2020, 2020, 3904947. [Google Scholar] [CrossRef]
- Zadra, G.; Ribeiro, C.F.; Chetta, P.; Ho, Y.; Cacciatore, S.; Gao, X.; Syamala, S.; Bango, C.; Photopoulos, C.; Huang, Y.; et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 631–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaytseva, Y.Y.; Rychahou, P.G.; Le, A.-T.; Scott, T.L.; Flight, R.M.; Kim, J.T.; Harris, J.; Liu, J.; Wang, C.; Morris, A.J.; et al. Preclinical evaluation of novel fatty acid synthase inhibitors in primary colorectal cancer cells and a patient-derived xenograft model of colorectal cancer. Oncotarget 2018, 9, 24787–24800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anselmino, N.; Bizzotto, J.; Sanchis, P.; Lage-Vickers, S.; Ortiz, E.; Valacco, P.; Paez, A.; Labanca, E.; Meiss, R.; Navone, N.; et al. HO-1 Interactors Involved in the Colonization of the Bone Niche: Role of ANXA2 in Prostate Cancer Progression. Biomolecules 2020, 10, 467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paez, A.V.; Pallavicini, C.; Schuster, F.; Valacco, M.P.; Giudice, J.; Ortiz, E.G.; Anselmino, N.; Labanca, E.; Binaghi, M.; Salierno, M.; et al. Heme oxygenase-1 in the forefront of a multi-molecular network that governs cell-cell contacts and filopodia-induced zippering in prostate cancer. Cell Death Dis. 2016, 7, e2570. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28032857 (accessed on 30 November 2021). [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Meyer, C.A.; Wang, Q.; Liu, J.S.; Shirley Liu, X.; Zhang, Y. GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012, 28, 2782–2788. [Google Scholar] [CrossRef] [Green Version]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. 2021, 2, 100141. Available online: https://www.sciencedirect.com/science/article/pii/S2666675821000667 (accessed on 30 November 2021). [CrossRef] [PubMed]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. 2021. Available online: https://bioconductor.org/packages/release/bioc/html/enrichplot.html (accessed on 30 November 2021).
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingenuity Pathway Analysis. Available online: https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis (accessed on 30 November 2021).
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med. 2016, 22, 369–378. [Google Scholar] [CrossRef]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Mundt, F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package. 2020. Available online: https://cran.r-project.org/package=factoextra (accessed on 30 November 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pheatmap: Pretty Heatmaps. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 30 November 2021).
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef] [Green Version]
- Lochner, A.; Moolman, J.A. The many faces of H89: A review. Cardiovasc. Drug Rev. 2006, 24, 261–274. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Palanisamy, N.; Yang, J.; Shepherd, P.D.A.; Li-Ning-Tapia, E.M.; Labanca, E.; Manyam, G.C.; Ravoori, M.K.; Kundra, V.; Araujo, J.C.; Efstathiou, E.; et al. The MD Anderson Prostate Cancer Patient-derived Xenograft Series (MDA PCa PDX) Captures the Molecular Landscape of Prostate Cancer and Facilitates Marker-driven Therapy Development. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 4933–4946. [Google Scholar] [CrossRef]
- Labanca, E.; Yang, J.; Shepherd, P.D.A.; Wan, X.; Starbuck, M.W.; Guerra, L.D.; Anselmino, N.; Bizzotto, J.A.; Dong, J.; Chinnaiyan, A.M.; et al. Fibroblast Growth Factor Receptor 1 Drives the Metastatic Progression of Prostate Cancer. Eur. Urol. Oncol. 2021, S2588-9311(21)00182-6. [Google Scholar] [CrossRef]
- Li, Z.G.; Mathew, P.; Yang, J.; Starbuck, M.W.; Zurita, A.J.; Liu, J.; Sikes, C.; Multani, A.S.; Efstathiou, E.; Lopez, A.; et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J. Clin. Investig. 2008, 118, 2697–2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labanca, E.; Bizzotto, J.; Sanchis, P.; Anselmino, N.; Yang, J.; Shepherd, P.D.A.; Paez, A.; Antico-Arciuch, V.; Lage-Vickers, S.; Hoang, A.G.; et al. Prostate cancer castrate resistant progression usage of non-canonical androgen receptor signaling and ketone body fuel. Oncogene 2021, 40, 6284–6298. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using “ggplot2” 2019. Available online: https://CRAN.R-project.org/package=survminer (accessed on 1 November 2021).
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76. Available online: https://www.sciencedirect.com/science/article/pii/S1550413119306175 (accessed on 30 November 2021). [CrossRef] [PubMed]
- Fu, Y.; Zou, T.; Shen, X.; Nelson, P.J.; Li, J.; Wu, C.; Yang, J.; Zheng, Y.; Bruns, C.; Zhao, Y.; et al. Lipid metabolism in cancer progression and therapeutic strategies. MedComm 2021, 2, 27–59. [Google Scholar] [CrossRef]
- Fernández, L.P.; Gómez de Cedrón, M.; Ramírez de Molina, A. Alterations of Lipid Metabolism in Cancer: Implications in Prognosis and Treatment. Front. Oncol. 2020, 10, 577420. [Google Scholar] [CrossRef]
- Thysell, E.; Surowiec, I.; Hörnberg, E.; Crnalic, S.; Widmark, A.; Johansson, A.I.; Stattin, P.; Bergh, A.; Moritz, T.; Antti, H.; et al. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. PLoS ONE 2010, 5, e14175. [Google Scholar] [CrossRef] [Green Version]
- Sena, L.A.; Denmeade, S.R. Fatty Acid Synthesis in Prostate Cancer: Vulnerability or Epiphenomenon? Cancer Res. 2021, 81, 4385–4393. [Google Scholar] [CrossRef]
- Loftus, T.M.; Jaworsky, D.E.; Frehywot, G.L.; Townsend, C.A.; Ronnett, G.V.; Lane, M.D.; Kuhajda, F.P. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science 2000, 288, 2379–2381. [Google Scholar] [CrossRef]
- McKenzie, A.J.; Svec, K.V.; Williams, T.F.; Howe, A.K. Protein kinase A activity is regulated by actomyosin contractility during cell migration and is required for durotaxis. Mol. Biol. Cell 2020, 31, 45–58. [Google Scholar] [CrossRef]
- Deeble, P.D.; Murphy, D.J.; Parsons, S.J.; Cox, M.E. Interleukin-6- and cyclic AMP-mediated signaling potentiates neuroendocrine differentiation of LNCaP prostate tumor cells. Mol. Cell. Biol. 2001, 21, 8471–8482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.E.; Deeble, P.D.; Bissonette, E.A.; Parsons, S.J. Activated 3′,5′-cyclic AMP-dependent protein kinase is sufficient to induce neuroendocrine-like differentiation of the LNCaP prostate tumor cell line. J. Biol. Chem. 2000, 275, 13812–13818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payen, V.L.; Hsu, M.Y.; Rädecke, K.S.; Wyart, E.; Vazeille, T.; Bouzin, C.; Porporato, P.E.; Sonveaux, P. Monocarboxylate Transporter MCT1 Promotes Tumor Metastasis Independently of Its Activity as a Lactate Transporter. Cancer Res. 2017, 77, 5591–5601. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchis, P.; Anselmino, N.; Lage-Vickers, S.; Sabater, A.; Lavignolle, R.; Labanca, E.; Shepherd, P.D.A.; Bizzotto, J.; Toro, A.; Mitrofanova, A.; et al. Bone Progenitors Pull the Strings on the Early Metabolic Rewiring Occurring in Prostate Cancer Cells. Cancers 2022, 14, 2083. https://doi.org/10.3390/cancers14092083
Sanchis P, Anselmino N, Lage-Vickers S, Sabater A, Lavignolle R, Labanca E, Shepherd PDA, Bizzotto J, Toro A, Mitrofanova A, et al. Bone Progenitors Pull the Strings on the Early Metabolic Rewiring Occurring in Prostate Cancer Cells. Cancers. 2022; 14(9):2083. https://doi.org/10.3390/cancers14092083
Chicago/Turabian StyleSanchis, Pablo, Nicolas Anselmino, Sofia Lage-Vickers, Agustina Sabater, Rosario Lavignolle, Estefania Labanca, Peter D. A. Shepherd, Juan Bizzotto, Ayelen Toro, Antonina Mitrofanova, and et al. 2022. "Bone Progenitors Pull the Strings on the Early Metabolic Rewiring Occurring in Prostate Cancer Cells" Cancers 14, no. 9: 2083. https://doi.org/10.3390/cancers14092083
APA StyleSanchis, P., Anselmino, N., Lage-Vickers, S., Sabater, A., Lavignolle, R., Labanca, E., Shepherd, P. D. A., Bizzotto, J., Toro, A., Mitrofanova, A., Valacco, M. P., Navone, N., Vazquez, E., Cotignola, J., & Gueron, G. (2022). Bone Progenitors Pull the Strings on the Early Metabolic Rewiring Occurring in Prostate Cancer Cells. Cancers, 14(9), 2083. https://doi.org/10.3390/cancers14092083







