Comparative Proteomic and Transcriptomic Analysis of the Impact of Androgen Stimulation and Darolutamide Inhibition
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Origin and Authentication of Cell Lines
2.3. Preparation of Samples for Further Analyses
2.3.1. CETSA Samples
2.3.2. Transcriptomic Samples
2.3.3. Proteomic Samples
2.3.4. AR ChIP-Seq and HiChIP Samples
2.4. AR CETSA HT Assay
2.5. Transcriptomic Data Generation and Analysis
2.6. Proteomic Data Generation and Analysis
2.7. Statistical Analyses of Proteomic and Transcriptomic Data
2.8. ELISA Methods
2.9. AR ChIP-Seq Analysis
2.10. AR HiChIP Analysis
3. Results
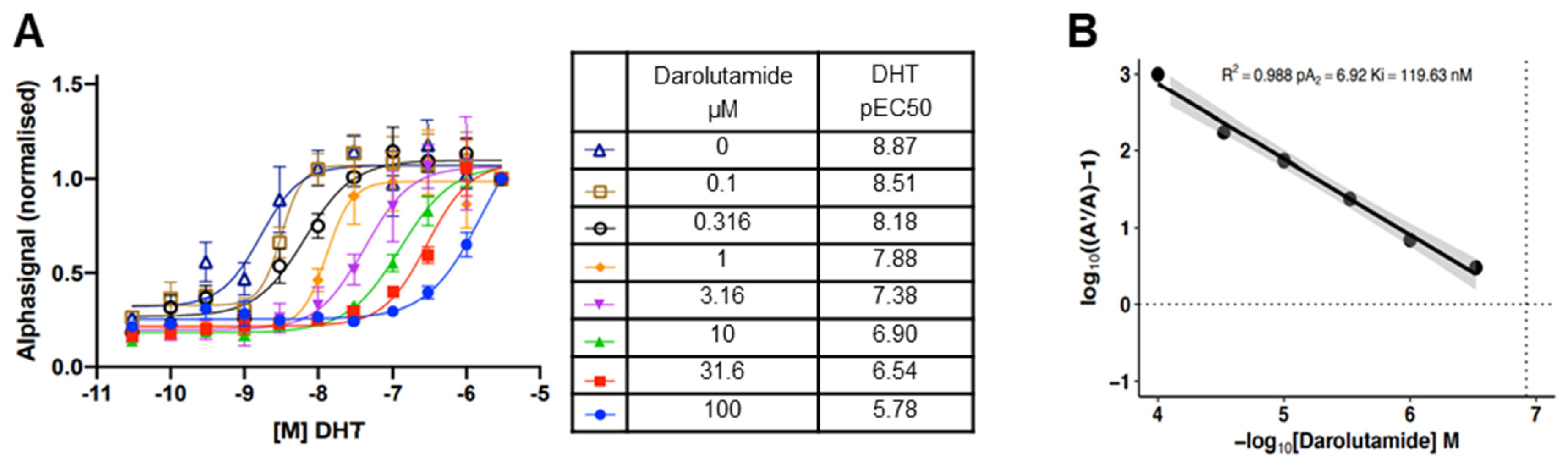
3.1. Darolutamide Is a Potent Antagonist of DHT Binding to the Human AR in the Cellular Environment
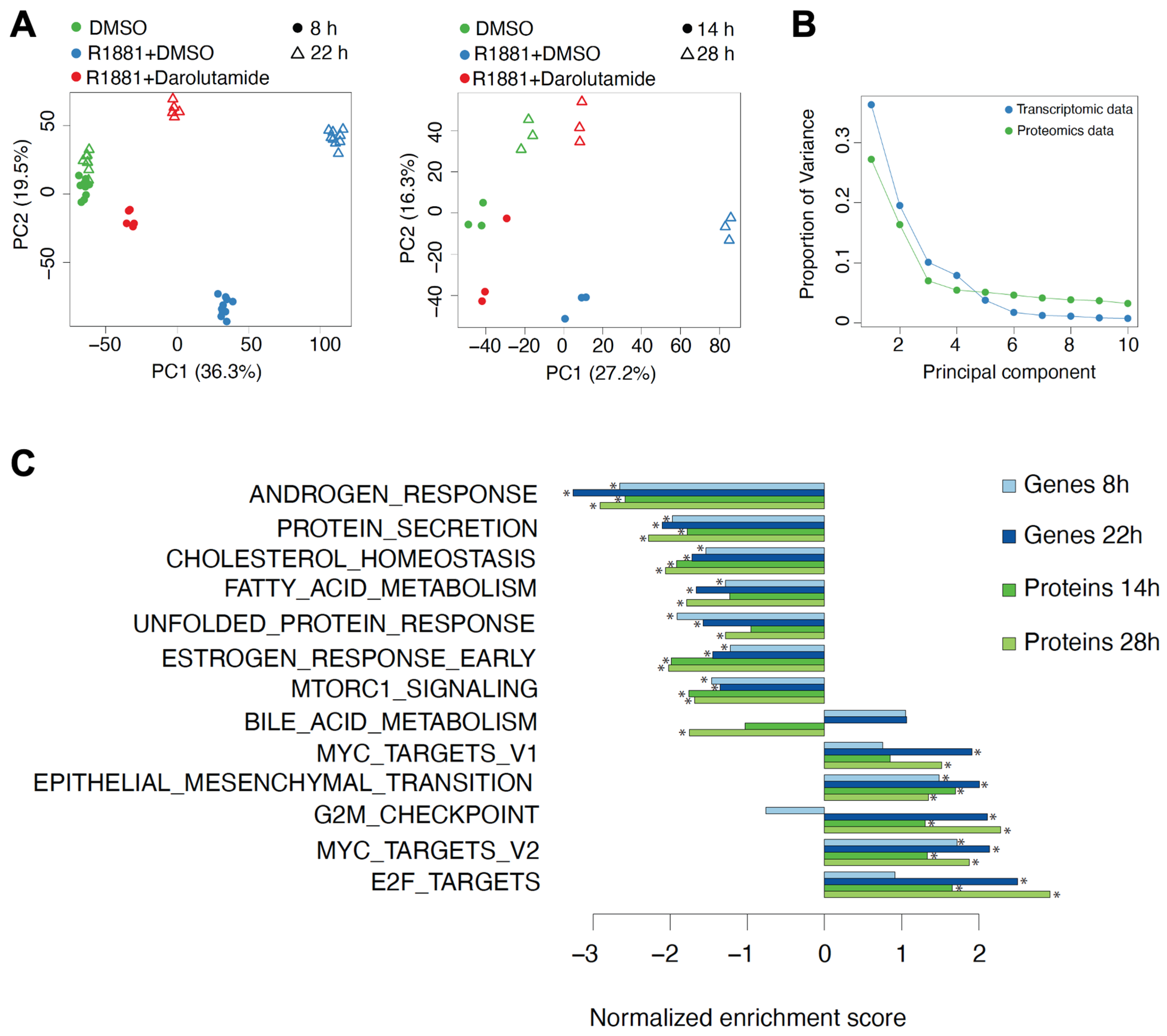
3.2. Most Expressed Genes Are Also Detected on Protein Level
3.3. Androgen Stimulation and Darolutamide Inhibition Induce Largely Concordant Changes at the Gene and Protein Levels
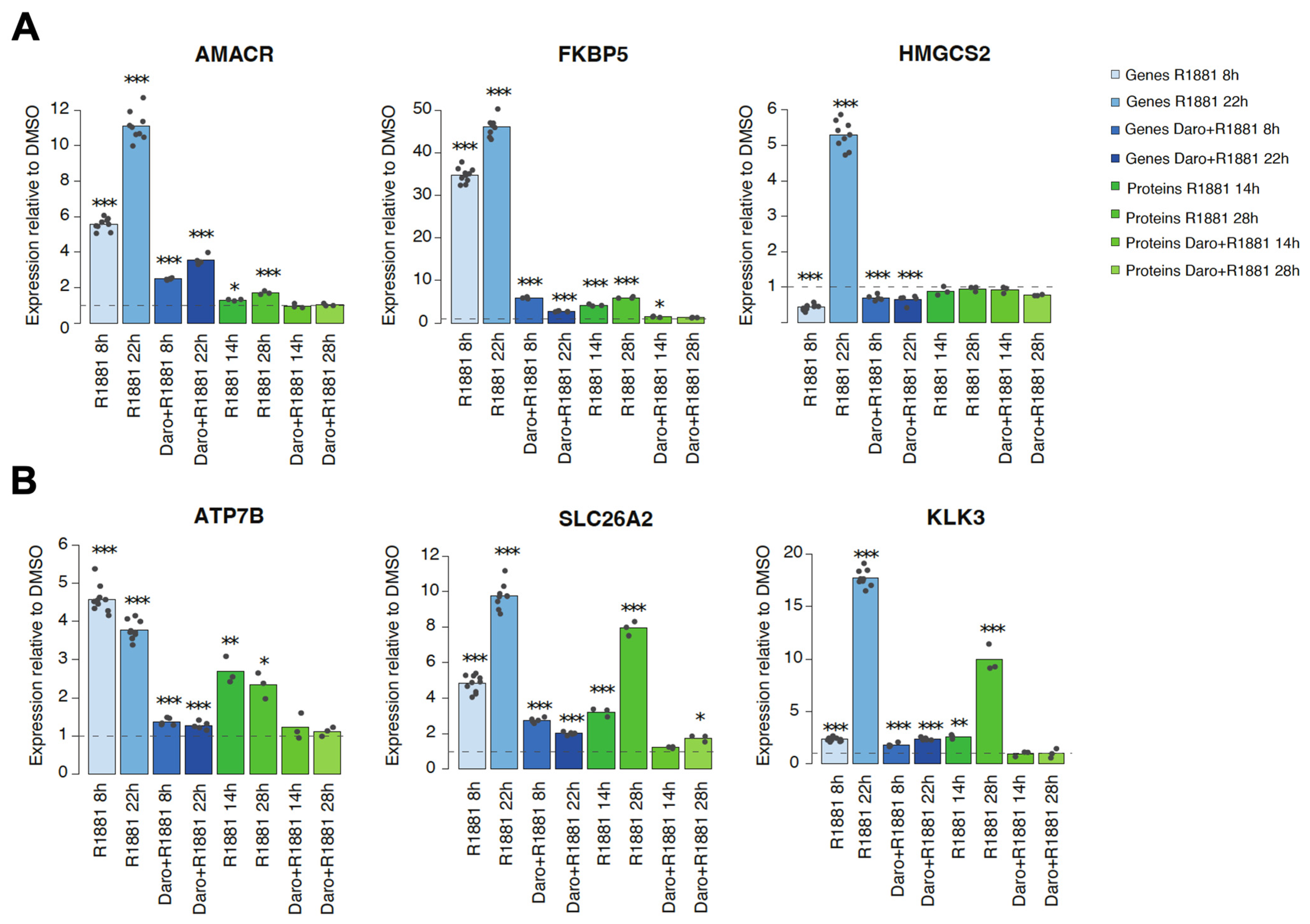
3.4. A Discordant Impact of Darolutamide on Gene and Protein Levels Is Observed
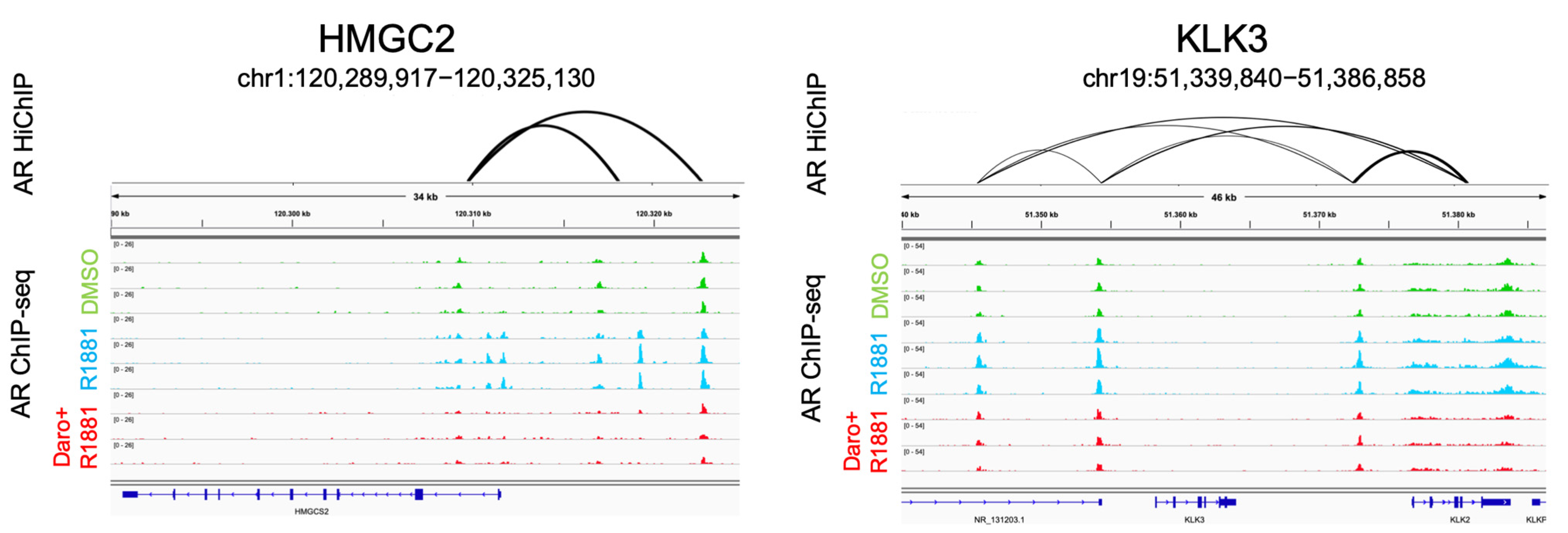
3.5. AR Binding Peaks and AR Loops Are Found in Androgen-Regulated Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.H.; Zoubeidi, A. Targeting androgen receptor signaling: A historical perspective. Endocr. Relat. Cancer 2021, 28, T11-8. [Google Scholar] [CrossRef] [PubMed]
- Spratt, D.E.; Shore, N.; Sartor, O.; Rathkopf, D.; Olivier, K. Treating the patient and not just the cancer: Therapeutic burden in prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Nevedomskaya, E.; Baumgart, S.J.; Haendler, B. Recent advances in prostate cancer treatment and drug discovery. Int. J. Mol. Sci. 2018, 19, 1359. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.A.; Malhotra, S.V.; Stoyanova, T. Second-generation antiandrogens: From discovery to standard of care in castration resistant prostate cancer. Front. Oncol. 2019, 9, 801. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.J.; Corey, E.; Guise, T.A.; Gulley, J.L.; Kevin Kelly, W.; Quinn, D.I.; Scholz, A.; Sgouros, G. Radium-223 mechanism of action: Implications for use in treatment combinations. Nat. Rev. Urol. 2019, 16, 745–756. [Google Scholar] [CrossRef]
- Ehsani, M.; David, F.O.; Baniahmad, A. Androgen receptor-dependent mechanisms mediating drug resistance in prostate cancer. Cancers 2021, 13, 1534. [Google Scholar] [CrossRef]
- Stelloo, S.; Nevedomskaya, E.; van der Poel, H.G.; de Jong, J.; van Leenders, G.J.; Jenster, G.; Wessels, L.F.; Bergman, A.M.; Zwart, W. Androgen receptor profiling predicts prostate cancer outcome. EMBO Mol. Med. 2015, 7, 1450–1464. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Li, F.; Takeda, D.Y.; Lenci, R.; Chonkar, A.; Chabot, M.; Cejas, P.; Vazquez, F.; Cook, J.; Shivdasani, R.A.; et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015, 47, 1346–1351. [Google Scholar] [CrossRef]
- Baumgart, S.J.; Nevedomskaya, E.; Haendler, B. Dysregulated transcriptional control in prostate cancer. Int. J. Mol. Sci. 2019, 20, 2883. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Qiu, X.; Zhu, Y.; Takeda, D.Y.; Pan, W.; Baca, S.C.; Gusev, A.; Korthauer, K.D.; Severson, T.M.; Ha, G.; et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020, 52, 790–799. [Google Scholar] [CrossRef]
- Hankey, W.; Chen, Z.; Wang, Q. Shaping chromatin states in prostate cancer by pioneer transcription factors. Cancer Res. 2020, 80, 2427–2436. [Google Scholar] [CrossRef]
- Baumgart, S.J.; Nevedomskaya, E.; Lesche, R.; Newman, R.; Mumberg, D.; Haendler, B. Darolutamide antagonizes androgen signaling by blocking enhancer and super-enhancer activation. Mol. Oncol. 2020, 14, 2022–2039. [Google Scholar] [CrossRef]
- Azhagiya Singam, E.R.; Tachachartvanich, P.; La Merrill, M.A.; Smith, M.T.; Durkin, K.A. Structural dynamics of agonist and antagonist binding to the androgen receptor. J. Phys. Chem. B 2019, 123, 7657–7666. [Google Scholar] [CrossRef]
- Takeda, K.; Hara, N.; Nishiyama, T.; Tasaki, M.; Ishizaki, F.; Tomita, Y. Corepressive function of nuclear receptor coactivator 2 in androgen receptor of prostate cancer cells treated with antiandrogen. BMC Cancer 2016, 16, 332. [Google Scholar] [CrossRef]
- Sugawara, T.; Baumgart, S.J.; Nevedomskaya, E.; Reichert, K.; Steuber, H.; Lejeune, P.; Mumberg, D.; Haendler, B. Darolutamide is a potent androgen receptor antagonist with strong efficacy in prostate cancer models. Int. J. Cancer 2019, 145, 1382–1394. [Google Scholar] [CrossRef]
- Teng, M.; Zhou, S.; Cai, C.; Lupien, M.; He, H.H. Pioneer of prostate cancer: Past, present and the future of FOXA1. Protein Cell 2021, 12, 29–38. [Google Scholar] [CrossRef]
- Verma, S.; Shankar, E.; Kalayci, F.N.C.; Mukunda, A.; Alassfar, M.; Singh, V.; Chan, E.R.; MacLennan, G.T.; Gupta, S. Androgen deprivation induces transcriptional reprogramming in prostate cancer cells to develop stem cell-like characteristics. Int. J. Mol. Sci. 2020, 21, 9568. [Google Scholar] [CrossRef]
- Simon, I.; Perales, S.; Casado-Medina, L.; Rodriguez-Martinez, A.; Garrido-Navas, M.D.C.; Puche-Sanz, I.; Diaz-Mochon, J.J.; Alaminos, C.; Lupianez, P.; Lorente, J.A.; et al. Cross-resistance to abiraterone and enzalutamide in castration resistance prostate cancer cellular models is mediated by AR transcriptional reactivation. Cancers 2021, 13, 1483. [Google Scholar] [CrossRef]
- Liu, Y.; Horn, J.L.; Banda, K.; Goodman, A.Z.; Lim, Y.; Jana, S.; Arora, S.; Germanos, A.A.; Wen, L.; Hardin, W.R.; et al. The androgen receptor regulates a druggable translational regulon in advanced prostate cancer. Sci. Transl. Med. 2019, 11, eaaw4993. [Google Scholar] [CrossRef]
- Mirzakhani, K.; Baniahmad, A. Protein translation controlled by the androgen receptor in prostate cancer: A novel therapeutic option? Transl. Cancer Res. 2020, 9, 2171–2174. [Google Scholar] [CrossRef] [PubMed]
- Latonen, L.; Afyounian, E.; Jylha, A.; Nattinen, J.; Aapola, U.; Annala, M.; Kivinummi, K.K.; Tammela, T.T.L.; Beuerman, R.W.; Uusitalo, H.; et al. Integrative proteomics in prostate cancer uncovers robustness against genomic and transcriptomic aberrations during disease progression. Nat. Commun. 2018, 9, 1176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Zhang, M.; Shi, C.; Sun, L.; Shan, L.; Zhang, H.; Song, Y. An overview of advances in multi-omics analysis in prostate cancer. Life Sci. 2020, 260, 118376. [Google Scholar] [CrossRef] [PubMed]
- Fizazi, K.; Shore, N.; Tammela, T.L.; Ulys, A.; Vjaters, E.; Polyakov, S.; Jievaltas, M.; Luz, M.; Alekseev, B.; Kuss, I.; et al. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 2019, 380, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Hussain, M.; Saad, F.; Fizazi, K.; Sternberg, C.N.; Crawford, E.D.; Kopyltsov, E.; Park, C.H.; Alekseev, B.; Montesa-Pino, A.; et al. Darolutamide and survival in metastatic, hormone-sensitive prostate cancer. N. Engl. J. Med. 2022, 386, 1132–1142. [Google Scholar] [CrossRef]
- Velluz, L. Current trends in steroid synthesis. Ann. Pharm. Fr. 1963, 21, 59–71. [Google Scholar]
- Dehm, S.M.; Schmidt, L.J.; Heemers, H.V.; Vessella, R.L.; Tindall, D.J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008, 68, 5469–5477. [Google Scholar] [CrossRef]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 1259038. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Mumbach, M.R.; Granja, J.M.; Flynn, R.A.; Roake, C.M.; Satpathy, A.T.; Rubin, A.J.; Qi, Y.; Jiang, Z.; Shams, S.; Louie, B.H.; et al. HiChIRP reveals RNA-associated chromosome conformation. Nat. Methods 2019, 16, 489–492. [Google Scholar] [CrossRef]
- Shaw, J.; Leveridge, M.; Norling, C.; Karen, J.; Molina, D.M.; O'Neill, D.; Dowling, J.E.; Davey, P.; Cowan, S.; Dabrowski, M.; et al. Determining direct binders of the androgen receptor using a high-throughput cellular thermal shift assay. Sci. Rep. 2018, 8, 163. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Li, Y.; Chan, S.C.; Brand, L.J.; Hwang, T.H.; Silverstein, K.A.; Dehm, S.M. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013, 73, 483–489. [Google Scholar] [CrossRef]
- Hieronymus, H.; Lamb, J.; Ross, K.N.; Peng, X.P.; Clement, C.; Rodina, A.; Nieto, M.; Du, J.; Stegmaier, K.; Raj, S.M.; et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006, 10, 321–330. [Google Scholar] [CrossRef]
- Moilanen, A.M.; Riikonen, R.; Oksala, R.; Ravanti, L.; Aho, E.; Wohlfahrt, G.; Nykanen, P.S.; Tormakangas, O.P.; Palvimo, J.J.; Kallio, P.J. Discovery of ODM-201, a new-generation androgen receptor inhibitor targeting resistance mechanisms to androgen signaling-directed prostate cancer therapies. Sci. Rep. 2015, 5, 12007. [Google Scholar] [CrossRef]
- Seashore-Ludlow, B.; Axelsson, H.; Lundback, T. Perspective on CETSA literature: Toward more quantitative data interpretation. SLAS Discov. 2020, 25, 118–126. [Google Scholar] [CrossRef]
- Lapek, J.D., Jr.; Greninger, P.; Morris, R.; Amzallag, A.; Pruteanu-Malinici, I.; Benes, C.H.; Haas, W. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat. Biotechnol. 2017, 35, 983–989. [Google Scholar] [CrossRef]
- Kustatscher, G.; Grabowski, P.; Schrader, T.A.; Passmore, J.B.; Schrader, M.; Rappsilber, J. Co-regulation map of the human proteome enables identification of protein functions. Nat. Biotechnol. 2019, 37, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Buccitelli, C.; Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Eraslan, B.; Wieland, T.; Hallstrom, B.; Hopf, T.; Zolg, D.P.; Zecha, J.; Asplund, A.; Li, L.H.; Meng, C.; et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019, 15, e8503. [Google Scholar] [CrossRef]
- Eraslan, B.; Wang, D.; Gusic, M.; Prokisch, H.; Hallstrom, B.M.; Uhlen, M.; Asplund, A.; Ponten, F.; Wieland, T.; Hopf, T.; et al. Quantification and discovery of sequence determinants of protein-per-mRNA amount in 29 human tissues. Mol. Syst. Biol. 2019, 15, e8513. [Google Scholar] [CrossRef]
- Buszczak, M.; Signer, R.A.; Morrison, S.J. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 2014, 159, 242–251. [Google Scholar] [CrossRef]
- Meissner, F.; Scheltema, R.A.; Mollenkopf, H.J.; Mann, M. Direct proteomic quantification of the secretome of activated immune cells. Science 2013, 340, 475–478. [Google Scholar] [CrossRef]
- Fizazi, K.; Shore, N.; Tammela, T.L.; Ulys, A.; Vjaters, E.; Polyakov, S.; Jievaltas, M.; Luz, M.; Alekseev, B.; Kuss, I.; et al. Nonmetastatic, castration-resistant prostate cancer and survival with darolutamide. N. Engl. J. Med. 2020, 383, 1040–1049. [Google Scholar] [CrossRef]
- Maeda, K.; Habara, M.; Kawaguchi, M.; Matsumoto, H.; Hanaki, S.; Masaki, T.; Sato, Y.; Matsuyama, H.; Kunieda, K.; Nakagawa, H.; et al. FKBP51 and FKBP52 regulate androgen receptor dimerization and proliferation in prostate cancer cells. Mol. Oncol. 2022, 16, 940–956. [Google Scholar] [CrossRef]
- Febbo, P.G.; Lowenberg, M.; Thorner, A.R.; Brown, M.; Loda, M.; Golub, T.R. Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. J. Urol. 2005, 173, 1772–1777. [Google Scholar] [CrossRef]
- Wyatt, A.W.; Gleave, M.E. Targeting the adaptive molecular landscape of castration-resistant prostate cancer. EMBO Mol. Med. 2015, 7, 878–894. [Google Scholar] [CrossRef]
- Alinezhad, S.; Vaananen, R.M.; Ochoa, N.T.; Vertosick, E.A.; Bjartell, A.; Bostrom, P.J.; Taimen, P.; Pettersson, K. Global expression of AMACR transcripts predicts risk for prostate cancer—A systematic comparison of AMACR protein and mRNA expression in cancerous and noncancerous prostate. BMC Urol. 2016, 16, 10. [Google Scholar] [CrossRef]
- Bader, D.A.; McGuire, S.E. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat. Rev. Urol. 2020, 17, 214–231. [Google Scholar] [CrossRef]
- Wilson, B.A.; Wang, H.; Nacev, B.A.; Mease, R.C.; Liu, J.O.; Pomper, M.G.; Isaacs, W.B. High-throughput screen identifies novel inhibitors of cancer biomarker alpha-methylacyl coenzyme A racemase (AMACR/P504S). Mol. Cancer Ther. 2011, 10, 825–838. [Google Scholar] [CrossRef]
- Kuefer, R.; Varambally, S.; Zhou, M.; Lucas, P.C.; Loeffler, M.; Wolter, H.; Mattfeldt, T.; Hautmann, R.E.; Gschwend, J.E.; Barrette, T.R.; et al. alpha-Methylacyl-CoA racemase: Expression levels of this novel cancer biomarker depend on tumor differentiation. Am. J. Pathol. 2002, 161, 841–848. [Google Scholar] [CrossRef]
- Zha, S.; Ferdinandusse, S.; Denis, S.; Wanders, R.J.; Ewing, C.M.; Luo, J.; De Marzo, A.M.; Isaacs, W.B. Alpha-methylacyl-CoA racemase as an androgen-independent growth modifier in prostate cancer. Cancer Res. 2003, 63, 7365–7376. [Google Scholar]
- Wan, S.; Xi, M.; Zhao, H.B.; Hua, W.; Liu, Y.L.; Zhou, Y.L.; Zhuo, Y.J.; Liu, Z.Z.; Cai, Z.D.; Wan, Y.P.; et al. HMGCS2 functions as a tumor suppressor and has a prognostic impact in prostate cancer. Pathol. Res. Pract. 2019, 215, 152464. [Google Scholar] [CrossRef]
- Brooke, G.N.; Gamble, S.C.; Hough, M.A.; Begum, S.; Dart, D.A.; Odontiadis, M.; Powell, S.M.; Fioretti, F.M.; Bryan, R.A.; Waxman, J.; et al. Antiandrogens act as selective androgen receptor modulators at the proteome level in prostate cancer cells. Mol. Cell Proteom. 2015, 14, 1201–1216. [Google Scholar] [CrossRef]
- Rowland, J.G.; Robson, J.L.; Simon, W.J.; Leung, H.Y.; Slabas, A.R. Evaluation of an in vitro model of androgen ablation and identification of the androgen responsive proteome in LNCaP cells. Proteomics 2007, 7, 47–63. [Google Scholar] [CrossRef]
- Liyanage, C.; Malik, A.; Abeysinghe, P.; Clements, J.; Batra, J. SWATH-MS based proteomic profiling of prostate cancer cells reveals adaptive molecular mechanisms in response to anti-androgen therapy. Cancers 2021, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, S.J.; Nevedomskaya, E.; Lesche, R.; Seidel, H.; Haendler, B. A detailed comparison berween second-generation AR antagonists reveals differences in the overall impact on gene regulation patterns in prostate cancer cells. Cancer Res. 2018, 78 (Suppl. 13), 1798. [Google Scholar] [CrossRef]
- Sinha, A.; Huang, V.; Livingstone, J.; Wang, J.; Fox, N.S.; Kurganovs, N.; Ignatchenko, V.; Fritsch, K.; Donmez, N.; Heisler, L.E.; et al. The proteogenomic landscape of curable prostate cancer. Cancer Cell 2019, 35, 414–427.e6. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Corey, E.; Gujral, T.S. Proteomic and transcriptomic profiling reveals mitochondrial oxidative phosphorylation as therapeutic vulnerability in androgen receptor pathway active prostate tumors. Cancers 2022, 14, 1739. [Google Scholar] [CrossRef] [PubMed]
- Nevedomskaya, E.; Haendler, B. From omics to multi-omics approaches for in-depth analysis of the molecular mechanisms of prostate cancer. Int. J. Mol. Sci. 2022, 23, 6281. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nevedomskaya, E.; Sugawara, T.; Baumgart, S.J.; Lesche, R.; Hahne, H.; Mumberg, D.; Haendler, B. Comparative Proteomic and Transcriptomic Analysis of the Impact of Androgen Stimulation and Darolutamide Inhibition. Cancers 2023, 15, 2. https://doi.org/10.3390/cancers15010002
Nevedomskaya E, Sugawara T, Baumgart SJ, Lesche R, Hahne H, Mumberg D, Haendler B. Comparative Proteomic and Transcriptomic Analysis of the Impact of Androgen Stimulation and Darolutamide Inhibition. Cancers. 2023; 15(1):2. https://doi.org/10.3390/cancers15010002
Chicago/Turabian StyleNevedomskaya, Ekaterina, Tatsuo Sugawara, Simon J. Baumgart, Ralf Lesche, Hannes Hahne, Dominik Mumberg, and Bernard Haendler. 2023. "Comparative Proteomic and Transcriptomic Analysis of the Impact of Androgen Stimulation and Darolutamide Inhibition" Cancers 15, no. 1: 2. https://doi.org/10.3390/cancers15010002
APA StyleNevedomskaya, E., Sugawara, T., Baumgart, S. J., Lesche, R., Hahne, H., Mumberg, D., & Haendler, B. (2023). Comparative Proteomic and Transcriptomic Analysis of the Impact of Androgen Stimulation and Darolutamide Inhibition. Cancers, 15(1), 2. https://doi.org/10.3390/cancers15010002







