Automatic Cancer Cell Taxonomy Using an Ensemble of Deep Neural Networks
Abstract
Simple Summary
Abstract
1. Introduction
- We proposed a deep-learning-based approach to prevent cross-contamination of several heterogeneous cancer cell lines.
- The experimental results showed that the proposed deep-learning-based approach identifies with an accuracy over 97%, demonstrating that our method can be a promising alternative approach to STR for the automated cancer cell taxonomy.
- We presented and discussed the effects of various design choices on the overall performance of CNN architectures for various clinical tasks that utilize microscopic images.
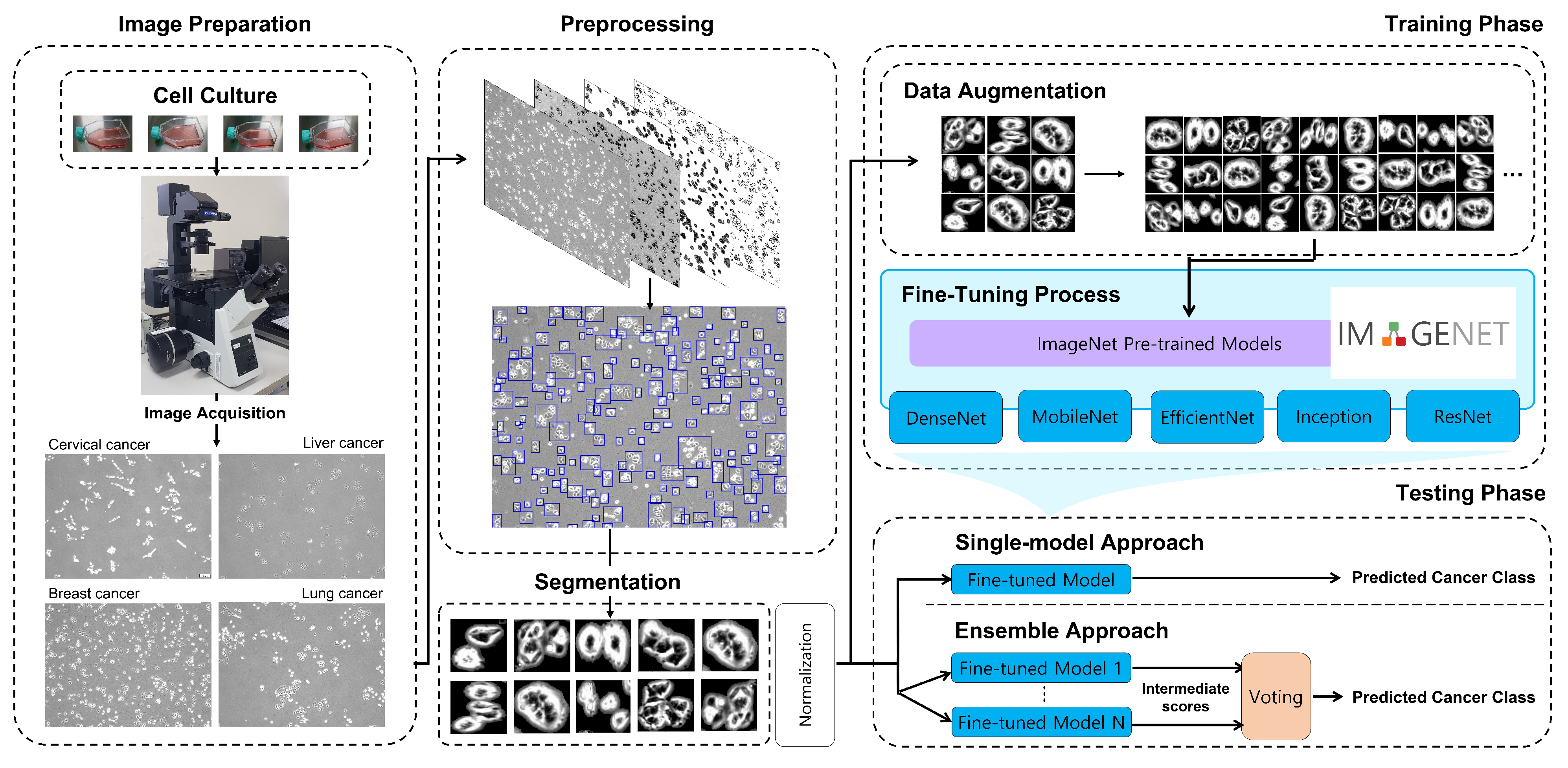
2. Method
2.1. Image Preparation
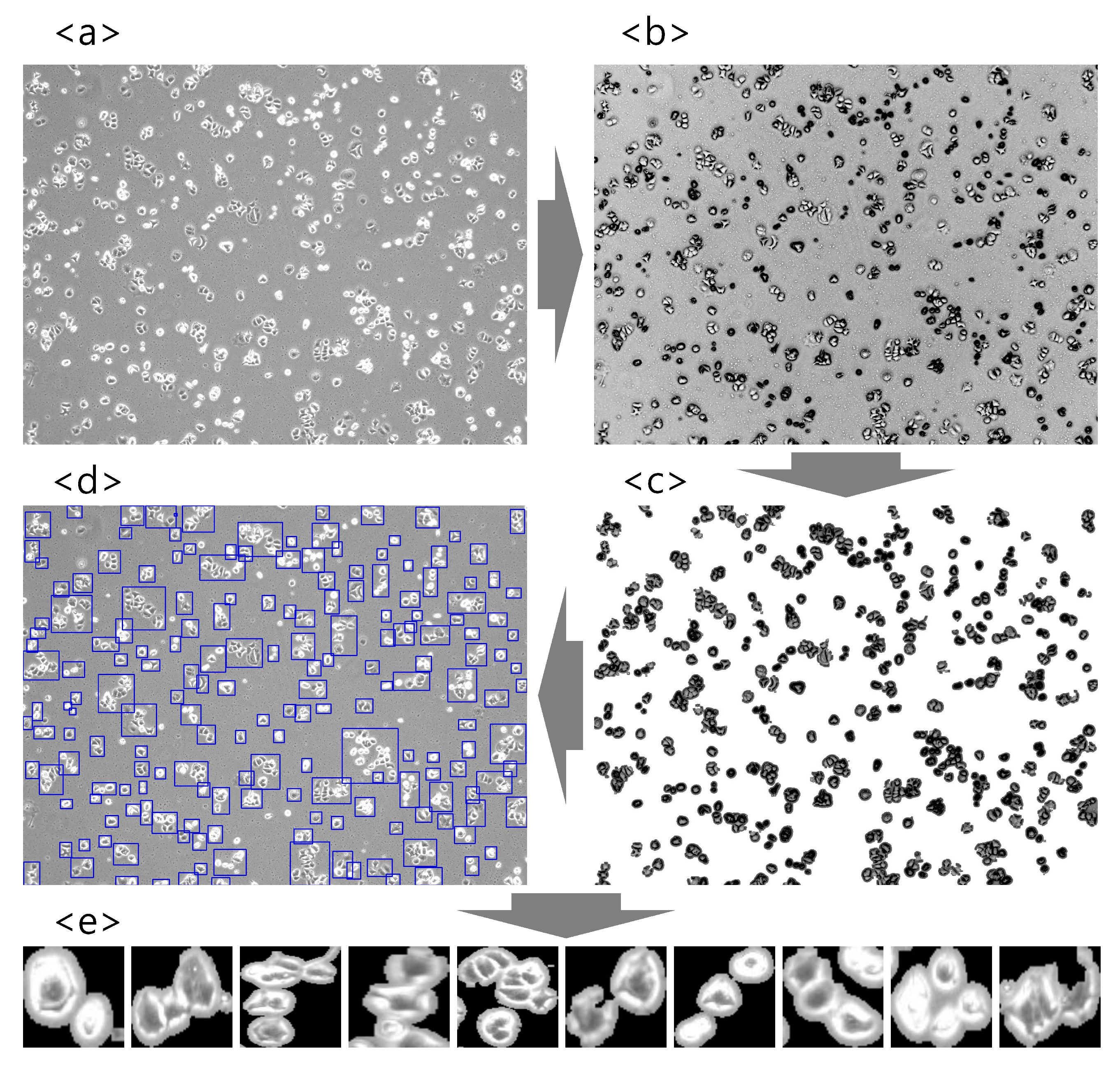
2.2. Image Preprocessing
2.3. Training CNNs for Cancer Classification
2.3.1. Data Augmentation
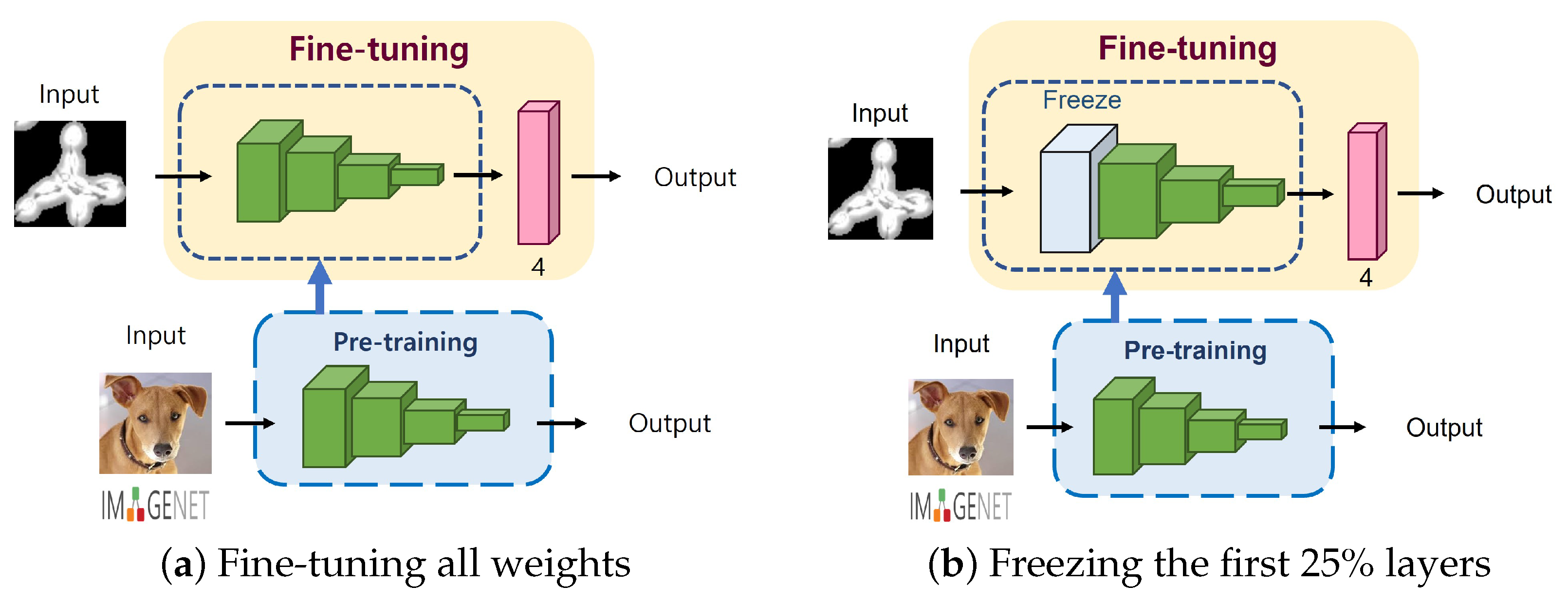
2.3.2. Degree of Fine-Tuning
2.3.3. Optimizer and Learning Rate Scheduler
2.4. Ensemble of CNNs
- -
- Single-architecture ensemble (single-arch, hereafter): As shown in Table 1, there are 16 available configurations for each CNN architecture. In this approach, we select the top-4, top-8, and top-16 best-performing configurations in terms of classification accuracy. Therefore, we can build three ensembles for each model, for a total of 15 single-arch ensemble prediction pipelines.
- -
- Multi-architecture ensemble (multi-arch, hereafter): In contrast to the single-arch pipeline, the multi-arch approach is composed of heterogeneous CNN architectures. To establish this pipeline, we select the top-1, top-2, and top-3 best-performing configurations from each model. Therefore, top-1, top-2, and top-3 multi-arch ensemble pipelines include 5, 10, and 15 individual classification models from different architectures, respectively.
3. Experimental Results
3.1. Experimental setup
3.2. Performance evaluation
4. Discussion
4.1. Performance of Deep-Learning-Based Approaches
4.2. Network Design Choice
4.3. Comparison with Previous Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Position Paper on Mammography Screening; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [CrossRef] [PubMed]
- Kainz, P.; Pfeiffer, M.; Urschler, M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ 2017, 2017, e3874. [Google Scholar] [CrossRef]
- Aubreville, M.; Knipfer, C.; Oetter, N.; Jaremenko, C.; Rodner, E.; Denzler, J.; Bohr, C.; Neumann, H.; Stelzle, F.; Maier, A. Automatic Classification of Cancerous Tissue in Laserendomicroscopy Images of the Oral Cavity using Deep Learning. Sci. Rep. 2017, 7, 11979. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated Classification of Lung Cancer Types from Cytological Images Using Deep Convolutional Neural Networks. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef]
- Khan, S.U.; Islam, N.; Jan, Z.; Ud Din, I.; Rodrigues, J.J. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Chekkoury, A.; Khurd, P.; Ni, J.; Bahlmann, C.; Kamen, A.; Patel, A.; Grady, L.; Singh, M.; Groher, M.; Navab, N.; et al. Automated Malignancy Detection in Breast Histopathological Images; Medical Imaging 2012: Computer-Aided Diagnosis; van Ginneken, B., Novak, C.L., Eds.; SPIE: San Diego, CA, USA, 2012; Volume 8315, p. 831515. [Google Scholar] [CrossRef]
- Pollanen, I.; Braithwaite, B.; Ikonen, T.; Niska, H.; Haataja, K.; Toivanen, P.; Tolonen, T. Computer-aided breast cancer histopathological diagnosis: Comparative analysis of three DTOCS-based features: SW-DTOCS, SW-WDTOCS and SW-3-4-DTOCS. In Proceedings of the 2014 4th International Conference on Image Processing Theory, Tools and Applications, IPTA, Paris, France, 14–17 October 2014. [Google Scholar] [CrossRef]
- Toratani, M.; Konno, M.; Asai, A.; Koseki, J.; Kawamoto, K.; Tamari, K.; Li, Z.; Sakai, D.; Kudo, T.; Satoh, T.; et al. A convolutional neural network uses microscopic images to differentiate between mouse and human cell lines and their radioresistant clones. Cancer Res. 2018, 78, 6703–6707. [Google Scholar] [CrossRef]
- Xing, F.; Yang, L. Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: A comprehensive review. IEEE Rev. Biomed. Eng. 2016, 9, 234–263. [Google Scholar] [CrossRef]
- Xing, F.; Xie, Y.; Su, H.; Liu, F.; Yang, L. Deep Learning in Microscopy Image Analysis: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 4550–4568. [Google Scholar] [CrossRef]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for nuclei detection, segmentation, and classification in digital histopathology: A review-current status and future potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; He, S.; Lee, Y.J.; He, Y.; Kong, E.M.; Li, H.; Anastasio, M.A.; Popescu, G.; Anastasio, M. Live-dead assay on unlabeled cells using phase imaging with computational specificity. Nat. Commun. 2022, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Noble, J.A.; Zisserman, A. Microscopy cell counting and detection with fully convolutional regression networks. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 283–292. [Google Scholar] [CrossRef]
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36, 829–838. [Google Scholar] [CrossRef]
- O’Connor, T.; Hawxhurst, C.; Shor, L.M.; Javidi, B. Red blood cell classification in lensless single random phase encoding using convolutional neural networks. Opt. Express 2020, 28, 33504. [Google Scholar] [CrossRef]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.J.; Thürlimann, B.; Senn, H.J.; André, F.; Baselga, J.; et al. Tailoring therapies-improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef]
- Solnica-Krezel, L. Conserved patterns of cell movements during vertebrate gastrulation. Curr. Biol. 2005, 15, R213–R228. [Google Scholar] [CrossRef]
- Gartler, S.M. Apparent HeLa cell contamination of human heteroploid cell lines. Nature 1968, 217, 750–751. [Google Scholar] [CrossRef]
- Lande, R. Natural Selection and Random Genetic Drift in Phenotypic Evolution. Evolution 1976, 30, 314. [Google Scholar] [CrossRef]
- Capes-Davis, A.; Theodosopoulos, G.; Atkin, I.; Drexler, H.G.; Kohara, A.; MacLeod, R.A.; Masters, J.R.; Nakamura, Y.; Reid, Y.A.; Reddel, R.R.; et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer 2010, 127, 1–8. [Google Scholar] [CrossRef]
- Neimark, J. Line of attack. Science 2015, 347, 938–940. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, D.; Kang, D.; Guo, X.; Guo, C.; Dongye, M.; Zhu, Y.; Chen, C.; Zhang, X.; Long, E.; et al. An artificial intelligent platform for live cell identification and the detection of cross-contamination. Ann. Transl. Med. 2020, 8, 697. [Google Scholar] [CrossRef] [PubMed]
- Lorsch, B.J.R.; Collins, F.S.; Lippincott-schwartz, J. Fixing problems with cell lines. Science 2014, 346, 1452–1453. [Google Scholar] [CrossRef] [PubMed]
- Masters, J.R. Cell-line authentication: End the scandal of false cell lines. Nature 2012, 492, 186. [Google Scholar] [CrossRef]
- Bian, X.; Yang, Z.; Feng, H.; Sun, H.; Liu, Y. A Combination of Species Identification and STR Profiling Identifies Cross-contaminated Cells from 482 Human Tumor Cell Lines. Sci. Rep. 2017, 7, 9774. [Google Scholar] [CrossRef]
- Almeida, J.L.; Cole, K.D.; Plant, A.L. Standards for Cell Line Authentication and Beyond. PLoS Biol. 2016, 14, e1002476. [Google Scholar] [CrossRef]
- Masters, J.R.; Thomson, J.A.; Daly-Burns, B.; Reid, Y.A.; Dirks, W.G.; Packer, P.; Toji, L.H.; Ohno, T.; Tanabe, H.; Arlett, C.F.; et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 8012–8017. [Google Scholar] [CrossRef]
- Poetsch, M.; Petersmann, A.; Woenckhaus, C.; Protzel, C.; Dittberner, T.; Lignitz, E.; Kleist, B. Evaluation of allelic alterations in short tandem repeats in different kinds of solid tumors - Possible pitfalls in forensic casework. Forensic Sci. Int. 2004, 145, 1–6. [Google Scholar] [CrossRef]
- Lohar, P.S. Textbook of Biotechnology; MJP Publisher: Chennai, India, 2019. [Google Scholar]
- Rubin, M.; Stein, O.; Turko, N.A.; Nygate, Y.; Roitshtain, D.; Karako, L.; Barnea, I.; Giryes, R.; Shaked, N.T. TOP-GAN: Stain-free cancer cell classification using deep learning with a small training set. Med. Image Anal. 2019, 57, 176–185. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Cai, H.; Tan, W.; Jin, C.; Li, L. Discrimination of Breast Cancer with Microcalcifications on Mammography by Deep Learning. Sci. Rep. 2016, 6, 27327. [Google Scholar] [CrossRef] [PubMed]
- Ayana, G.; Dese, K.; Choe, S.W. Transfer Learning in Breast Cancer Diagnoses via Ultrasound Imaging. Cancers 2021, 13, 738. [Google Scholar] [CrossRef] [PubMed]
- Meng, N.; Lam, E.Y.; Tsia, K.K.; So, H.K.H. Large-Scale Multi-Class Image-Based Cell Classification with Deep Learning. IEEE J. Biomed. Health Inform. 2019, 23, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Oei, R.W.; Hou, G.; Liu, F.; Zhong, J.; Zhang, J.; An, Z.; Xu, L.; Yang, Y. Convolutional neural network for cell classification using microscope images of intracellular actin networks. PLoS ONE 2019, 14, e0213626. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.W.; Terman, D.S.; Rivers, A.E.; Rivera, J.; Lottenberg, R.; Sorg, B.S. Drug-loaded sickle cells programmed ex vivo for delayed hemolysis target hypoxic tumor microvessels and augment tumor drug delivery. J. Control. Release 2013, 171, 184–192. [Google Scholar] [CrossRef]
- Cho, K.; Seo, J.H.; Heo, G.; Choe, S.W. An Alternative Approach to Detecting Cancer Cells by Multi-Directional Fluorescence Detection System Using Cost-Effective LED and Photodiode. Sensors 2019, 19, 2301. [Google Scholar] [CrossRef]
- Nelissen, B.G.L.; van Herwaarden, J.A.; Moll, F.L.; van Diest, P.J.; Pasterkamp, G. SlideToolkit: An Assistive Toolset for the Histological Quantification of Whole Slide Images. PLoS ONE 2014, 9, e110289. [Google Scholar] [CrossRef]
- Choe, S.W.; Choi, H. Suppression technique of hela cell proliferation using ultrasonic power amplifiers integrated with a series-diode linearizer. Sensors 2018, 18, 4248. [Google Scholar] [CrossRef]
- Choi, H.; Ryu, J.-M.; Choe, S.-W. A novel therapeutic instrument using an ultrasound-light-emitting diode with an adjustable telephoto lens for suppression of tumor cell proliferation. Measurement 2019, 147, 106865. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October, 2018; Springer: Rhodes, Greece, 2018; Volume 11141, pp. 270–279. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 July 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2016, arXiv:1609.04747. [Google Scholar]
- Duchi, J.; Hazan, E.; Singer, Y. Adaptive Subgradient Methods for Online Learning and Stochastic Optimization. J. Mach. Learn. Res. 2011, 12, 2121–2159. [Google Scholar]
- Cao, Y.; Geddes, T.A.; Yang, J.Y.H.; Yang, P. Ensemble deep learning in bioinformatics. Nat. Mach. Intell. 2020, 2, 500–508. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Bartlett, P.; Freund, Y.; Lee, W.S.; Schapire, R.E. Boosting the margin: A new explanation for the effectiveness of voting methods. Ann. Stat. 1998, 26, 1651–1686. [Google Scholar] [CrossRef]
- David, H.W. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Sophea, P.; Handayani, D.O.D.; Boursier, P. Abnormal cervical cell detection using hog descriptor and SVM classifier. In Proceedings of the 2018 Fourth International Conference on Advances in Computing, Communication & Automation (ICACCA), Subang Jaya, Malaysia, 26–28 October 2018; pp. 1–6. [Google Scholar]
- Kumar, R.; Srivastava, R.; Srivastava, S. Detection and classification of cancer from microscopic biopsy images using clinically significant and biologically interpretable features. J. Med. Eng. 2015, 2015, 457906. [Google Scholar] [CrossRef] [PubMed]
- Follen, M.; Richards-Kortum, R. Emerging Technologies and Cervical Cancer. JNCI J. Natl. Cancer Inst. 2000, 92, 363–365. [Google Scholar] [CrossRef][Green Version]
- Shi, J.; Wang, R.; Zheng, Y.; Jiang, Z.; Zhang, H.; Yu, L. Cervical cell classification with graph convolutional network. Comput. Methods Programs Biomed. 2021, 198, 105807. [Google Scholar] [CrossRef]
- Chankong, T.; Theera-Umpon, N.; Auephanwiriyakul, S. Automatic cervical cell segmentation and classification in Pap smears. Comput. Methods Programs Biomed. 2014, 113, 539–556. [Google Scholar] [CrossRef]
- Sharma, M.; Singh, S.K.; Agrawal, P.; Madaan, V. Classification of clinical dataset of cervical cancer using KNN. Indian J. Sci. Technol. 2016, 9, 1–5. [Google Scholar] [CrossRef]
- Gençtav, A.; Aksoy, S.; Önder, S. Unsupervised segmentation and classification of cervical cell images. Pattern Recognit. 2012, 45, 4151–4168. [Google Scholar] [CrossRef]
- Marinakis, Y.; Dounias, G.; Jantzen, J. Pap smear diagnosis using a hybrid intelligent scheme focusing on genetic algorithm based feature selection and nearest neighbor classification. Comput. Biol. Med. 2009, 39, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Linh, T.D.; Nhi, H.L.; Toan, B.T.; Vuong, M.N.; Phuong, T.N. Detection of tuberculosis from chest X-ray images: Boosting the performance with vision transformer and transfer learning. Expert Syst. Appl. 2021, 184, 115519. [Google Scholar] [CrossRef]
- Beal, J.; Kim, E.; Tzeng, E.; Park, D.H.; Zhai, A.; Kislyuk, D. Toward transformer-based object detection. arXiv 2020, arXiv:2012.09958. [Google Scholar]
- Ayush, K.; Uzkent, B.; Meng, C.; Tanmay, K.; Burke, M.; Lobell, D.; Ermon, S. Geography-Aware Self-Supervised Learning. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; pp. 10181–10190. [Google Scholar]
- Yan, K.; Cai, J.; Jin, D.; Miao, S.; Harrison, A.P.; Guo, D.; Tang, Y.; Xiao, J.; Lu, J.; Lu, L. Self-supervised learning of pixel-wise anatomical embeddings in radiological images. arXiv 2020, arXiv:2012.02383. [Google Scholar] [CrossRef]
- Lin, L.; Song, S.; Yang, W.; Liu, J. Ms2l: Multi-task self-supervised learning for skeleton based action recognition. In Proceedings of the 28th ACM International Conference on Multimedia, Seattle, WA, USA, 12–16 October 2020; pp. 2490–2498. [Google Scholar]
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the Knowledge in a Neural Network. arXiv 2015, arXiv:1503.02531. [Google Scholar]
- Lan, X.; Zhu, X.; Gong, S. Knowledge Distillation by On-the-Fly Native Ensemble. In Advances in Neural Information Processing Systems; Bengio, S., Wallach, H., Larochelle, H., Grauman, K., Cesa-Bianchi, N., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2018; Volume 31. [Google Scholar]

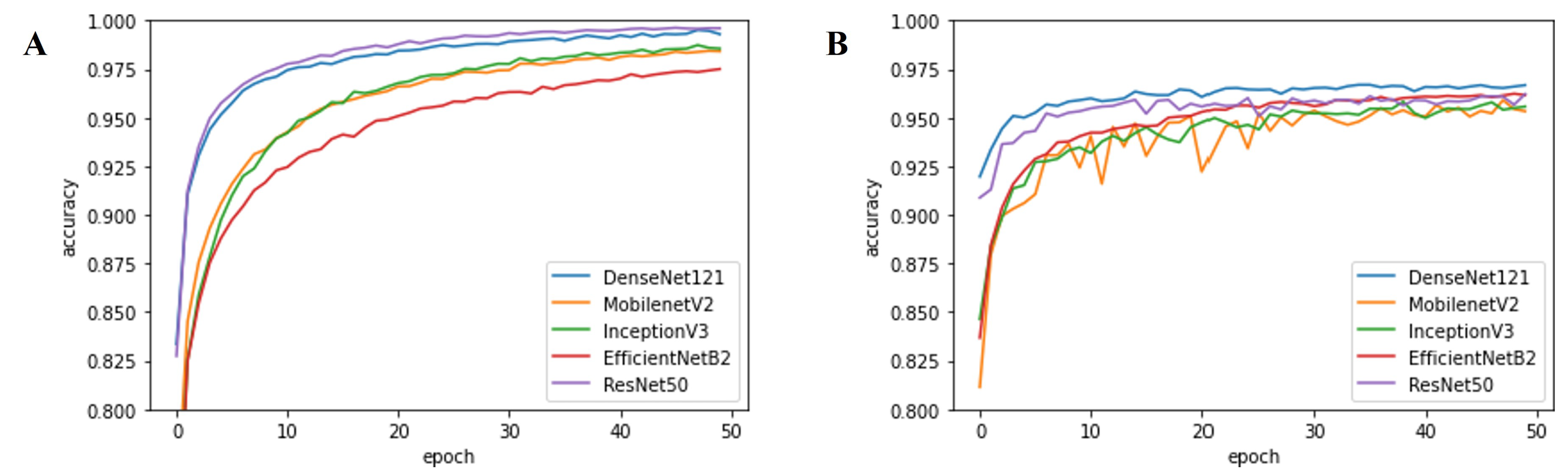
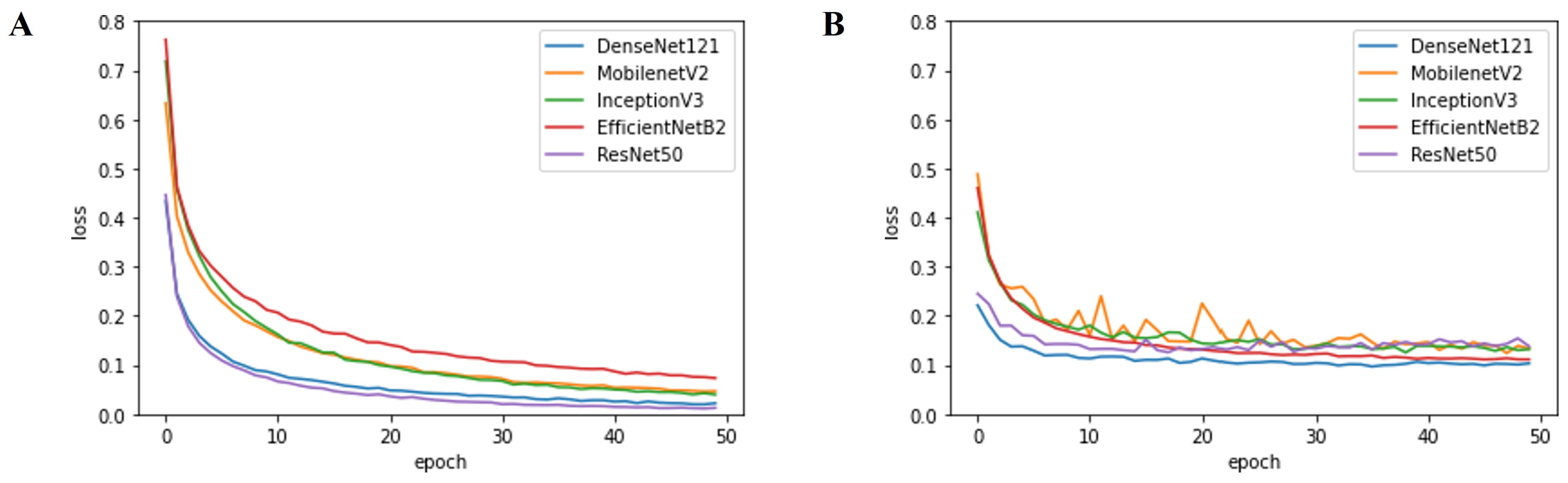
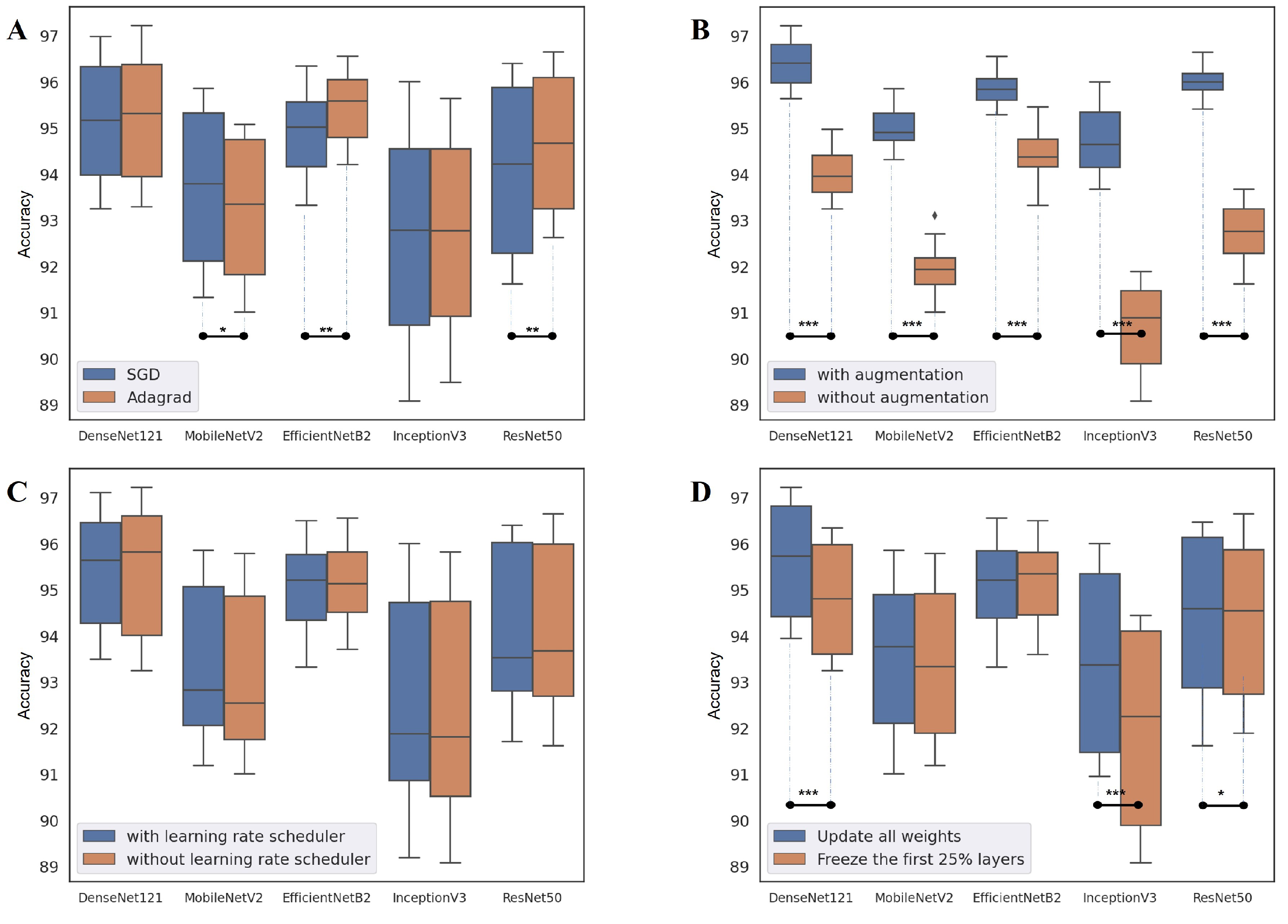
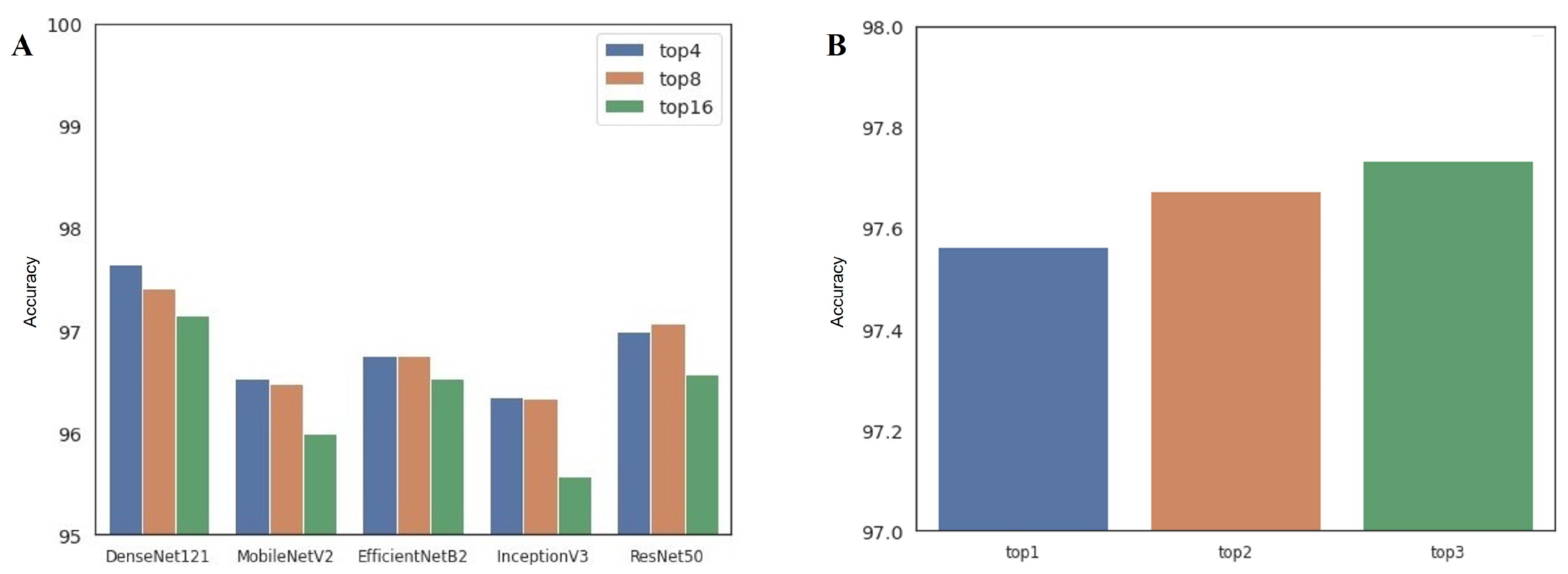
| Parameter | Option | Note |
|---|---|---|
| Data augmentation | O | Rotation, translation, and vertical flip |
| X | Without any augmentation | |
| Fine-tuning | Without freeze | All weights are updated |
| 25% freeze | Only 75% of weights are updated | |
| Optimizer | SGD | Stochastic gradient descent |
| AdaGrad | Adaptive gradient-based optimization | |
| Learning rate scheduler | O | Exponential decay |
| X | Learning rate is fixed to 0.001 |
| Algorithm | Model | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| Machine Learning | SVM | 58.7 ± 0.74 | 58.34 ± 0.76 | 58.7 ± 0.74 | 58.52 ± 0.75 |
| RF | 49.55 ± 0.32 | 49.01 ± 0.33 | 49.55 ± 0.32 | 49.3 ± 0.32 | |
| LDA | 46.26 ± 0.98 | 44.81 ± 0.98 | 45.26 ± 0.98 | 45.03 ± 0.98 | |
| KNN | 44.05 ± 0.92 | 45.86 ± 1.1 | 44.05 ± 0.92 | 44.93 ± 0.94 | |
| Average | 49.39 ± 5.94 | 49.51 ± 5.52 | 49.39 ± 5.94 | 49.44 ± 5.72 | |
| Deep Learning | DenseNet121 | 96.915 ± 0.072 | 96.916 ± 0.077 | 96.915 ± 0.072 | 96.915 ± 0.075 |
| EfficientNetB2 | 96.195 ± 0.23 | 96.23 ± 0.272 | 96.176 ± 0.194 | 96.203 ± 0.232 | |
| ResNet50 | 96.265 ± 0.138 | 96.274 ± 0.13 | 96.265 ± 0.138 | 96.269 ± 0.134 | |
| InceptionV3 | 95.57 ± 0.322 | 95.604 ± 0.376 | 95.556 ± 0.298 | 95.58 ± 0.336 | |
| MobileNetV2 | 95.412 ± 0.223 | 95.446 ± 0.229 | 95.412 ± 0.224 | 95.429 ± 0.226 | |
| Average | 96.071 ± 0.584 | 96.1 ± 0.58 | 96.06 ± 0.581 | 96.08 ± 0.58 | |
| Ensemble (Single-architecture) | DenseNet121 | 97.64 ± 0.16 | 97.643 ± 0.16 | 97.64 ± 0.16 | 97.641 ± 0.16 |
| EfficientNetB2 | 96.757 ± 0.202 | 96.763 ± 0.294 | 96.757 ± 0.294 | 96.76 ± 0.294 | |
| ResNet50 | 97.066 ± 0.148 | 97.073 ± 0.145 | 97.066 ± 0.148 | 96.07 ± 0.147 | |
| InceptionV3 | 96.342 ± 0.196 | 96.345 ± 0.202 | 96.342 ± 0.196 | 96.343 ± 0.199 | |
| MobileNetV2 | 96.533 ± 0.209 | 96.55 ± 0.226 | 96.533 ± 0.209 | 96.541 ± 0.217 | |
| Average | 96.868 ± 0.5 | 96.875 ± 0.5 | 96.868 ± 0.5 | 96.871 ± 0.5 | |
| Ensemble (Multi-architecture) | Top-1 | 97.563 ± 0.145 | 97.568 ± 0.145 | 97.563 ± 0.145 | 97.565 ± 0.145 |
| Top-2 | 97.673 ± 0.122 | 97.677 ± 0.124 | 97.673 ± 0.122 | 97.675 ± 0.123 | |
| Top-3 | 97.735 ± 0.132 | 97.74 ± 0.14 | 97.74 ± 0.132 | 97.74 ± 0.134 | |
| Average | 97.657 ± 0.144 | 97.661 ± 0.149 | 97.657 ± 0.144 | 97.659 ± 0.144 |
| Algorithm | Data Augmentation | Degree of Fine-Tuning | Optimizer | Learning Rate Scheduler |
|---|---|---|---|---|
| DenseNet121 | O | All weights | SGD | X |
| EfficientNetB2 | O | All weights | AdaGrad | X |
| ResNet50 | O | All weights | AdaGrad | X |
| InceptionV3 | O | All weights | SGD | O |
| MobileNetV2 | O | Freeze the early 25% layers | SGD | O |
| Degree of Fine-Tuning | All Weights | Freeze the First 25% Layers | |
|---|---|---|---|
| Model | |||
| DenseNet121 | 6,957,956 | 6,716,740 | |
| MobileNetV2 | 2,228,996 | 2,197,060 | |
| EfficientNetB2 | 7,706,630 | 7,700,858 | |
| InceptionV3 | 21,776,548 | 21,348,836 | |
| ResNet50 | 23,542,788 | 23,315,972 | |
| Ref. | Task | Image Acquisition | Method | Num. of Classes | Metric | Performance | Feature |
|---|---|---|---|---|---|---|---|
| Rubin et al. [33] | Cancer cell classification | Low-coherence off-axis holography without statining | GAN-based approach | 4 classes (healthy skin, melanoma cells, colorectal adenocarcinoma colon cells, metastatic colorectal adenocarcinoma cells) | Accuracy | 90–99% | CNN feature |
| Oei et al. [38] | Breast cancer cell detection | Confocal immunofluorescence microscopy images with staining | CNN | 2 classes (breast normal cells and cancer cells) | Accuracy | 97.2% | CNN feature |
| Kumar et al. [59] | Cervical cancer cell detection | Microscopic biopsy images with staining | RF, SVM, KNN, fuzzy KNN | 2 classese (noncancerous, cancerous) | Accuracy | 92.19% | Texture features, morphology and shape features, HOG, wavelet features, etc. |
| Shi et al. [61] | Cervical cancer cell classification | Microscopic images of Pap smear slides with staining | Graph neural network | 5 types of cervical cancer cells (superficial–intermediate, parabasal, koilocytotic, dyskeratotic, and metaplastic cells) | Accuracy | 94.93% | CNN feature |
| Sophea et al. [58] | HOG + SVM | 2 classes (normal and abnormal) | Accuracy | 94.7% | HOG | ||
| Chankong et al. [62] | Bayes, LDA, KNN, ANN, SVM | 7 classes (superficial squamous, intermediate squamous, columnar, mild dysplasia, moderate dysplasia, severe dysplasia, and carcinoma in situ) | Accuracy | 93.78% | Hand-crafted features (area of cucleus, nucleus-to-cytoplasm ratio, etc.) | ||
| Sharma et al. [63] | KNN | Accuracy | 82.9% | ||||
| Gençtav et al. [64] | Bayesian, decision tree, SVM | Precision | 91.7% | ||||
| Marinakis et al. [65] | GA | Accuracy | 96.73% | ||||
| Our proposed method | Cancer cell classification | Microscopic images of cell culture flask without staining | CNN ensemble | 4 classes of cell culture flask (HeLa, MCF-7, Huh7, and NCI-H1299) | Accuracy | 97.735% | CNN feature |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choe, S.-w.; Yoon, H.-Y.; Jeong, J.-Y.; Park, J.; Jeong, J.-W. Automatic Cancer Cell Taxonomy Using an Ensemble of Deep Neural Networks. Cancers 2022, 14, 2224. https://doi.org/10.3390/cancers14092224
Choe S-w, Yoon H-Y, Jeong J-Y, Park J, Jeong J-W. Automatic Cancer Cell Taxonomy Using an Ensemble of Deep Neural Networks. Cancers. 2022; 14(9):2224. https://doi.org/10.3390/cancers14092224
Chicago/Turabian StyleChoe, Se-woon, Ha-Yeong Yoon, Jae-Yeop Jeong, Jinhyung Park, and Jin-Woo Jeong. 2022. "Automatic Cancer Cell Taxonomy Using an Ensemble of Deep Neural Networks" Cancers 14, no. 9: 2224. https://doi.org/10.3390/cancers14092224
APA StyleChoe, S.-w., Yoon, H.-Y., Jeong, J.-Y., Park, J., & Jeong, J.-W. (2022). Automatic Cancer Cell Taxonomy Using an Ensemble of Deep Neural Networks. Cancers, 14(9), 2224. https://doi.org/10.3390/cancers14092224







