Inhibiting ALK2/ALK3 Signaling to Differentiate and Chemo-Sensitize Medulloblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Sphere-Forming Assays
2.3. Transient and Lentivirus Transfections
2.4. Immunoblot
2.5. Flow Cytometry Analyzes
2.6. MTT Assays
2.7. qRT-PCR
2.8. RT2 Profiler PCR Arrays
2.9. Analysis of Real-Time PCR Array Data
2.10. Statistical Analysis of PCR Array Data
3. Results
3.1. Signaling Pathways Regulating Pluripotency of Stem Cells Are Induced in MB Tumorsphere(s)
3.2. The Pluripotent Gene OCT4A Is Induced in MB Tumorsphere(s)
3.3. Oct4A Enhances Tumor-Initiating Capability and Increases Resistance to Standard Drugs Treatment
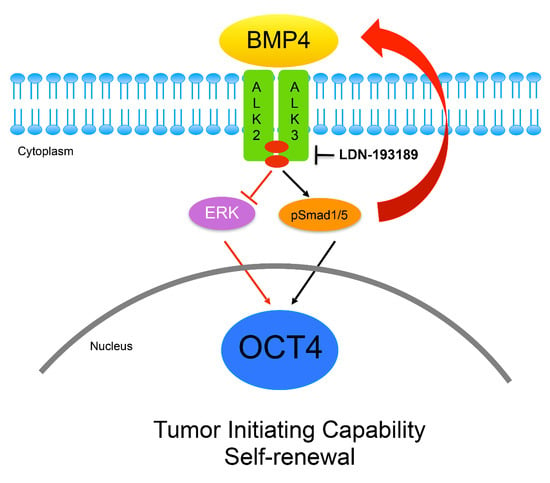
3.4. OCT4A Induces BMP4-ALK2/3 Signaling Pathway in MBs
3.5. Inhibition of ALK2/ALK3 Signaling Blocked Migration, Reduced TICs, and Increased Cytotoxicity to Chemotherapy in MB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Werbowetski-Ogilvie, T.E. From sorting to sequencing in the molecular era: The evolution of the cancer stem cell model in medulloblastoma. FEBS J. 2021, 289, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Dhanyamraju, P.K.; Patel, T.N.; Dovat, S. Medulloblastoma: “Onset of the molecular era”. Mol. Biol. Rep. 2020, 47, 9931–9937. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Grobner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754 e736. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Archer, T.C.; Mahoney, E.L.; Pomeroy, S.L. Medulloblastoma: Molecular Classification-Based Personal Therapeutics. Neurotherapeutics 2017, 14, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Ramaswamy, V.; Taylor, M.D. Medulloblastoma: From Myth to Molecular. J. Clin. Oncol. 2017, 35, 2355–2363. [Google Scholar] [CrossRef]
- Tallman, M.M.; Zalenski, A.A.; Venere, M. Cancer Stem Cells in Pediatric Brain Tumors; Gliomas; Debinski, W., Ed.; Exon Publications: Brisbane, Australia, 2021. [Google Scholar] [CrossRef]
- Huang, G.H.; Xu, Q.F.; Cui, Y.H.; Li, N.; Bian, X.W.; Lv, S.Q. Medulloblastoma stem cells: Promising targets in medulloblastoma therapy. Cancer Sci. 2016, 107, 583–589. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Morrison, L.C.; McClelland, R.; Aiken, C.; Bridges, M.; Liang, L.; Wang, X.; Di Curzio, D.; Del Bigio, M.R.; Taylor, M.D.; Werbowetski-Ogilvie, T.E. Deconstruction of medulloblastoma cellular heterogeneity reveals differences between the most highly invasive and self-renewing phenotypes. Neoplasia 2013, 15, 384–398. [Google Scholar] [CrossRef] [Green Version]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; Rampazzo, E.; Persano, L.; Abbadi, S.; Frasson, C.; Denaro, L.; D’Avella, D.; Panchision, D.M.; Della Puppa, A.; Scienza, R.; et al. Interaction of hypoxia-inducible factor-1alpha and Notch signaling regulates medulloblastoma precursor proliferation and fate. Stem Cells 2010, 28, 1918–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerabek, S.; Merino, F.; Scholer, H.R.; Cojocaru, V. OCT4: Dynamic DNA binding pioneers stem cell pluripotency. Biochim. Biophys. Acta 2014, 1839, 138–154. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, V.; Rezende, N.C.; Scotland, K.B.; Shaffer, S.M.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009, 18, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, M.K.; Singer, D.B. Embryonic stem cell biology. Adv. Pediatr. 2008, 55, 43–77. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Scholer, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. The Role and Specific Mechanism of OCT4 in Cancer Stem Cells: A Review. Int. J. Stem Cells 2020, 13, 312–325. [Google Scholar] [CrossRef]
- Murakami, S.; Ninomiya, W.; Sakamoto, E.; Shibata, T.; Akiyama, H.; Tashiro, F. SRY and OCT4 Are Required for the Acquisition of Cancer Stem Cell-Like Properties and Are Potential Differentiation Therapy Targets. Stem Cells 2015, 33, 2652–2663. [Google Scholar] [CrossRef]
- Chen, Y.C.; Hsu, H.S.; Chen, Y.W.; Tsai, T.H.; How, C.K.; Wang, C.Y.; Hung, S.C.; Chang, Y.L.; Tsai, M.L.; Lee, Y.Y.; et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE 2008, 3, e2637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, P.B.G.; Teixeira Dos Santos, M.C.; Rodini, C.O.; Kaid, C.; Pereira, M.C.L.; Furukawa, G.; da Cruz, D.S.G.; Goldfeder, M.B.; Rocha, C.R.R.; Rosenberg, C.; et al. High OCT4A levels drive tumorigenicity and metastatic potential of medulloblastoma cells. Oncotarget 2017, 8, 19192–19204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, I.L.; Montero-Montero, L.; Martin-Villar, E.; Martin-Perez, J.; Sainz, B.; Renart, J.; Toscano Simoes, R.; Soares Veloso, E.; Salviano Teixeira, C.; de Oliveira, M.C.; et al. Reduced expression of the murine HLA-G homolog Qa-2 is associated with malignancy, epithelial-mesenchymal transition and stemness in breast cancer cells. Sci. Rep. 2017, 7, 6276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmberg, J.; He, X.; Peredo, I.; Orrego, A.; Hesselager, G.; Ericsson, C.; Hovatta, O.; Oba-Shinjo, S.M.; Marie, S.K.; Nister, M.; et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS ONE 2011, 6, e18454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodini, C.O.; Suzuki, D.E.; Saba-Silva, N.; Cappellano, A.; de Souza, J.E.; Cavalheiro, S.; Toledo, S.R.; Okamoto, O.K. Expression analysis of stem cell-related genes reveal OCT4 as a predictor of poor clinical outcome in medulloblastoma. J. Neurooncol. 2012, 106, 71–79. [Google Scholar] [CrossRef]
- Mohiuddin, I.S.; Wei, S.J.; Kang, M.H. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165432. [Google Scholar] [CrossRef]
- Saigusa, S.; Tanaka, K.; Toiyama, Y.; Yokoe, T.; Okugawa, Y.; Ioue, Y.; Miki, C.; Kusunoki, M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann. Surg. Oncol. 2009, 16, 3488–3498. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal. Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Wu, D.; Wang, Y.; Wang, Z.; Zou, C.; Dai, Y.; Ng, C.F.; Teoh, J.Y.; Chan, F.L. Development of a novel and economical agar-based non-adherent three-dimensional culture method for enrichment of cancer stem-like cells. Stem Cell Res. Ther. 2018, 9, 243. [Google Scholar] [CrossRef]
- Hayon, T.; Dvilansky, A.; Shpilberg, O.; Nathan, I. Appraisal of the MTT-based assay as a useful tool for predicting drug chemosensitivity in leukemia. Leuk. Lymphoma 2003, 44, 1957–1962. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arikawa, E.; Sun, Y.; Wang, J.; Zhou, Q.; Ning, B.; Dial, S.L.; Guo, L.; Yang, J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genom. 2008, 9, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenkiewicz, M.; Li, N.; Singh, S.K. Culture and isolation of brain tumor initiating cells. Curr. Protoc. Stem Cell Biol. 2009, 11, 3.3.1–3.3.10. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.A.; Lee, J.Y.; Phi, J.H.; Wang, K.C.; Park, C.K.; Park, S.H.; Kim, S.K. Identification of brain tumour initiating cells using the stem cell marker aldehyde dehydrogenase. Eur. J. Cancer 2014, 50, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Carvalho, D.; Taylor, K.R.; Olaciregui, N.G.; Molinari, V.; Clarke, M.; Mackay, A.; Ruddle, R.; Henley, A.; Valenti, M.; Hayes, A.; et al. ALK2 inhibitors display beneficial effects in preclinical models of ACVR1 mutant diffuse intrinsic pontine glioma. Commun. Biol. 2019, 2, 156. [Google Scholar] [CrossRef]
- Zhou, K.; Shi, X.; Huo, J.; Liu, W.; Yang, D.; Yang, T.; Qin, T.; Wang, C. Bone morphogenetic protein 4 is overexpressed in and promotes migration and invasion of drug-resistant cancer cells. Int. J. Biol. Macromol. 2017, 101, 427–437. [Google Scholar] [CrossRef]
- Cheng, Y.; Liao, S.; Xu, G.; Hu, J.; Guo, D.; Du, F.; Contreras, A.; Cai, K.Q.; Peri, S.; Wang, Y.; et al. NeuroD1 Dictates Tumor Cell Differentiation in Medulloblastoma. Cell Rep. 2020, 31, 107782. [Google Scholar] [CrossRef]
- Casciati, A.; Tanori, M.; Manczak, R.; Saada, S.; Tanno, B.; Giardullo, P.; Porcu, E.; Rampazzo, E.; Persano, L.; Viola, G.; et al. Human Medulloblastoma Cell Lines: Investigating on Cancer Stem Cell-Like Phenotype. Cancers 2020, 12, 226. [Google Scholar] [CrossRef] [Green Version]
- Zanini, C.; Ercole, E.; Mandili, G.; Salaroli, R.; Poli, A.; Renna, C.; Papa, V.; Cenacchi, G.; Forni, M. Medullospheres from DAOY, UW228 and ONS-76 cells: Increased stem cell population and proteomic modifications. PLoS ONE 2013, 8, e63748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, W.; Li, K.; Wang, F.; Qin, Y.R.; Fan, Q.X. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J. Gastroenterol. 2012, 18, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zeng, Q.; Luo, H.; Zou, J.; Cao, C.; Liang, J.; Wu, D.; Liu, L. Increased expression of OCT4 is associated with low differentiation and tumor recurrence in human hepatocellular carcinoma. Pathol. Res. Pract. 2012, 208, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Atlasi, Y.; Mowla, S.J.; Ziaee, S.A.; Bahrami, A.R. OCT-4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int. J. Cancer 2007, 120, 1598–1602. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Rebollo, M.; Mateo, F.; Franke, K.; Huen, M.S.; Lopitz-Otsoa, F.; Rodriguez, M.S.; Plans, V.; Thomson, T.M. Nucleolar exit of RNF8 and BRCA1 in response to DNA damage. Exp. Cell Res. 2012, 318, 2365–2376. [Google Scholar] [CrossRef]
- Filipponi, D.; Emelyanov, A.; Muller, J.; Molina, C.; Nichols, J.; Bulavin, D.V. DNA Damage Signaling-Induced Cancer Cell Reprogramming as a Driver of Tumor Relapse. Mol. Cell 2019, 74, 651–663.e8. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Conserved POU/OCT- and GATA-binding sites in 5’-flanking promoter region of mammalian WNT8B orthologs. Int. J. Oncol. 2007, 30, 1273–1277. [Google Scholar] [CrossRef] [Green Version]
- Katoh, M. Network of WNT and other regulatory signaling cascades in pluripotent stem cells and cancer stem cells. Curr. Pharm. Biotechnol. 2011, 12, 160–170. [Google Scholar] [CrossRef]
- Bach, D.H.; Park, H.J.; Lee, S.K. The Dual Role of Bone Morphogenetic Proteins in Cancer. Mol. Ther. Oncolytics 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Huang, J.; Gong, J. Bone morphogenetic protein 4 (BMP4) is required for migration and invasion of breast cancer. Mol. Cell Biochem. 2012, 363, 179–190. [Google Scholar] [CrossRef]
- Cao, Y.; Slaney, C.Y.; Bidwell, B.N.; Parker, B.S.; Johnstone, C.N.; Rautela, J.; Eckhardt, B.L.; Anderson, R.L. BMP4 inhibits breast cancer metastasis by blocking myeloid-derived suppressor cell activity. Cancer Res. 2014, 74, 5091–5102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.; Yu, J.; Park, A.; Dubon, M.J.; Do, J.; Kim, Y.; Nam, D.; Noh, J.; Park, K.S. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via Notch signaling. Sci. Rep. 2019, 9, 11724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccirillo, S.G.; Reynolds, B.A.; Zanetti, N.; Lamorte, G.; Binda, E.; Broggi, G.; Brem, H.; Olivi, A.; Dimeco, F.; Vescovi, A.L. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 2006, 444, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Wang, Y.; Cheng, Y.; Liao, S.; Hu, J.; Du, F.; Xu, G.; Liu, Y.; Cai, K.Q.; Cheung, M.; et al. Tumor cells generate astrocyte-like cells that contribute to SHH-driven medulloblastoma relapse. J. Exp. Med. 2021, 218, e20202350. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filipponi, D.; Pagnuzzi-Boncompagni, M.; Pagès, G. Inhibiting ALK2/ALK3 Signaling to Differentiate and Chemo-Sensitize Medulloblastoma. Cancers 2022, 14, 2095. https://doi.org/10.3390/cancers14092095
Filipponi D, Pagnuzzi-Boncompagni M, Pagès G. Inhibiting ALK2/ALK3 Signaling to Differentiate and Chemo-Sensitize Medulloblastoma. Cancers. 2022; 14(9):2095. https://doi.org/10.3390/cancers14092095
Chicago/Turabian StyleFilipponi, Doria, Marina Pagnuzzi-Boncompagni, and Gilles Pagès. 2022. "Inhibiting ALK2/ALK3 Signaling to Differentiate and Chemo-Sensitize Medulloblastoma" Cancers 14, no. 9: 2095. https://doi.org/10.3390/cancers14092095
APA StyleFilipponi, D., Pagnuzzi-Boncompagni, M., & Pagès, G. (2022). Inhibiting ALK2/ALK3 Signaling to Differentiate and Chemo-Sensitize Medulloblastoma. Cancers, 14(9), 2095. https://doi.org/10.3390/cancers14092095