Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC
Abstract
:Simple Summary
Abstract
1. Introduction
2. Companion Diagnostics
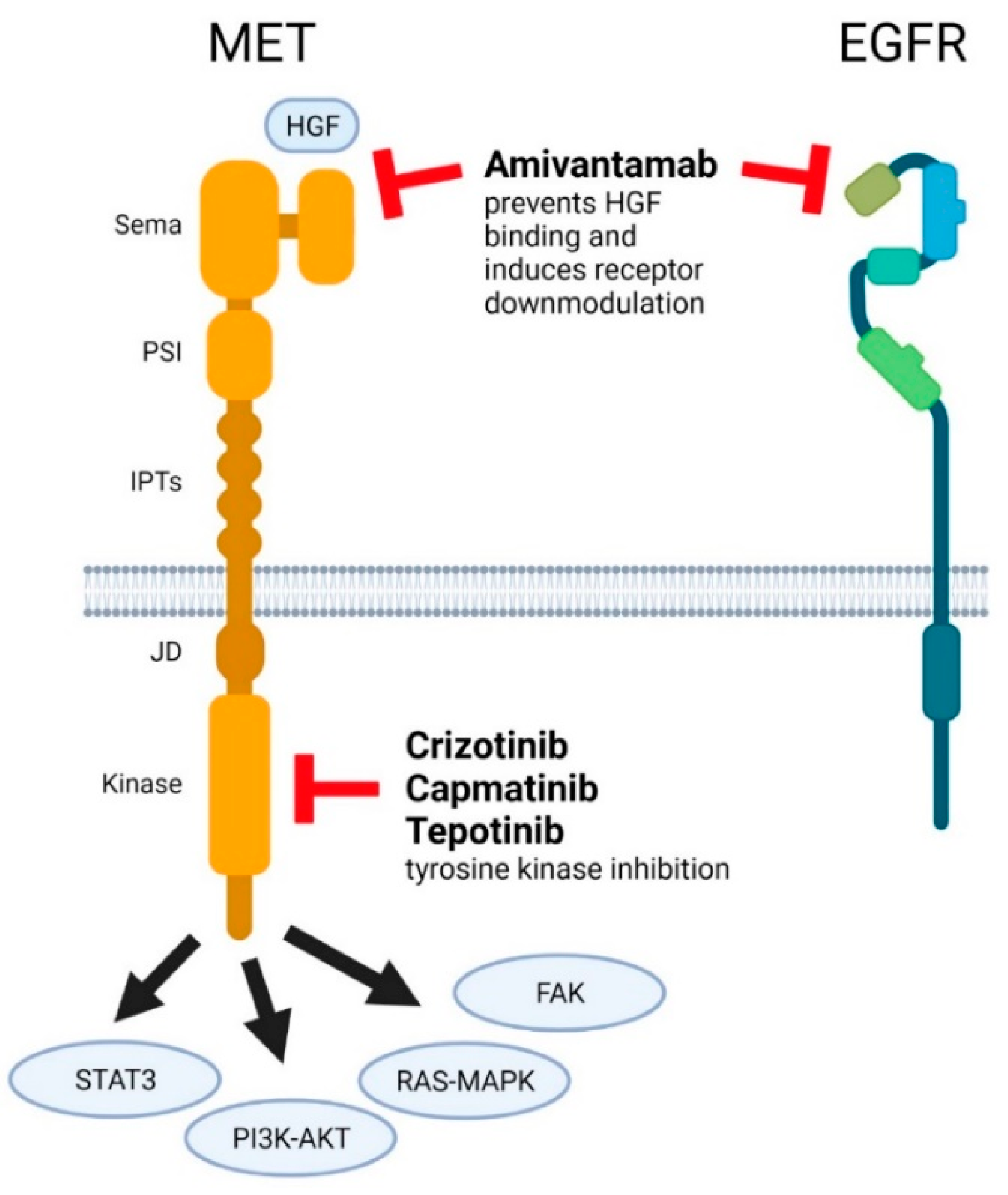
3. MET-Targeted Therapy and NSCLC
3.1. Crizotinib
3.2. Capmatinib
3.3. Tepotinib
3.4. Amivantamab
4. Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Jørgensen, J.T.; Winther, H.; Askaa, J.; Andresen, L.; Olsen, D.; Mollerup, J. A Companion Diagnostic with Significant Clinical Impact in Treatment of Breast and Gastric Cancer. J. Front. Oncol. 2021, 11, 676939. [Google Scholar] [CrossRef]
- Jørgensen, J.T. Oncology Drug-Companion Diagnostic Combinations. Cancer Treat. Res. Commun. 2021, 29, 100492. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.S.; Park, M.; Blair, D.G.; Tainsky, M.A.; Huebner, K.; Croce, C.M.; Vande Woude, G. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984, 311, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L.; Vigna, E.; Narsimhan, R.P.; Gaudino, G.; Zarnegar, R.; Michalopoulos, G.K.; Comoglio, P.M. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene 1991, 6, 501–504. [Google Scholar]
- Matsumoto, K.; Umitsu, M.; De Silva, D.M.; Roy, A.; Bottaro, D.P. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 2017, 108, 296–307. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Luo, J.; Chang, J.; Rekhtman, N.; Arcila, M.; Drilon, A. MET-dependent solid tumours-molecular diagnosis and targeted therapy. Nat. Rev. Clin. Oncol. 2020, 17, 569–587. [Google Scholar] [CrossRef]
- Recondo, G.; Che, J.; Jänne, P.A.; Awad, M.M. Targeting MET Dysregulation in Cancer. Cancer Discov. 2020, 10, 922–934. [Google Scholar] [CrossRef] [PubMed]
- FDA. Capmatinib (Tabrecta) Full Prescribing Information. Revised: 5/2020. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/213591s000lbl.pdf (accessed on 3 December 2021).
- FDA. List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools). Update 1 December 2021. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools (accessed on 2 December 2021).
- Hortobagyi, G.N. Opportunities and challenges in the development of targeted therapies. Semin. Oncol. 2004, 31, 21–27. [Google Scholar] [CrossRef]
- Simon, R.; Maitournam, A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clin. Cancer Res. 2004, 10, 6759–6763, Erratum in Clin. Cancer Res. 2006, 12, 3229. [Google Scholar] [CrossRef] [Green Version]
- FDA. Guidance for Industry and Food and Drug Administration Staff. In Vitro Companion Diagnostic Devices. 2014. Available online: https://www.fda.gov/media/81309/download (accessed on 6 December 2021).
- Regulation (EU) 2017/746 of the European Parliament and of the Council of 5 April 2017 on In Vitro Diagnostic Medical Devices and Repealing Directive 98/79/EC and Commission Decision. 2010/227/EU. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32017R0746&from=EN (accessed on 6 December 2021).
- Regulation 2021/0323 of the European Parliament and of the Council of 14 October 2021. Transitional Provisions for Certain In Vitro Diagnostic Medical Devices and Deferred Application of Requirements for In-House Devices. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:52021PC0627&from=EN (accessed on 6 December 2021).
- Coleman, N.; Hong, L.; Zhang, J.; Heymach, J.; Hong, D.; Le, X. Beyond epidermal growth factor receptor: MET amplification as a general resistance driver to targeted therapy in oncogene-driven non-small-cell lung cancer. ESMO Open 2021, 6, 100319. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Zhang, L.; Kim, D.-W.; Liu, X.; Lee, D.H.; Yang, J.C.-H.; Ahn, M.-J.; Vansteenkiste, J.F.; Su, W.-C.; Felip, E.; et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, MET Factor-Dysregulated Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 3101–3109. [Google Scholar] [CrossRef]
- FDA. Crizotinib (Xalkori) Full Prescribing Information. Revised 9/2021. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/202570s031lbl.pdf (accessed on 8 December 2021).
- FDA. Tepotinib (Tepmetko) Full Prescribing Information. Revised 2/2021. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/214096s000lbl.pdf (accessed on 14 December 2021).
- FDA. Amivantamab (Rybrevant) Full Prescribing Information. Revised 5/2021. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/761210s000lbl.pdf (accessed on 18 December 2021).
- Kwak, E.L.; Bang, Y.-J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.-H.I.; Dezube, B.J.; Jänne, P.A.; Costa, D.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.T.; Kim, D.W.; Nakagawa, K.; Seto, T.; Crinó, L.; Ahn, M.J.; De Pas, T.; Besse, B.; Solomon, B.J.; Blackhall, F.; et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 2013, 368, 2385–2394. [Google Scholar] [CrossRef] [Green Version]
- Shaw, A.T.; Ou, S.H.; Bang, Y.J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef] [Green Version]
- Moro-Sibilot, D.; Cozic, N.; Pérol, M.; Mazières, J.; Otto, J.; Souquet, P.J.; Bahleda, R.; Wislez, M.; Zalcman, G.; Guibert, S.D.; et al. Crizotinib in c-MET- or ROS1-positive NSCLC: Results of the AcSé phase II trial. Ann. Oncol. 2019, 30, 1985–1991. [Google Scholar] [CrossRef] [PubMed]
- Landi, L.; Chiari, R.; Tiseo, M.; D’Incà, F.; Dazzi, C.; Chella, A.; Delmonte, A.; Bonanno, L.; Giannarelli, D.; Cortinovis, D.; et al. Crizotinib in MET-Deregulated or ROS1-Rearranged Pretreated Non-Small Cell Lung Cancer (METROS): A Phase II, Prospective, Multicenter, Two-Arms Trial. Clin. Cancer Res. 2019, 25, 7312–7319. [Google Scholar] [CrossRef] [Green Version]
- Drilon, A.; Clark, J.W.; Weiss, J.; Ou, S.I.; Camidge, D.R.; Solomon, B.J.; Otterson, G.A.; Villaruz, L.C.; Riely, G.J.; Heist, R.S.; et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 2020, 26, 47–51. [Google Scholar] [CrossRef]
- Schuler, M.; Berardi, R.; Lim, W.T.; de Jonge, M.; Bauer, T.M.; Azaro, A.; Gottfried, M.; Han, J.Y.; Lee, D.H.; Wollner, M.; et al. Molecular correlates of response to capmatinib in advanced non-small-cell lung cancer: Clinical and biomarker results from a phase I trial. Ann. Oncol. 2020, 31, 789–797. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.Y.; Reguart, N.; Garon, E.B.; Groen, H.J.M.; Tan, D.S.W.; Hida, T.; de Jonge, M.; Orlov, S.V.; et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 944–957. [Google Scholar] [CrossRef]
- Wu, Y.L.; Cheng, Y.; Zhou, J.; Lu, S.; Zhang, Y.; Zhao, J.; Kim, D.W.; Soo, R.A.; Kim, S.W.; Pan, H.; et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): An open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir. Med. 2020, 8, 1132–1143. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef] [PubMed]
- FDA. Summary of Safety and Effectiveness Data. FoundationOne CDx. 2020. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019S011B.pdf (accessed on 14 December 2021).
- Socinski, M.A.; Pennell, N.A.; Davies, K.D. MET Exon 14 Skipping Mutations in Non-Small-Cell Lung Cancer: An Overview of Biology, Clinical Outcomes, and Testing Considerations. JCO Precis. Oncol. 2021, 5, 653–663. [Google Scholar] [CrossRef]
- Brazel, D.; Nagasaka, M. Spotlight on Amivantamab (JNJ-61186372) for EGFR Exon 20 Insertions Positive Non-Small Cell Lung Cancer. Lung Cancer 2021, 12, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Haura, E.B.; Leighl, N.B.; Mitchell, P.; Shu, C.A.; Girard, N.; Viteri, S.; Han, J.Y.; Kim, S.W.; Lee, C.K.; et al. Amivantamab in EGFR Exon 20 Insertion-Mutated Non-Small-Cell Lung Cancer Progressing on Platinum Chemotherapy: Initial Results from the CHRYSALIS Phase I Study. J. Clin. Oncol. 2021, 39, 3391–3402. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.P.; Hudson, R.; Wang, M.H. Progress and challenge in development of biotherapeutics targeting MET receptor for treatment of advanced cancer. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188425. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Brodrick, P. Targeted Therapy Approaches for MET Abnormalities in Non-Small Cell Lung Cancer. Drugs 2021, 81, 547–554. [Google Scholar] [CrossRef]
- Spigel, D.R.; Edelman, M.J.; O’Byrne, K.; Paz-Ares, L.; Mocci, S.; Phan, S.; Shames, D.S.; Smith, D.; Yu, W.; Paton, V.E.; et al. Results from the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non-Small-Cell Lung Cancer: METLung. J. Clin. Oncol. 2017, 35, 412–420. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, C.; Schulz, A.A.; Winkelmann, R.; Smith, K.; Wild, P.J.; Demes, M. Comparison of MET gene amplification analysis by next-generation sequencing and fluorescence in situ hybridization. Oncotarget 2021, 12, 2273–2282. [Google Scholar] [CrossRef]
- Peng, L.X.; Jie, G.L.; Li, A.N.; Liu, S.Y.; Sun, H.; Zheng, M.M.; Zhou, J.Y.; Zhang, J.T.; Zhang, X.C.; Zhou, Q.; et al. MET amplification identified by next-generation sequencing and its clinical relevance for MET inhibitors. Exp. Hematol. Oncol. 2021, 10, 52. [Google Scholar] [CrossRef]
- Schubart, C.; Stöhr, R.; Tögel, L.; Fuchs, F.; Sirbu, H.; Seitz, G.; Seggewiss-Bernhardt, R.; Leistner, R.; Sterlacci, W.; Vieth, M.; et al. MET Amplification in Non-Small Cell Lung Cancer (NSCLC)-A Consecutive Evaluation Using Next-Generation Sequencing (NGS) in a Real-World Setting. Cancers 2021, 13, 5023. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Karaszewska, B.; Kang, Y.K.; Chung, H.C.; Shankaran, V.; Siena, S.; Go, N.F.; Yang, H.; Schupp, M.; Cunningham, D. A Multicenter Phase II Study of AMG 337 in Patients with MET-Amplified Gastric/Gastroesophageal Junction/Esophageal Adenocarcinoma and Other MET-Amplified Solid Tumors. Clin. Cancer Res. 2019, 25, 2414–2423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jørgensen, J.T.; Mollerup, J.; Yang, H.; Go, N.; Nielsen, K.B. MET deletion is a frequent event in gastric/gastroesophageal junction/esophageal cancer: A cross-sectional analysis of gene status and signal distribution in 1580 patients. Ann. Transl. Med. 2021, 9, 225. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Morimoto, M.; Moriguchi, M.; Izumi, N.; Takayama, T.; Yoshiji, H.; Hino, K.; Oikawa, T.; Chiba, T.; Motomura, K.; et al. A randomized, double-blind, placebo-controlled, phase 3 study of tivantinib in Japanese patients with MET-high hepatocellular carcinoma. Cancer Sci. 2020, 111, 3759–3769. [Google Scholar] [CrossRef]
- Decaens, T.; Barone, C.; Assenat, E.; Wermke, M.; Fasolo, A.; Merle, P.; Blanc, J.F.; Grando, V.; Iacobellis, A.; Villa, E.; et al. Phase 1b/2 trial of tepotinib in sorafenib pretreated advanced hepatocellular carcinoma with MET overexpression. Br. J. Cancer 2021, 125, 190–199. [Google Scholar] [CrossRef] [PubMed]
| Drug | Drug Class | Approved Indication(s) | FDA Approved CDx Assay(s) | |
|---|---|---|---|---|
| Crizotinib | Small molecule inhibitor | Treatment of patients with metastatic NSCLC whose tumors are ALK or ROS1-positive as detected by an FDA-approved test | ALK | FoundationOne CDx VENTANA ALK (D5F3) CDx Assay Vysis ALK Break Apart FISH Probe Kit ROS1 |
| ROS1 | Oncomine Dx Target Test | |||
| MET | No approved CDx available | |||
| Capmatinib | Small molecule inhibitor | Treatment of adult patients with NSCLC whose tumors have a mutation that leads to MET exon 14 skipping as detected by an FDA-approved test | MET | FoundationOne CDx |
| Tepotinib | Small molecule inhibitor | Treatment of adult patients with metastatic NSCLC harboring MET exon 14 skipping alterations | MET | No approved CDx available |
| Amivantamab | Bispecific antibody | Treatment of adult patients with locally advanced or metastatic NSCLC with EGFR exon 20 insertion mutations, as detected by an FDA-approved test, whose disease has progressed on or after platinum-based chemotherapy | EGFR exon 20 insertion | Guardant360® CDx |
| MET | No approved CDx available | |||
| Drug | Publication [Reference] | Method | Biomarker/CDx | N | Objective Response Rate |
|---|---|---|---|---|---|
| Crizotinib | Moro-Sibilot D et al. [24] | FISH NGS | MET GCN ≥ 6 | 25 | 16% |
| METex14 | 25 | 12% | |||
| Landi L et al. [25] | FISH NGS | MET/CEP7 > 2.2 | 16 | 31% | |
| METex14 | 10 | 20% | |||
| Drilon A et al. [26] | NGS | METex14 | 65 | 32% | |
| Capmatinib | Schuler M et al. [27] | FISH | MET GCN < 4 | 17 | 6% |
| 4 ≤ MET GCN < 6 | 12 | 25% | |||
| MET GCN ≥ 6 | 15 | 47% | |||
| MET/CEP7 ≥ 2.0 | 9 | 44% | |||
| MET/CEP7 < 2.0 | 32 | 22% | |||
| IHC | MET IHC2+ | 14 | 14% | ||
| MET IHC3+ | 37 | 27% | |||
| Wu YL et al. [17] | FISH | MET GCN < 4 | 41 | 12% | |
| 4 ≤ MET GCN < 6 | 18 | 22% | |||
| MET GCN ≥ 6 | 36 | 47% | |||
| IHC | MET IHC2+ | 16 | 19% | ||
| MET IHC3+ | 78 | 32% | |||
| Wolf J et al. [28] | NGS | METex14 | 69 (Previous treated) | 41% | |
| METex14 | 28 (Treatment naïve) | 64% | |||
| NGS | MET GCN < 4 | 30 (Previous treated) | 7% | ||
| MET GCN 4 or 5 | 54 (Previous treated) | 9% | |||
| MET GCN 6–9 | 42 (Previous treated) | 12% | |||
| MET GCN ≥ 10 | 69 (Previous treated) | 28% | |||
| MET GCN ≥ 10 | 15 (Treatment naïve) | 40% | |||
| Tepotinib | Wu YL et al. [29] | IHC | MET IHC3+ | 19 | 68% |
| FISH | MET/CEP7 ≥ 2.0 | 12 | 67% | ||
| Paik PK et al. [30] | NGS | METex14 | 99 | 46% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jørgensen, J.T.; Mollerup, J. Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC. Cancers 2022, 14, 2150. https://doi.org/10.3390/cancers14092150
Jørgensen JT, Mollerup J. Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC. Cancers. 2022; 14(9):2150. https://doi.org/10.3390/cancers14092150
Chicago/Turabian StyleJørgensen, Jan Trøst, and Jens Mollerup. 2022. "Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC" Cancers 14, no. 9: 2150. https://doi.org/10.3390/cancers14092150
APA StyleJørgensen, J. T., & Mollerup, J. (2022). Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC. Cancers, 14(9), 2150. https://doi.org/10.3390/cancers14092150







