Role of Amino Acid Transporter SNAT1/SLC38A1 in Human Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culturing of Melanoma Cell Lines
2.2. mRNA Expression Analysis
2.3. Protein Analysis with Western Blotting
2.4. siRNA Transfection
2.5. Immunohistochemical Analysis
2.6. Immunofluorescence Staining
2.7. XTT Viability Assay
2.8. Analysis of Cell Proliferation
2.9. Clonogenic Assay
2.10. Scratch Wound Healing Assay
2.11. Migration and Invasion Analysis with Boyden Chamber Assay
2.12. Apoptosis Analysis
2.13. Cell Cycle Analysis
2.14. Senesce-Associated β-Galactosidase Staining
2.15. Statistical Analysis
3. Results
3.1. SNAT1 Is Upregulated In Vitro and In Vivo in Melanoma
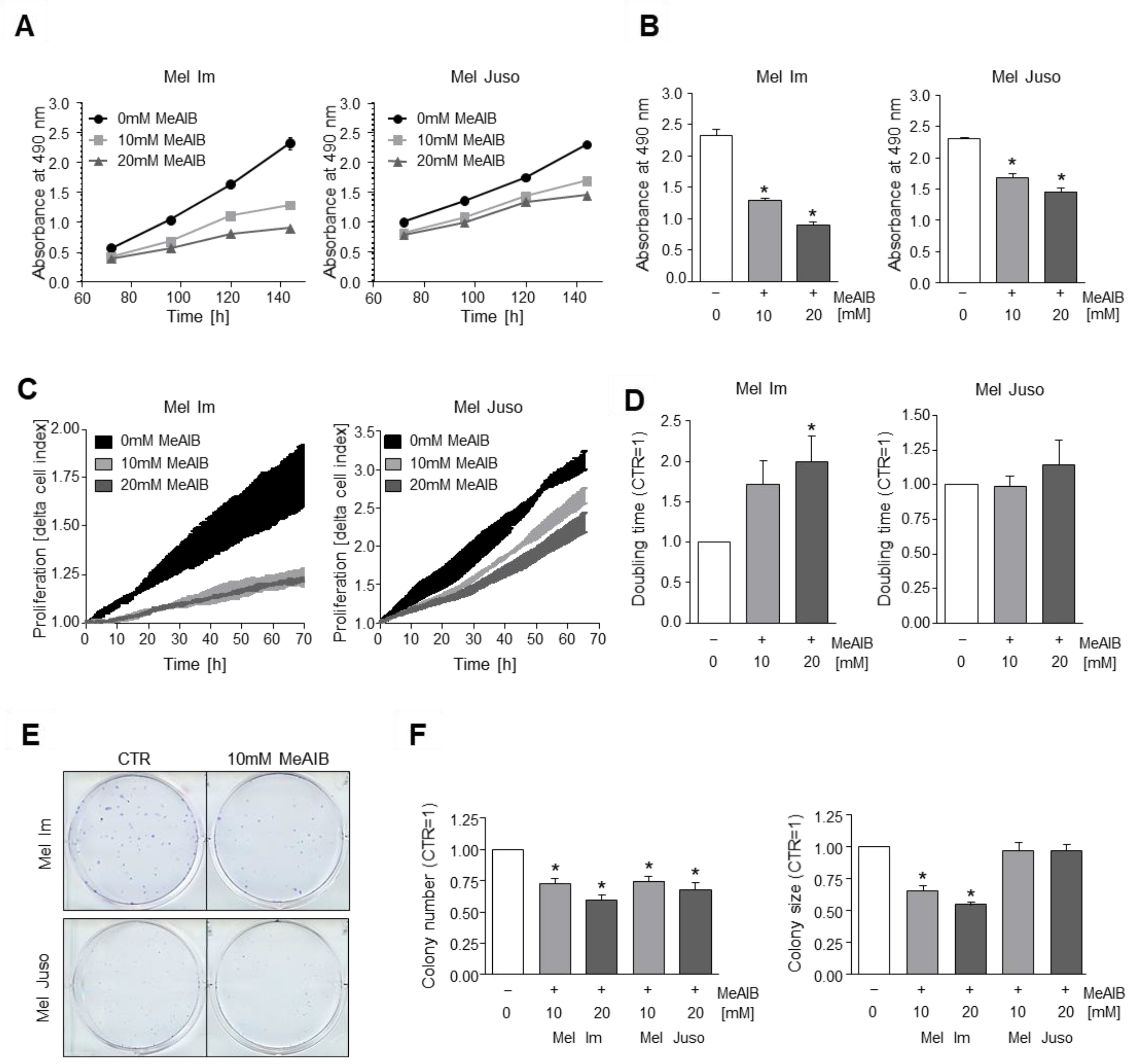
3.2. Competitive Inhibition of SNAT1 Reduces Proliferative Potential of Melanoma Cells In Vitro
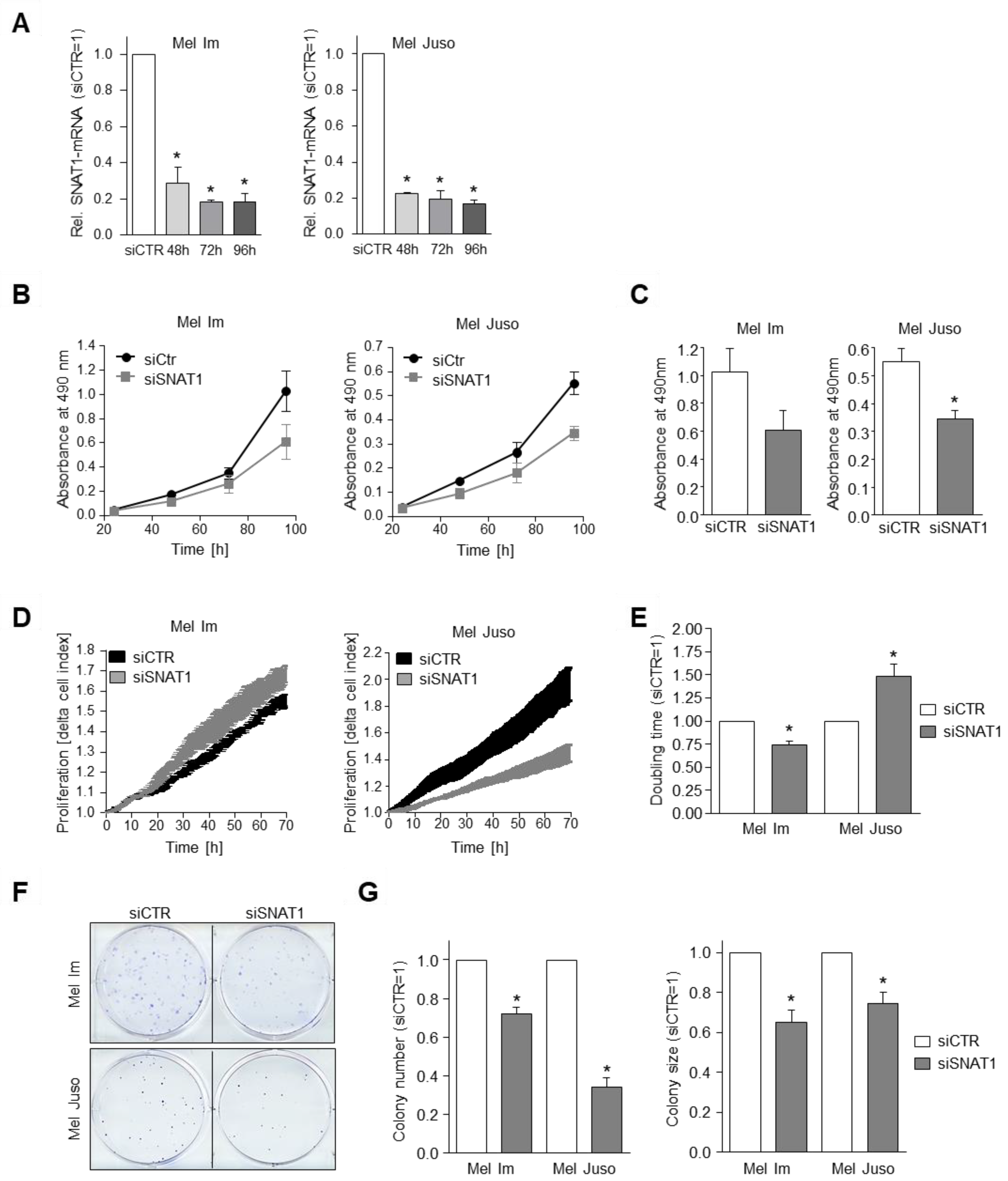
3.3. SNAT1 Knockdown Reduces Proliferation Rate of Melanoma Cells In Vitro
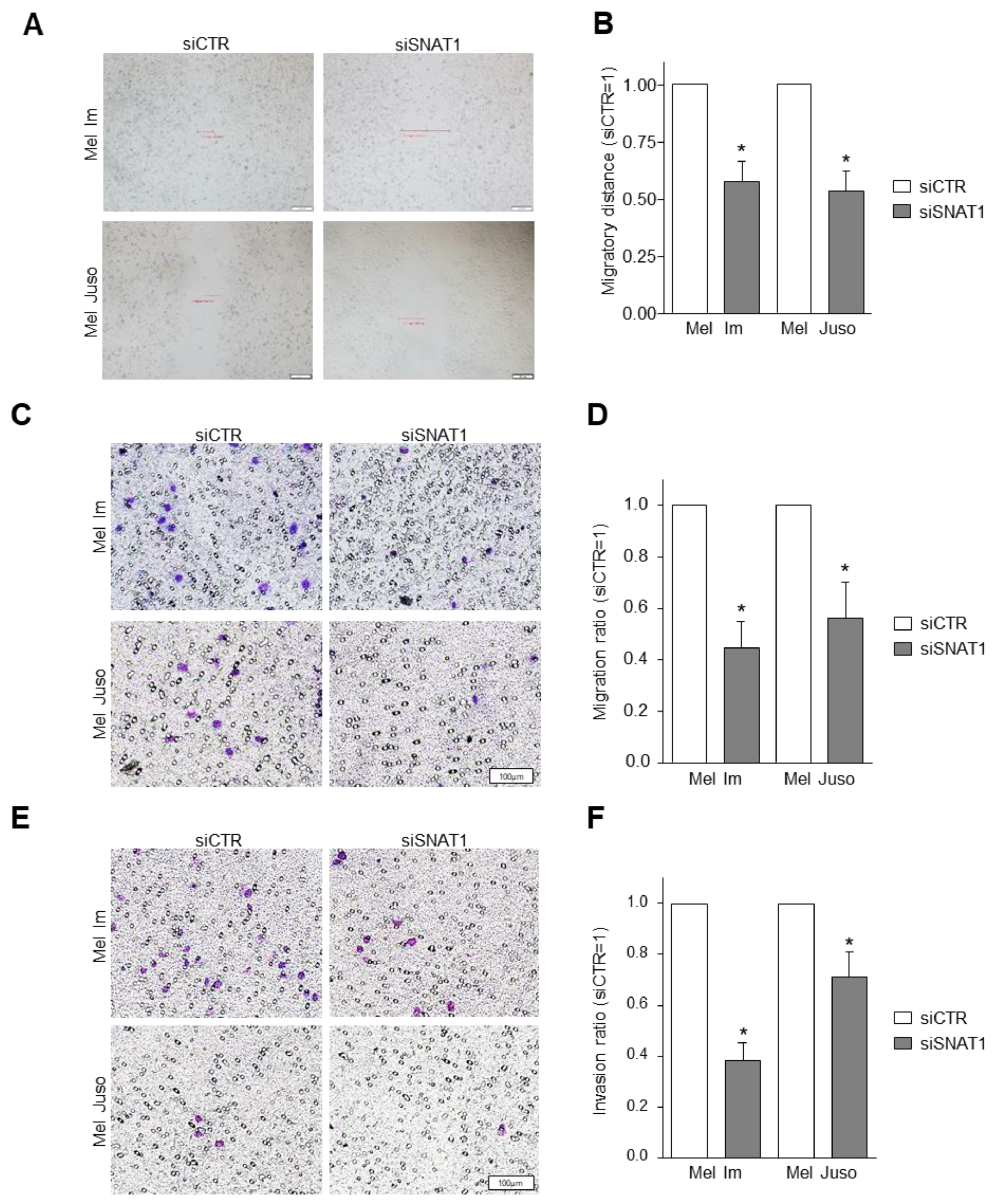
3.4. SNAT1 Downregulation Leads to Reduction of Cellular Migration and Invasion
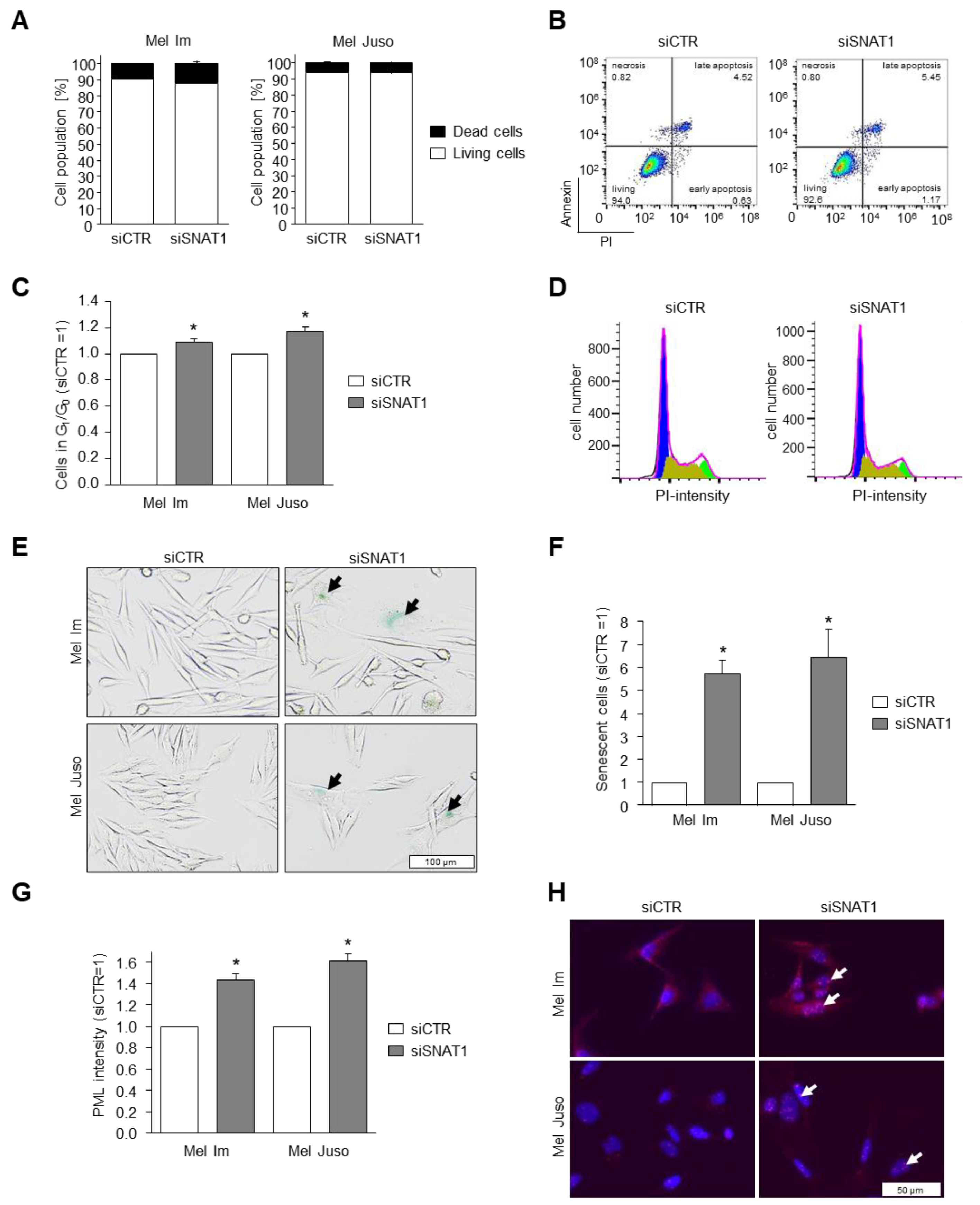
3.5. Reduction of SNAT1 Expression Induces Cell Cycle Arrest and Senescence in Melanoma Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Koch, A.; Lang, S.A.; Wild, P.J.; Gantner, S.; Mahli, A.; Spanier, G.; Berneburg, M.; Müller, M.; Bosserhoff, A.K.; Hellerbrand, C. Glucose transporter isoform 1 expression enhances metastasis of malignant melanoma cells. Oncotarget 2015, 6, 32748–32760. [Google Scholar] [CrossRef]
- Böhme, I.; Bosserhoff, A.K. Acidic tumor microenvironment in human melanoma. Pigment Cell Melanoma Res. 2016, 29, 508–523. [Google Scholar] [CrossRef]
- Böhme, I.; Bosserhoff, A. Extracellular acidosis triggers a senescence-like phenotype in human melanoma cells. Pigment Cell Melanoma Res. 2020, 33, 41–51. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Sugawara, M.; Huang, W.; Martindale, R.G.; Leibach, F.H.; Ganapathy, M.E.; Prasad, P.D.; Ganapathy, V. Structure, Function, and Tissue Expression Pattern of Human SN2, a Subtype of the Amino Acid Transport System N. Biochem. Biophys. Res. Commun. 2001, 281, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yang, T.; Peng, Z.; Xiao, H.; Jiang, N.; Zhang, L.; Ca, D.; Wu, P.; Pan, Q. SLC1A5 Silencing Inhibits Esophageal Cancer Growth via Cell Cycle Arrest and Apoptosis. Cell. Physiol. Biochem. 2018, 48, 397. [Google Scholar] [CrossRef]
- Chien, H.-C.; Colas, C.; Finke, K.; Springer, S.; Stoner, L.; Zur, A.A.; Venteicher, B.; Campbell, J.; Hall, C.; Flint, A.; et al. Reevaluating the Substrate Specificity of the L-Type Amino Acid Transporter (LAT1). J. Med. Chem. 2018, 61, 7358–7373. [Google Scholar] [CrossRef]
- Kaira, K.; Kawashima, O.; Endoh, H.; Imaizumi, K.; Goto, Y.; Kamiyoshihara, M.; Sugano, M.; Yamamoto, R.; Osaki, T.; Tanaka, S.; et al. Expression of amino acid transporter (LAT1 and 4F2hc) in pulmonary pleomorphic carcinoma. Hum. Pathol. 2019, 84, 142–149. [Google Scholar] [CrossRef]
- Bröer, A.; Rahimi, F.; Bröer, S. Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells. J. Biol. Chem. 2016, 291, 13194–13205. [Google Scholar] [CrossRef]
- Bröer, S. Amino Acid Transporters as Targets for Cancer Therapy: Why, Where, When, and How. Int. J. Mol. Sci. 2020, 21, 6156. [Google Scholar] [CrossRef] [PubMed]
- Yamada, D.; Kawabe, K.; Tosa, I.; Tsukamoto, S.; Nakazato, R.; Kou, M.; Fujikawa, K.; Nakamura, S.; Ono, M.; Oohashi, T.; et al. Inhibition of the glutamine transporter SNAT1 confers neuroprotection in mice by modulating the mTOR-autophagy system. Commun. Biol. 2019, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, Y.; Fang, W.; Liu, K.; Jiao, X.; Wang, Z.; Wang, J.; Zang, Y.-S. Increased SNAT1 is a marker of human osteosarcoma and potential therapeutic target. Oncotarget 2017, 8, 78930–78939. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Cao, F.; Fang, W.; Hu, Y.; Chen, Y.; Ding, H.; Yu, G. Activation of SNAT1/SLC38A1 in human breast cancer: Correlation with p-Akt overexpression. BMC Cancer 2013, 13, 343. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-Y.; Sohn, B.H.; Johnson, R.L.; Kang, M.-H.; Kim, S.B.; Shim, J.-J.; Mangala, L.S.; Kim, J.H.; Yoo, J.E.; Rodriguez-Aguayo, C.; et al. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 2016, 63, 159–172. [Google Scholar] [CrossRef]
- Zhou, F.; Xie, W.; Chen, S.; Wang, X.; Liu, Q.; Pan, X.; Su, F.; Feng, M. SLC38A1 promotes proliferation and migration of human colorectal cancer cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2017, 37, 30–36. [Google Scholar] [CrossRef]
- Kondoh, N.; Imazeki, N.; Arai, M.; Hada, A.; Hatsuse, K.; Matsuo, H.; Matsubara, O.; Ohkura, S.; Yamamoto, M. Activation of a system A amino acid transporter, ATA1/SLC38A1, in human hepatocellular carcinoma and preneoplastic liver tissues. Int. J. Oncol. 2007, 31, 81–87. [Google Scholar] [CrossRef][Green Version]
- Xie, J.; Li, P.; Gao, H.-F.; Qian, J.-X.; Yuan, L.-Y.; Wang, J.-J. Overexpression of SLC38A1 is associated with poorer prognosis in Chinese patients with gastric cancer. BMC Gastroenterol. 2014, 14, 70. [Google Scholar] [CrossRef]
- Li, Y.; Shao, H.; Da, Z.; Pan, J.; Fu, B. High expression of SLC38A1 predicts poor prognosis in patients with de novo acute myeloid leukemia. J. Cell. Physiol. 2019, 234, 20322–20328. [Google Scholar] [CrossRef]
- Md Fuzi, A.A.; Omar, S.Z.; Mohamed, Z.; Mat Adenan, N.A.; Mokhtar, N.M. High throughput silencing identifies novel genes in endometrioid endometrial cancer. Taiwan. J. Obstet. Gynecol. 2018, 57, 217–226. [Google Scholar] [CrossRef]
- Schulte, M.L.; Fu, A.; Zhao, P.; Li, J.; Geng, L.; Smith, S.T.; Kondo, J.; Coffey, R.J.; Johnson, M.O.; Rathmell, J.C.; et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 2018, 24, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Hannus, M.; Beitzinger, M.; Engelmann, J.C.; Weickert, M.-T.; Spang, R.; Hannus, S.; Meister, G. siPools: Highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014, 42, 8049–8061. [Google Scholar] [CrossRef] [PubMed]
- Arndt, S.; Wacker, E.; Li, Y.-F.; Shimizu, T.; Thomas, H.M.; Morfill, G.E.; Karrer, S.; Zimmermann, J.L.; Bosserhoff, A.-K. Cold atmospheric plasma, a new strategy to induce senescence in melanoma cells. Exp. Dermatol. 2013, 22, 284–289. [Google Scholar] [CrossRef]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Stoll, R.; Lodermeyer, S.; Bosserhoff, A.-K. Detailed analysis of MIA protein by mutagenesis. Biol. Chem. 2006, 387, 1601–1606. [Google Scholar] [CrossRef]
- Franchi-Gazzola, R.; Dall’Asta, V.; Sala, R.; Visigalli, R.; Bevilacqua, E.; Gaccioli, F.; Gazzola, G.C.; Bussolati, O. The role of the neutral amino acid transporter SNAT2 in cell volume regulation. Acta Physiol. 2006, 187, 273–283. [Google Scholar] [CrossRef]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef]
- Pommer, M.; Kuphal, S.; Bosserhoff, A.K. Amphiregulin Regulates Melanocytic Senescence. Cells 2021, 10, 326. [Google Scholar] [CrossRef]
- Salama, R.; Sadaie, M.; Hoare, M.; Narita, M. Cellular senescence and its effector programs. Genes Dev. 2014, 28, 99–114. [Google Scholar] [CrossRef]
- Zavorka Thomas, M.E.; Lu, X.; Talebi, Z.; Jeon, J.Y.; Buelow, D.R.; Gibson, A.A.; Uddin, M.E.; Brinton, L.T.; Nguyen, J.; Collins, M.; et al. Gilteritinib Inhibits Glutamine Uptake and Utilization in FLT3-ITD-Positive AML. Mol. Cancer Ther. 2021, 20, 2207–2217. [Google Scholar] [CrossRef]
- Wang, Q.; Beaumont, K.A.; Otte, N.J.; Font, J.; Bailey, C.G.; van Geldermalsen, M.; Sharp, D.M.; Tiffen, J.C.; Ryan, R.M.; Jormakka, M.; et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer 2014, 135, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.; Fenster, B.E.; Ito, H.; Takeda, K.; Bae, N.S.; Hirai, T.; Yu, Z.X.; Ferrans, V.J.; Howard, B.H.; Finkel, T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J. Biol. Chem. 1999, 274, 7936–7940. [Google Scholar] [CrossRef]
- Moiseeva, O.; Bourdeau, V.; Roux, A.; Deschênes-Simard, X.; Ferbeyre, G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell. Biol. 2009, 29, 4495–4507. [Google Scholar] [CrossRef] [PubMed]
- Gwangwa, M.V.; Joubert, A.M.; Visagie, M.H. Effects of glutamine deprivation on oxidative stress and cell survival in breast cell lines. Biol. Res. 2019, 52, 15. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hwang, S.; Kim, M.; Seo, S.B.; Lee, J.-H.; Jeong, S.M. Mitochondrial glutamine metabolism via GOT2 supports pancreatic cancer growth through senescence inhibition. Cell Death Dis. 2018, 9, 55. [Google Scholar] [CrossRef]
- Pacifico, F.; Badolati, N.; Mellone, S.; Stornaiuolo, M.; Leonardi, A.; Crescenzi, E. Glutamine promotes escape from therapy-induced senescence in tumor cells. Aging 2021, 13, 20962–20991. [Google Scholar] [CrossRef]
- Roesch, A.; Paschen, A.; Landsberg, J.; Helfrich, I.; Becker, J.C.; Schadendorf, D. Phenotypic tumour cell plasticity as a resistance mechanism and therapeutic target in melanoma. Eur. J. Cancer 2016, 59, 109–112. [Google Scholar] [CrossRef]
- Roesch, A.; Fukunaga-Kalabis, M.; Schmidt, E.C.; Zabierowski, S.E.; Brafford, P.A.; Vultur, A.; Basu, D.; Gimotty, P.; Vogt, T.; Herlyn, M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010, 141, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Dembinski, J.L.; Krauss, S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin. Exp. Metastasis 2009, 26, 611–623. [Google Scholar] [CrossRef]
- Kusumbe, A.P.; Bapat, S.A. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009, 69, 9245–9253. [Google Scholar] [CrossRef]
- Moore, N.; Houghton, J.; Lyle, S. Slow-cycling therapy-resistant cancer cells. Stem Cells Dev. 2012, 21, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A.; Vultur, A.; Bogeski, I.; Wang, H.; Zimmermann, K.M.; Speicher, D.; Körbel, C.; Laschke, M.W.; Gimotty, P.A.; Philipp, S.E.; et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 2013, 23, 811–825. [Google Scholar] [CrossRef] [PubMed]

| Cell Line | Origin | Mutation |
|---|---|---|
| NHEM | Healthy tissue | - |
| Sbcl2 | Primary tumor (radial growth phase) | N-RASQ61K |
| WM3211 | Primary tumor (vertical growth phase) | p53T724G |
| WM1366 | Primary tumor (vertical growth phase) | N-RASQ61L |
| Mel Juso | Primary tumor | N-RASQ61L |
| WM1158 | Metastasis | BRAFV600E, PTENDel/V343E |
| Mel Im | Metastasis | BRAFV600E |
| SKMel28 | Metastasis | BRAFV600E, PTENT167A, CDK4R24C, p53L145R |
| HEK293 | Embryonic kidney | - |
| Primer | Forward Primer 5′-3′ | Reverse Primer 5′-3′ | Annealing Temperature | Measurement Temperature |
|---|---|---|---|---|
| SLC38A1/ SNAT1 | GCTTTGGTTAAAGAGCGGGC | CTGAGGGTCA-CGAATCGGAG | 60 °C | 78 °C |
| SLC38A2/ SNAT2 | CTGAAGACGTCTGCGTGAGA | CCAAGGATTCCACTGCCCAC | 60 °C | 86 °C |
| SLC1A5/ ASCT2 | CTGGCTGGTAACCGCTACTC | TGTCCGAAAGCTGGGAGTTC | 60 °C | 86 °C |
| SLC7A5/ LAT1 | GGCCGAGGAGAAGGAAGAGG | CCTCCAGCATGTAGGCGTAG | 60 °C | 86 °C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Böhme-Schäfer, I.; Lörentz, S.; Bosserhoff, A.K. Role of Amino Acid Transporter SNAT1/SLC38A1 in Human Melanoma. Cancers 2022, 14, 2151. https://doi.org/10.3390/cancers14092151
Böhme-Schäfer I, Lörentz S, Bosserhoff AK. Role of Amino Acid Transporter SNAT1/SLC38A1 in Human Melanoma. Cancers. 2022; 14(9):2151. https://doi.org/10.3390/cancers14092151
Chicago/Turabian StyleBöhme-Schäfer, Ines, Sandra Lörentz, and Anja Katrin Bosserhoff. 2022. "Role of Amino Acid Transporter SNAT1/SLC38A1 in Human Melanoma" Cancers 14, no. 9: 2151. https://doi.org/10.3390/cancers14092151
APA StyleBöhme-Schäfer, I., Lörentz, S., & Bosserhoff, A. K. (2022). Role of Amino Acid Transporter SNAT1/SLC38A1 in Human Melanoma. Cancers, 14(9), 2151. https://doi.org/10.3390/cancers14092151







