Dynamics of the Tumor Immune Microenvironment during Neoadjuvant Chemotherapy of High-Grade Serous Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
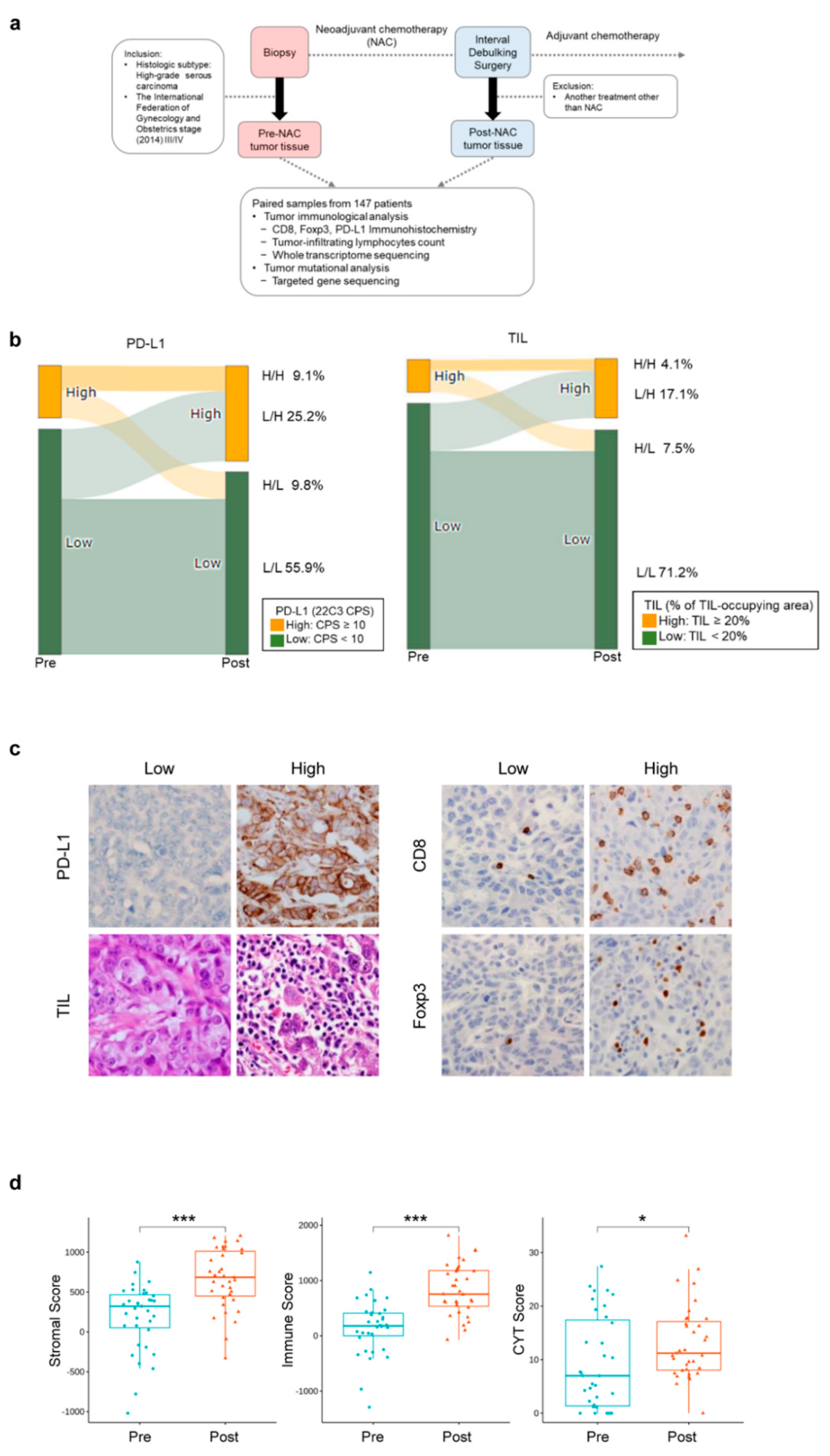
2.1. Study Design and Patient Samples
2.2. Treatment
2.3. Pathological Evaluation
2.4. Isolation of Genomic DNA and RNA
2.5. Whole Transcriptome Sequencing
2.6. Whole Transcriptome Data Analysis
2.7. Targeted-Gene Sequencing
2.8. Statistical Analysis
3. Results
3.1. Study Population
3.2. Changes in the Tumor Immune Microenvironment of HGSOCs before and after NAC
3.3. Genetic Alterations in HGSOCs
3.4. Patterns of Pre-NAC Immune Microenvironment and Their Association with Outcome
3.5. Profound Impact of NAC on the Immune Microenvironment
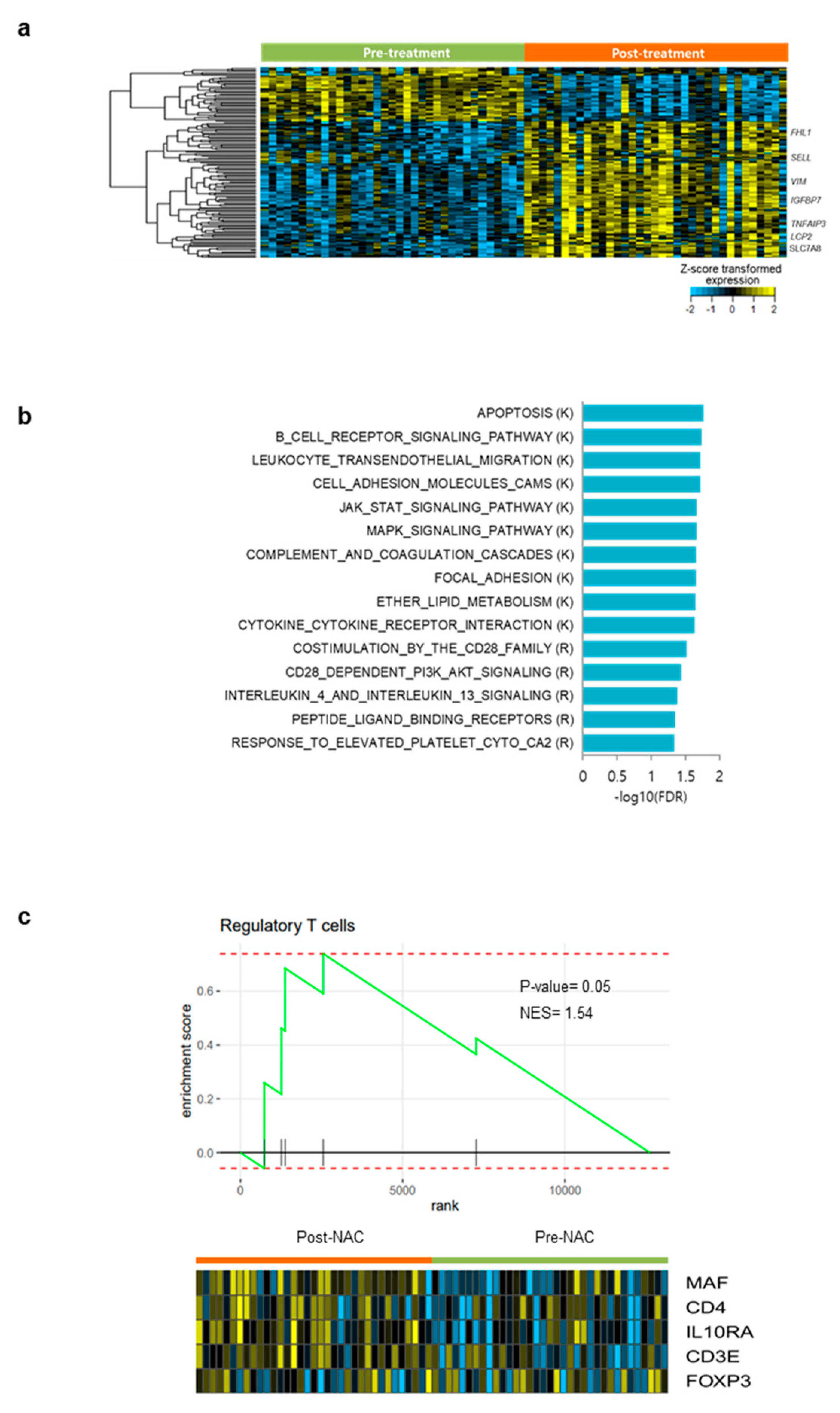
3.6. Differentially Expressed Gene Analysis and Gene Set Enrichment Analysis in Pre- and Post- NAC Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, J.Y.; Kim, S.; Kim, Y.T.; Lim, M.C.; Lee, B.; Jung, K.W.; Kim, J.W.; Park, S.Y.; Won, Y.J. Changes in ovarian cancer survival during the 20 years before the era of targeted therapy. BMC Cancer 2018, 18, 601. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.I.; Chang, H.K.; Park, S.J.; Lim, J.; Won, Y.J.; Lim, M.C. The incidence and survival of cervical, ovarian, and endometrial cancer in korea, 1999-2017: Korea central cancer registry. Obstet. Gynecol. Sci. 2021, 64, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, S.; Hook, J.; Nankivell, M.; Jayson, G.C.; Kitchener, H.; Lopes, T.; Luesley, D.; Perren, T.; Bannoo, S.; Mascarenhas, M.; et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (chorus): An open-label, randomised, controlled, non-inferiority trial. Lancet 2015, 386, 249–257. [Google Scholar] [CrossRef]
- Vergote, I.; Trope, C.G.; Amant, F.; Kristensen, G.B.; Ehlen, T.; Johnson, N.; Verheijen, R.H.; van der Burg, M.E.; Lacave, A.J.; Panici, P.B.; et al. Neoadjuvant chemotherapy or primary surgery in stage iiic or iv ovarian cancer. N. Engl. J. Med. 2010, 363, 943–953. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Lee, J.Y.; Nam, E.J.; Kim, S.W.; Kim, S.; Kim, Y.T. Rethinking radical surgery in interval debulking surgery for advanced-stage ovarian cancer patients undergoing neoadjuvant chemotherapy. J. Clin. Med. 2020, 9, 1235. [Google Scholar] [CrossRef]
- Monk, B.J.; Colombo, N.; Oza, A.M.; Fujiwara, K.; Birrer, M.J.; Randall, L.; Poddubskaya, E.V.; Scambia, G.; Shparyk, Y.V.; Lim, M.C.; et al. Chemotherapy with or without avelumab followed by avelumab maintenance versus chemotherapy alone in patients with previously untreated epithelial ovarian cancer (javelin ovarian 100): An open-label, randomised, phase 3 trial. Lancet Oncol. 2021, 22, 1275–1289. [Google Scholar] [CrossRef]
- Moore, K.N.; Bookman, M.; Sehouli, J.; Miller, A.; Anderson, C.; Scambia, G.; Myers, T.; Taskiran, C.; Robison, K.; Mäenpää, J.; et al. Atezolizumab, bevacizumab, and chemotherapy for newly diagnosed stage iii or iv ovarian cancer: Placebo-controlled randomized phase iii trial (imagyn050/gog 3015/engot-ov39). J. Clin. Oncol. 2021, 39, 1842–1855. [Google Scholar] [CrossRef]
- Opzoomer, J.W.; Sosnowska, D.; Anstee, J.E.; Spicer, J.F.; Arnold, J.N. Cytotoxic chemotherapy as an immune stimulus: A molecular perspective on turning up the immunological heat on cancer. Front. Immunol. 2019, 10, 1654. [Google Scholar] [CrossRef] [Green Version]
- Bracci, L.; Schiavoni, G.; Sistigu, A.; Belardelli, F. Immune-based mechanisms of cytotoxic chemotherapy: Implications for the design of novel and rationale-based combined treatments against cancer. Cell Death Differ. 2014, 21, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Kumar, A.B.; Finnes, H.; Markovic, S.N.; Park, S.; Dronca, R.S.; Dong, H. Combining immune checkpoint inhibitors with conventional cancer therapy. Front. Immunol. 2018, 9, 1739. [Google Scholar] [CrossRef] [Green Version]
- Mesnage, S.J.L.; Auguste, A.; Genestie, C.; Dunant, A.; Pain, E.; Drusch, F.; Gouy, S.; Morice, P.; Bentivegna, E.; Lhomme, C.; et al. Neoadjuvant chemotherapy (nact) increases immune infiltration and programmed death-ligand 1 (pd-l1) expression in epithelial ovarian cancer (eoc). Ann. Oncol. 2017, 28, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.S.; Sanii, S.; Kroeger, D.R.; Milne, K.; Talhouk, A.; Chiu, D.S.; Rahimi, K.; Shaw, P.A.; Clarke, B.A.; Nelson, B.H. Neoadjuvant chemotherapy of ovarian cancer results in three patterns of tumor-infiltrating lymphocyte response with distinct implications for immunotherapy. Clin. Cancer Res. 2017, 23, 925–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böhm, S.; Montfort, A.; Pearce, O.M.T.; Topping, J.; Chakravarty, P.; Everitt, G.L.A.; Clear, A.; McDermott, J.R.; Ennis, D.; Dowe, T.; et al. Neoadjuvant chemotherapy modulates the immune microenvironment in metastases of tubo-ovarian high-grade serous carcinoma. Clin. Cancer Res. 2016, 22, 3025–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pölcher, M.; Braun, M.; Friedrichs, N.; Rudlowski, C.; Bercht, E.; Fimmers, R.; Sauerwald, A.; Keyver-Paik, M.D.; Kübler, K.; Büttner, R.; et al. Foxp3(+) cell infiltration and granzyme b(+)/foxp3(+) cell ratio are associated with outcome in neoadjuvant chemotherapy-treated ovarian carcinoma. Cancer Immunol. Immunother. 2010, 59, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.H.; Lal, S.; Lee, J.E.; Choi, Y.L.; Wen, J.; Ram, S.; Ding, Y.; Lee, S.H.; Powell, E.; Lee, S.K.; et al. Chemotherapy induces dynamic immune responses in breast cancers that impact treatment outcome. Nat. Commun. 2020, 11, 6175. [Google Scholar] [CrossRef] [PubMed]
- Cree, I.A.; White, V.A.; Indave, B.I.; Lokuhetty, D. Revising the who classification: Female genital tract tumours. Histopathology 2020, 76, 151–156. [Google Scholar] [CrossRef]
- Bohm, S.; Faruqi, A.; Said, I.; Lockley, M.; Brockbank, E.; Jeyarajah, A.; Fitzpatrick, A.; Ennis, D.; Dowe, T.; Santos, J.L.; et al. Chemotherapy response score: Development and validation of a system to quantify histopathologic response to neoadjuvant chemotherapy in tubo-ovarian high-grade serous carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 2457–2463. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (tils) in breast cancer: Recommendations by an international tils working group 2014. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Matulonis, U.A.; Shapira-Frommer, R.; Santin, A.D.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Provencher, D.M.; et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase ii keynote-100 study. Ann. Oncol. 2019, 30, 1080–1087. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of rna-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, K.E.; Zhang, L.; Wang, J.; Smith, G.H.; Newman, S.; Schneider, T.M.; Pillai, R.N.; Kudchadkar, R.R.; Owonikoko, T.K.; Ramalingam, S.S.; et al. Clinical validation and implementation of a targeted next-generation sequencing assay to detect somatic variants in non-small cell lung, melanoma, and gastrointestinal malignancies. J. Mol. Diagn. 2016, 18, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Schroder, M.S.; Culhane, A.C.; Quackenbush, J.; Haibe-Kains, B. Survcomp: An r/bioconductor package for performance assessment and comparison of survival models. Bioinformatics 2011, 27, 3206–3208. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for pd-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral t cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial cd8+ tumor-infiltrating lymphocytes and a high cd8+/regulatory t cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543. [Google Scholar] [CrossRef] [Green Version]
- Clarke, B.; Tinker, A.V.; Lee, C.-H.; Subramanian, S.; van de Rijn, M.; Turbin, D.; Kalloger, S.; Han, G.; Ceballos, K.; Cadungog, M.G.; et al. Intraepithelial t cells and prognosis in ovarian carcinoma: Novel associations with stage, tumor type, and brca1 loss. Mod. Pathol. 2009, 22, 393–402. [Google Scholar] [CrossRef] [Green Version]
- Hamanishi, J.; Mandai, M.; Iwasaki, M.; Okazaki, T.; Tanaka, Y.; Yamaguchi, K.; Higuchi, T.; Yagi, H.; Takakura, K.; Minato, N.; et al. Programmed cell death 1 ligand 1 and tumor-infiltrating cd8+ t lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 3360–3365. [Google Scholar] [CrossRef] [Green Version]
- Wouters, M.C.A.; Komdeur, F.L.; Workel, H.H.; Klip, H.G.; Plat, A.; Kooi, N.M.; Wisman, G.B.A.; Mourits, M.J.E.; Arts, H.J.G.; Oonk, M.H.M.; et al. Treatment regimen, surgical outcome, and t-cell differentiation influence prognostic benefit of tumor-infiltrating lymphocytes in high-grade serous ovarian cancer. Clin. Cancer Res. 2016, 22, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.S.; Sahota, R.A.; Milne, K.; Kost, S.E.; Nesslinger, N.J.; Watson, P.H.; Nelson, B.H. Cd20+ tumor-infiltrating lymphocytes have an atypical cd27− memory phenotype and together with cd8+ t cells promote favorable prognosis in ovarian cancer. Clin. Cancer Res. 2012, 18, 3281–3292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeij, R.; de Bock, G.H.; Leffers, N.; ten Hoor, K.A.; Schulze, U.; Hollema, H.; van der Burg, S.H.; van der Zee, A.G.J.; Daemen, T.; Nijman, H.W. Tumor-infiltrating cytotoxic t lymphocytes as independent prognostic factor in epithelial ovarian cancer with wilms tumor protein 1 overexpression. J. Immunother. 2011, 34, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, J.; Mandai, M.; Abiko, K.; Matsumura, N.; Baba, T.; Yoshioka, Y.; Kosaka, K.; Konishi, I. The comprehensive assessment of local immune status of ovarian cancer by the clustering of multiple immune factors. Clin. Immunol. 2011, 141, 338–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 2015, 28, 690–714. [Google Scholar] [CrossRef] [Green Version]
- Leary, A.; Genestie, C.; Blanc-Durand, F.; Gouy, S.; Dunant, A.; Maulard, A.; Drusch, F.; Cheaib, B.; Michels, J.; Bentivegna, E.; et al. Neoadjuvant chemotherapy alters the balance of effector to suppressor immune cells in advanced ovarian cancer. Cancer Immunol. Immunother. 2021, 70, 519–531. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, J.Y.; Lee, Y.J.; Kim, S.H.; Lee, J.Y.; Nam, E.J.; Kim, S.; Kim, S.W.; Kim, Y.T. Expression of programmed cell death ligand 1 and immune checkpoint markers in residual tumors after neoadjuvant chemotherapy for advanced high-grade serous ovarian cancer. Gynecol. Oncol. 2018, 151, 414–421. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, D.; Shim, J.E.; Bae, S.J.; Jung, Y.J.; Kim, S.; Lee, H.; Kim, S.H.; Jo, S.B.; Lee, J.Y.; et al. Genomic profiling of the residual disease of advanced high-grade serous ovarian cancer after neoadjuvant chemotherapy. Int. J. Cancer 2020, 146, 1851–1861. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, J.W.; Lim, M.C.; Kim, S.; Kim, H.S.; Choi, C.H.; Yi, J.Y.; Park, S.Y.; Kim, B.G. A phase ii study of neoadjuvant chemotherapy plus durvalumab and tremelimumab in advanced-stage ovarian cancer: A korean gynecologic oncology group study (kgog 3046), tru-d. J. Gyneco.l Oncol. 2019, 30, e112. [Google Scholar] [CrossRef] [Green Version]


| Variables | Number (%) or Median (Range) |
|---|---|
| Age (years) | 58 (39–81) |
| FIGO stage | |
| III | 72 (49.0) |
| IV | 75 (51.0) |
| ASA score before NAC | |
| 1 | 8 (5.4) |
| 2 | 80 (54.4) |
| 3 | 58 (39.5) |
| Not applicable | 1 (0.7) |
| Germline BRCA status | |
| Wild | 97 (66.0) |
| BRCA1/2 mutation | 36 (24.5) |
| Not applicable | 14 (9.5) |
| Serum CA-125 level (U/mL) | 1509.0 (94.2–21,994.7) |
| Cycles of NAC | 3 (3–4) |
| Chemotherapy response score | |
| 1 | 4 (2.7) |
| 2 | 98 (66.7) |
| 3 | 45 (30.6) |
| Residual disease after IDS | |
| No gross tumor | 73 (49.7) |
| ≤0.5 cm | 53 (36.0) |
| >0.5 cm and ≤1.0 cm | 21 (14.3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.J.; Woo, H.Y.; Kim, Y.-N.; Park, J.; Nam, E.J.; Kim, S.W.; Kim, S.; Kim, Y.T.; Park, E.; Joung, J.-G.; et al. Dynamics of the Tumor Immune Microenvironment during Neoadjuvant Chemotherapy of High-Grade Serous Ovarian Cancer. Cancers 2022, 14, 2308. https://doi.org/10.3390/cancers14092308
Lee YJ, Woo HY, Kim Y-N, Park J, Nam EJ, Kim SW, Kim S, Kim YT, Park E, Joung J-G, et al. Dynamics of the Tumor Immune Microenvironment during Neoadjuvant Chemotherapy of High-Grade Serous Ovarian Cancer. Cancers. 2022; 14(9):2308. https://doi.org/10.3390/cancers14092308
Chicago/Turabian StyleLee, Yong Jae, Ha Young Woo, Yoo-Na Kim, Junsik Park, Eun Ji Nam, Sang Wun Kim, Sunghoon Kim, Young Tae Kim, Eunhyang Park, Je-Gun Joung, and et al. 2022. "Dynamics of the Tumor Immune Microenvironment during Neoadjuvant Chemotherapy of High-Grade Serous Ovarian Cancer" Cancers 14, no. 9: 2308. https://doi.org/10.3390/cancers14092308






