Epimutations and Their Effect on Chromatin Organization: Exciting Avenues for Cancer Treatment
Simple Summary
Abstract
1. Introduction
2. Techniques to Study Chromatin Interactions
2.1. Chromosome Conformation Capture (3C)
2.2. Chromosome Conformation Capture-on-Chip (4C)
2.3. Chromosome Conformation Capture Carbon Copy (5C)
2.4. Hi-C
2.5. Micro-C
2.6. Chromatin Interaction Analysis Paired-End Tag Sequencing (ChIA-PET)
2.7. Capture-C
3. Epigenetic Factors Involved in Regulating Chromatin Organization
3.1. DNA Methylation
3.2. Histone Acetylation
3.3. Histone Methylation
3.4. Histone Phosphorylation
3.5. Ubiquitination
3.6. SUMOylation
3.7. Citrullination
3.8. ADP Ribosylation
3.9. Histone Glycosylation
3.10. Proline Isomerization
4. Epimutations, TADs and Cancer
4.1. Writes, Readers, and Erasers
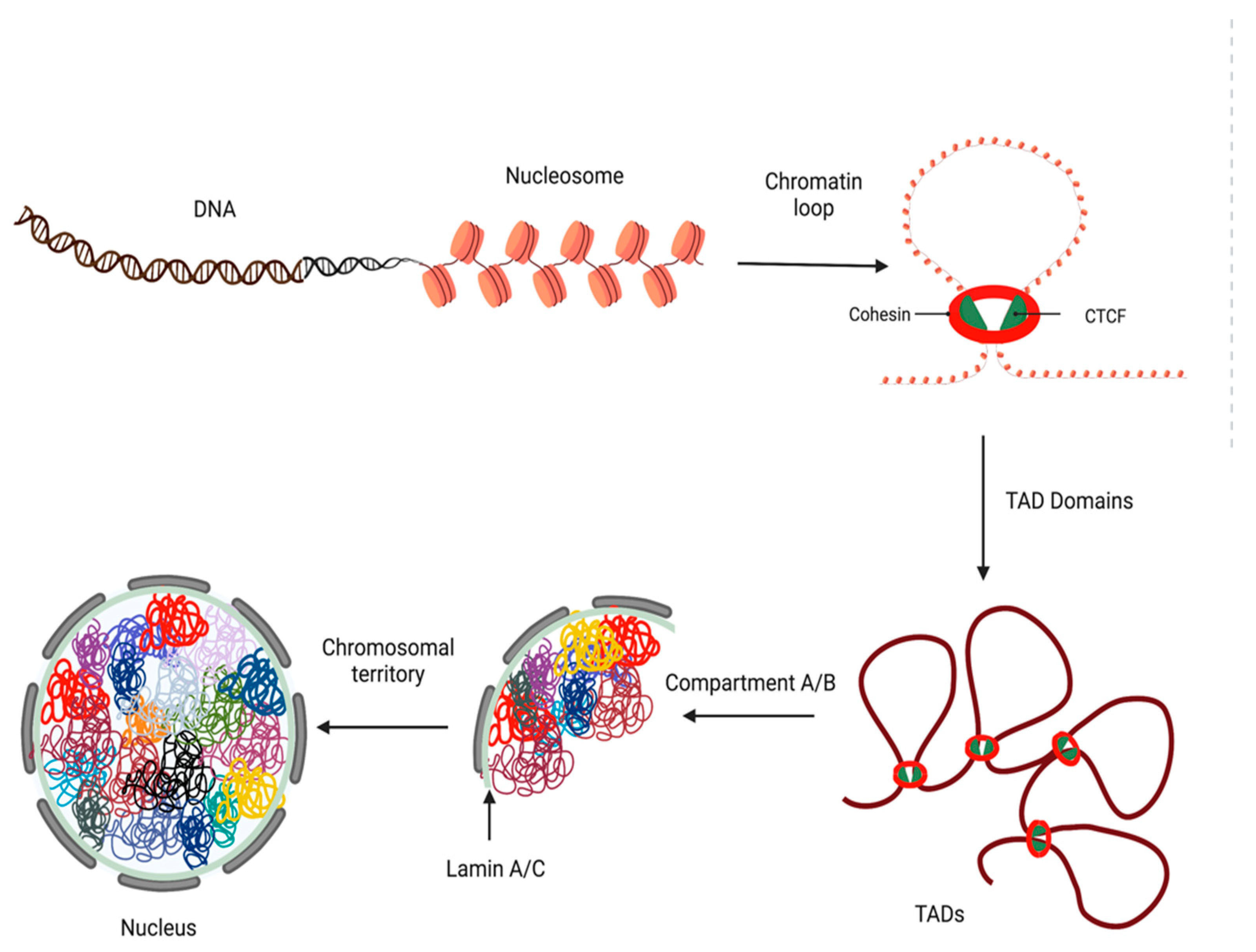
4.2. Loops, TADs, and Insulators
4.3. Epigenetic Inhibitors
4.3.1. DNA Modification Inhibitors
4.3.2. Histone Modification Inhibitors
5. Future Perspective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3C | chromosome conformation capture |
| 3D | three-dimensional |
| 4C | chromosome conformation capture on a chip |
| 53BP1 | tumor protein p53 binding protein 1 |
| 5C | chromosome conformation capture carbon copy |
| 5mC | 5-methylcytosine |
| AML | acute myeloid leukemia |
| APLF | aprataxin and PNKP like factor |
| AURKB | Aurora kinase B |
| BARD1 | BRCA1 associated RING domain 1 |
| BAZ1B | bromodomain adjacent to zinc finger domain 1B |
| BET | bromodomain and extra-terminal |
| BIR | Baculovirus IAP repeat |
| BRCA1 | breast cancer gene 1 |
| BRCC36 | BRCA1/BRCA2-containing complex subunit 3 |
| BRCT | BRCA1 C terminus |
| BRD2/3/4/T | bromodomain containing 2/3/4/T |
| BRPF1 | BRD and PHD finger-containing protein 1 |
| CREBBP/CBP | CREB-binding protein |
| CHFR | checkpoint with forkhead and RING finger domains |
| ChIA-PET | chromatin interaction analysis paired-end tag sequencing |
| ChIP | Chromatin immunoprecipitation |
| CHi-C | Capture Hi-C |
| CLL | chronic lymphocytic leukemia |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| Cas9 | CRISPR-associated endonuclease |
| CRISPRa | CRISPR activation |
| CRISPRi | CRISPR inactivation |
| CT | chromosome territory |
| CTCF | CCCTC-binding factor |
| CTCL | cutaneous T-cell lymphoma |
| DLBCL | diffuse large B-cell lymphoma |
| DNMT1/3a/3b | DNA methyltransferase 1/3a/3b |
| DNMTi | DNA methylatransferase inhibitor |
| DNMTs | DNA methyltransferases |
| DOT1L | DOT1 like histone lysine methyltransferase |
| EP300 | E1A binding protein p300 |
| EYA1/3 | eyes absent homolog 1/3 |
| EZH1/2 | enhancer of Zeste homolog 1/2 |
| G9a/EHMT2 | euchromatic histone lysine methyltransferase 2 |
| GNATs | Gcn5-related N-acetyltransferases |
| GLP/EHMT1 | euchromatic histone lysine methyltransferase 1 |
| HAT | histone acetyltransferase |
| HATi | histone acetylatransferase inhibitor |
| HATs | histone acetyltransferases |
| HDAC | histone deacetylase |
| HDACi | histone deacetylase inhibitor |
| HDACs | histone deacetylases |
| HDM | histone demethylase |
| HDMs | histone demethylases |
| HMT | histone methyltransferase |
| HMTi | histone methyltransferase inhibitor |
| HMTs | histone methyltransferases |
| HoxA9 | Homeobox A9 |
| HP1 | heterochromatin protein 1 |
| IDH | isocitrate dehydrogenase |
| JmjC | Jumonji C |
| KAT5 | lysine acetyltransferase 5 |
| KDM1/1A/1B/2A/2B/3A/3B/4/5B | lysine demethylase 1/1A/1B/2A/2B/3A/3B/4/5B |
| Kilobases | Kb |
| Kilodaltons | kDa |
| KMT4/7 | lysine N-methyltransferase4/7 |
| LSD1 | lysine-specific demethylase 1 |
| MBD | methyl-CpG binding domain |
| MBT | malignant brain tumor |
| MDS | myelodysplastic syndrome |
| Megabases | Mb |
| MM | Multiple myeloma |
| MORF | monocytic leukemia zinc finger protein-related factor |
| MRG15 | MORF4-related gene on chromosome 15 |
| MYST | MOZ, Ybf2,Sas2, TIP60 |
| NAD | nicotinamide adenine dinucleotide |
| NBS1 | Nijmegen breakage syndrome 1 |
| NET | neutrophil extracellular traps |
| NG | next-generation |
| NGS | next-generation sequencing |
| NSCLC | non-small cell lung cancer |
| NSD1-3 | nuclear receptor binding SET domain proteins 1-3 |
| O-GlcNAc | O-linked N-acetylglucosamine |
| OTUB1/2 | OTU domain-containing ubiquitin aldehyde-binding protein 1/2 |
| p21 | Cyclin dependent kinase inhibitor 1A |
| p300 | E1A binding protein p300 |
| PAD | peptidyl arginine deiminase |
| PARG | poly(ADP-ribose) glycohydrolase |
| PARP1 | poly(ADP-ribose) polymerase 1 |
| PBZ | PAR-binding zinc finger |
| PCR | polymerase chain reaction |
| PETs | paired-end tags |
| PHF | plant homeodomain (PHD) finger |
| PHF1/19 | PHD finger protein 1/19 |
| PP1 | protein phosphatase 1 |
| PP2 | protein phosphatase 2 |
| PPARgamma | peroxisome proliferator activated receptor gamma |
| PPIases | peptidylprolyl isomerases |
| PPP1/2CA | protein phosphatase 1/2 catalytic subunit alpha |
| PRMT1/2/4/5/6/7 | protein arginine methyltransferase 1/2/4/5/6/7 |
| PTCL | peripheral T-cell lymphoma |
| PWWP | Pro-Trp-Trp-Pro |
| Rad21 | RAD21 cohesin complex component |
| RNF146 | RING finger protein 146 |
| SCC | squamous cell carcinoma |
| ssDNA | single-stranded DNA |
| SIRT | Sirtuin 1 |
| SMC1/3 | structural maintenance of chromosomes 1/3 |
| STAG1/3 | stromal antigen 1/3 |
| SUMO | small ubiquitin-like modified proteins |
| SUV39H1/2 | suppressor of variegation 3-9 homolog 1/2 |
| TAC | transactivation domain |
| TAD | topologically associated domain |
| TAF3 | TATA-Box Binding Protein Associated Factor 3 |
| TBD | To be determined |
| TDRD3 | Tudor domain containing proteins 3 |
| TET1/2 | ten-eleven translocation 1/2 |
| TP53 | tumor protein 53 |
| USP3/16/26/44 | ubiquitin specific peptidase 3/16/26/44 |
| WSTF | Williams Syndrome transcription factor |
| XRCC1 | X-ray repair cross complementing 1 |
References
- Felsenfeld, G.; Groudine, M. Controlling the double helix. Nature 2003, 421, 448–453. [Google Scholar] [CrossRef]
- Maeshima, K.; Ide, S.; Babokhov, M. Dynamic chromatin organization without the 30-nm fiber. Curr. Opin. Cell Biol. 2019, 58, 95–104. [Google Scholar] [CrossRef]
- Ou, H.D.; Phan, S.; Deerinck, T.J.; Thor, A.; Ellisman, M.H.; O’Shea, C.C. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357, eaag0025. [Google Scholar] [CrossRef]
- Dekker, J.; Misteli, T. Long-Range Chromatin Interactions. Cold Spring Harb. Perspect. Biol. 2015, 7, a019356. [Google Scholar] [CrossRef]
- Carter, D.; Chakalova, L.; Osborne, C.S.; Dai, Y.-f.; Fraser, P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002, 32, 623–626. [Google Scholar] [CrossRef]
- Tolhuis, B.; Palstra, R.J.; Splinter, E.; Grosveld, F.; de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 2002, 10, 1453–1465. [Google Scholar] [CrossRef]
- Szabo, Q.; Bantignies, F.; Cavalli, G. Principles of genome folding into topologically associating domains. Sci. Adv. 2019, 5, eaaw1668. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing chromosome conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef]
- Denker, A.; de Laat, W. The second decade of 3C technologies: Detailed insights into nuclear organization. Genes Dev. 2016, 30, 1357–1382. [Google Scholar] [CrossRef]
- Hagège, H.; Klous, P.; Braem, C.; Splinter, E.; Dekker, J.; Cathala, G.; de Laat, W.; Forné, T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007, 2, 1722–1733. [Google Scholar] [CrossRef]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006, 38, 1348–1354. [Google Scholar] [CrossRef]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef]
- Dostie, J.; Dekker, J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat. Protoc. 2007, 2, 988–1002. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- van Berkum, N.L.; Lieberman-Aiden, E.; Williams, L.; Imakaev, M.; Gnirke, A.; Mirny, L.A.; Dekker, J.; Lander, E.S. Hi-C: A method to study the three-dimensional architecture of genomes. J. Vis. Exp. 2010, 39, e1869. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Weiner, A.; Lajoie, B.; Dekker, J.; Friedman, N.; Rando, O.J. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell 2015, 162, 108–119. [Google Scholar] [CrossRef]
- Li, X.; Luo, O.J.; Wang, P.; Zheng, M.; Wang, D.; Piecuch, E.; Zhu, J.J.; Tian, S.Z.; Tang, Z.; Li, G.; et al. Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions. Nat. Protoc. 2017, 12, 899–915. [Google Scholar] [CrossRef]
- Hughes, J.R.; Roberts, N.; McGowan, S.; Hay, D.; Giannoulatou, E.; Lynch, M.; De Gobbi, M.; Taylor, S.; Gibbons, R.; Higgs, D.R. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014, 46, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Mifsud, B.; Tavares-Cadete, F.; Young, A.N.; Sugar, R.; Schoenfelder, S.; Ferreira, L.; Wingett, S.W.; Andrews, S.; Grey, W.; Ewels, P.A.; et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015, 47, 598–606. [Google Scholar] [CrossRef]
- Davies, J.O.J.; Telenius, J.M.; McGowan, S.J.; Roberts, N.A.; Taylor, S.; Higgs, D.R.; Hughes, J.R. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 2016, 13, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Gereige, L.-M.; Mikkola, H.K.A. DNA methylation is a guardian of stem cell self-renewal and multipotency. Nat. Genet. 2009, 41, 1164–1166. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Pradhan, S.; Bacolla, A.; Wells, R.D.; Roberts, R.J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999, 274, 33002–33010. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome-biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef]
- Marmorstein, R.; Zhou, M.M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef]
- Di Cerbo, V.; Schneider, R. Cancers with wrong HATs: The impact of acetylation. Brief. Funct. Genom. 2013, 12, 231–243. [Google Scholar] [CrossRef]
- Cang, S.; Feng, J.; Konno, S.; Han, L.; Liu, K.; Sharma, S.C.; Choudhury, M.; Chiao, J.W. Deficient histone acetylation and excessive deacetylase activity as epigenomic marks of prostate cancer cells. Int. J. Oncol. 2009, 35, 1417–1422. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Porter, N.J.; Christianson, D.W. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr. Opin. Struct. Biol. 2019, 59, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.S. The protein arginine methyltransferase family: An update about function, new perspectives and the physiological role in humans. Cell. Mol. Life Sci. 2009, 66, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Whetstine, J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 2007, 25, 1–14. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Ciccone, D.N.; Su, H.; Hevi, S.; Gay, F.; Lei, H.; Bajko, J.; Xu, G.; Li, E.; Chen, T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009, 461, 415–418. [Google Scholar] [CrossRef]
- Højfeldt, J.W.; Agger, K.; Helin, K. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discov. 2013, 12, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Cloos, P.A.; Christensen, J.; Agger, K.; Helin, K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008, 22, 1115–1140. [Google Scholar] [CrossRef]
- Banerjee, T.; Chakravarti, D. A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 2011, 31, 4858–4873. [Google Scholar] [CrossRef] [PubMed]
- North, J.A.; Javaid, S.; Ferdinand, M.B.; Chatterjee, N.; Picking, J.W.; Shoffner, M.; Nakkula, R.J.; Bartholomew, B.; Ottesen, J.J.; Fishel, R.; et al. Phosphorylation of histone H3(T118) alters nucleosome dynamics and remodeling. Nucleic Acids Res. 2011, 39, 6465–6474. [Google Scholar] [CrossRef] [PubMed]
- Loury, R.; Sassone-Corsi, P. Histone phosphorylation: How to proceed. Methods 2003, 31, 40–48. [Google Scholar] [CrossRef]
- Cohen, P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 1989, 58, 453–508. [Google Scholar] [CrossRef]
- Hirota, T.; Lipp, J.J.; Toh, B.-H.; Peters, J.-M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 2005, 438, 1176–1180. [Google Scholar] [CrossRef]
- Cohen, P. The origins of protein phosphorylation. Nat. Cell. Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef]
- Ubersax, J.A.; Ferrell, J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 530–541. [Google Scholar] [CrossRef]
- Cohen, P. Protein kinases--the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002, 1, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arab. Book/Am. Soc. Plant Biol. 2014, 12, e0174. [Google Scholar] [CrossRef]
- Kimura, Y.; Tanaka, K. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 2010, 147, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Jason, L.J.; Moore, S.C.; Lewis, J.D.; Lindsey, G.; Ausió, J. Histone ubiquitination: A tagging tail unfolds? Bioessays 2002, 24, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhai, L.; Xu, J.; Joo, H.Y.; Jackson, S.; Erdjument-Bromage, H.; Tempst, P.; Xiong, Y.; Zhang, Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 2006, 22, 383–394. [Google Scholar] [CrossRef]
- Vaughan, R.M.; Kupai, A.; Rothbart, S.B. Chromatin Regulation through Ubiquitin and Ubiquitin-like Histone Modifications. Trends Biochem. Sci. 2021, 46, 258–269. [Google Scholar] [CrossRef]
- Hoeller, D.; Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 2009, 458, 438–444. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, J. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 2003, 23, 5113–5121. [Google Scholar] [CrossRef]
- Witus, S.R.; Stewart, M.D.; Klevit, R.E. The BRCA1/BARD1 ubiquitin ligase and its substrates. Biochem. J. 2021, 478, 3467–3483. [Google Scholar] [CrossRef]
- Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.R.; Hochstrasser, M. SUMO-1: Ubiquitin gains weight. Trends Cell Biol. 1997, 7, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Rosonina, E.; Akhter, A.; Dou, Y.; Babu, J.; Sri Theivakadadcham, V.S. Regulation of transcription factors by sumoylation. Transcription 2017, 8, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Ryu, H.Y.; Ahn, S.H.; Hochstrasser, M. SUMO and cellular adaptive mechanisms. Exp. Mol. Med. 2020, 52, 931–939. [Google Scholar] [CrossRef]
- Rogers, G.E.; Simmonds, D.H. Content of citrulline and other amino-acids in a protein of hair follicles. Nature 1958, 182, 186–187. [Google Scholar] [CrossRef]
- Saiki, M.; Watase, M.; Matsubayashi, H.; Hidaka, Y. Recognition of the N-terminal histone H2A and H3 peptides by peptidylarginine deiminase IV. Protein Pept. Lett. 2009, 16, 1012–1016. [Google Scholar] [CrossRef]
- Wang, Y.; Wysocka, J.; Sayegh, J.; Lee, Y.H.; Perlin, J.R.; Leonelli, L.; Sonbuchner, L.S.; McDonald, C.H.; Cook, R.G.; Dou, Y.; et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004, 306, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Christophorou, M.A.; Castelo-Branco, G.; Halley-Stott, R.P.; Oliveira, C.S.; Loos, R.; Radzisheuskaya, A.; Mowen, K.A.; Bertone, P.; Silva, J.C.R.; Zernicka-Goetz, M.; et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 2014, 507, 104–108. [Google Scholar] [CrossRef]
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone deimination antagonizes arginine methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Zhang, M.; Li, T.; Muth, A.; Thompson, P.R.; Coonrod, S.A.; Zhang, X. Peptidylarginine deiminase 1-catalyzed histone citrullination is essential for early embryo development. Sci. Rep. 2016, 6, 38727. [Google Scholar] [CrossRef]
- Maronek, M.; Gardlik, R. The Citrullination-Neutrophil Extracellular Trap Axis in Chronic Diseases. J. Innate Immun. 2022, 14, 393–417. [Google Scholar] [CrossRef]
- Leidecker, O.; Bonfiglio, J.J.; Colby, T.; Zhang, Q.; Atanassov, I.; Zaja, R.; Palazzo, L.; Stockum, A.; Ahel, I.; Matic, I. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 2016, 12, 998–1000. [Google Scholar] [CrossRef]
- Laing, S.; Unger, M.; Koch-Nolte, F.; Haag, F. ADP-ribosylation of arginine. Amino Acids 2011, 41, 257–269. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Ding, M.; Yu, Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 2013, 10, 981–984. [Google Scholar] [CrossRef]
- Liu, C.; Vyas, A.; Kassab, M.A.; Singh, A.K.; Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 2017, 45, 8129–8141. [Google Scholar] [CrossRef]
- Brustel, J.; Muramoto, T.; Fumimoto, K.; Ellins, J.; Pears, C.J.; Lakin, N.D. Linking DNA repair and cell cycle progression through serine ADP-ribosylation of histones. Nat. Commun. 2022, 13, 185. [Google Scholar] [CrossRef]
- Messner, S.; Hottiger, M.O. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011, 21, 534–542. [Google Scholar] [CrossRef]
- Zhang, S.; Roche, K.; Nasheuer, H.-P.; Lowndes, N.F. Modification of Histones by Sugar β-N-Acetylglucosamine (GlcNAc) Occurs on Multiple Residues, Including Histone H3 Serine 10, and Is Cell Cycle-regulated. J. Biol. Chem. 2011, 286, 37483–37495. [Google Scholar] [CrossRef]
- Fong, J.J.; Nguyen, B.L.; Bridger, R.; Medrano, E.E.; Wells, L.; Pan, S.; Sifers, R.N. β-N-Acetylglucosamine (O-GlcNAc) Is a Novel Regulator of Mitosis-specific Phosphorylations on Histone H3. J. Biol. Chem. 2012, 287, 12195–12203. [Google Scholar] [CrossRef]
- Sakabe, K.; Hart, G.W. O-GlcNAc Transferase Regulates Mitotic Chromatin Dynamics. J. Biol. Chem. 2010, 285, 34460–34468. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Vocadlo, D.J.; Vosseller, K. Hyper-O-GlcNAcylation Is Anti-apoptotic and Maintains Constitutive NF-κB Activity in Pancreatic Cancer Cells. J. Biol. Chem. 2013, 288, 15121–15130. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, Y.; Bian, C.; Fujiki, R.; Yu, X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 2013, 493, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, X.; Gao, W.; Li, P.; Hou, J.; Li, J.; Wong, J. Differential Regulation of the Ten-Eleven Translocation (TET) Family of Dioxygenases by O-Linked β-N-Acetylglucosamine Transferase (OGT). J. Biol. Chem. 2014, 289, 5986–5996. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.P.; Zhang, K.; Wu, J.; Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015, 356, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.J.; Santos-Rosa, H.; Kouzarides, T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 2006, 126, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Howe, F.S.; Boubriak, I.; Sale, M.J.; Nair, A.; Clynes, D.; Grijzenhout, A.; Murray, S.C.; Woloszczuk, R.; Mellor, J. Lysine Acetylation Controls Local Protein Conformation by Influencing Proline Isomerization. Mol. Cell 2014, 55, 733–744. [Google Scholar] [CrossRef][Green Version]
- Wainwright, E.N.; Scaffidi, P. Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity. Trends Cancer 2017, 3, 372–386. [Google Scholar] [CrossRef]
- Easwaran, H.; Tsai, H.C.; Baylin, S.B. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef]
- Bates, S.E. Epigenetic Therapies for Cancer. N. Engl. J. Med. 2020, 383, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Mio, C.; Bulotta, S.; Russo, D.; Damante, G. Reading Cancer: Chromatin Readers as Druggable Targets for Cancer Treatment. Cancers 2019, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yu, N.K.; Kaang, B.K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015, 47, e166. [Google Scholar] [CrossRef]
- Nikolaev, L.G.; Akopov, S.B.; Didych, D.A.; Sverdlov, E.D. Vertebrate Protein CTCF and its Multiple Roles in a Large-Scale Regulation of Genome Activity. Curr. Genom. 2009, 10, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, A.B.; Schmitz, U.; Bailey, C.G.; Rasko, J.E.J. CTCF as a regulator of alternative splicing: New tricks for an old player. Nucleic Acids Res. 2021, 49, 7825–7838. [Google Scholar] [CrossRef]
- Jia, Z.; Li, J.; Ge, X.; Wu, Y.; Guo, Y.; Wu, Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol. 2020, 21, 75. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, M.; Pallotta, M.M.; Musio, A. The multifaceted roles of cohesin in cancer. J. Exp. Clin. Cancer Res. 2022, 41, 96. [Google Scholar] [CrossRef]
- Baranello, L.; Kouzine, F.; Levens, D. CTCF and cohesin cooperate to organize the 3D structure of the mammalian genome. Proc. Natl. Acad. Sci. USA 2014, 111, 889–890. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320. [Google Scholar] [CrossRef]
- Peters, J.M.; Tedeschi, A.; Schmitz, J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008, 22, 3089–3114. [Google Scholar] [CrossRef]
- Oh, S.; Oh, C.; Yoo, K.H. Functional roles of CTCF in breast cancer. BMB Rep. 2017, 50, 445–453. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, N.; Pati, D. Cohesin subunit RAD21: From biology to disease. Gene 2020, 758, 144966. [Google Scholar] [CrossRef] [PubMed]
- Sarogni, P.; Palumbo, O.; Servadio, A.; Astigiano, S.; D’Alessio, B.; Gatti, V.; Cukrov, D.; Baldari, S.; Pallotta, M.M.; Aretini, P.; et al. Overexpression of the cohesin-core subunit SMC1A contributes to colorectal cancer development. J. Exp. Clin. Cancer Res. 2019, 38, 108. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.K.; Kim, J.S.; Waldman, T. Cohesin mutations in human cancer. Biochim. Biophys. Acta 2016, 1866, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, T.; Luo, H.; Liu, Y.; Liu, D. Targeting Epigenetic Regulatory Enzymes for Cancer Therapeutics: Novel Small-Molecule Epidrug Development. Front. Oncol. 2022, 12, 848221. [Google Scholar] [CrossRef]
- Liu, N.; Zhao, R.; Ma, Y.; Wang, D.; Yan, C.; Zhou, D.; Yin, F.; Li, Z. The Development of Epigenetics and Related Inhibitors for Targeted Drug Design in Cancer Therapy. Curr. Top. Med. Chem. 2018, 18, 2380–2394. [Google Scholar] [CrossRef]
- Bennett, R.L.; Licht, J.D. Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 187–207. [Google Scholar] [CrossRef]
- Kantarjian, H.; Issa, J.P.; Rosenfeld, C.S.; Bennett, J.M.; Albitar, M.; DiPersio, J.; Klimek, V.; Slack, J.; de Castro, C.; Ravandi, F.; et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer 2006, 106, 1794–1803. [Google Scholar] [CrossRef]
- Silverman, L.R.; Demakos, E.P.; Peterson, B.L.; Kornblith, A.B.; Holland, J.C.; Odchimar-Reissig, R.; Stone, R.M.; Nelson, D.; Powell, B.L.; DeCastro, C.M.; et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J. Clin. Oncol. 2002, 20, 2429–2440. [Google Scholar] [CrossRef]
- Marquez, V.E.; Barchi, J.J., Jr.; Kelley, J.A.; Rao, K.V.; Agbaria, R.; Ben-Kasus, T.; Cheng, J.C.; Yoo, C.B.; Jones, P.A. Zebularine: A unique molecule for an epigenetically based strategy in cancer chemotherapy. The magic of its chemistry and biology. Nucleosides Nucleotides Nucleic Acids 2005, 24, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Rau, R.E.; Rodriguez, B.A.; Luo, M.; Jeong, M.; Rosen, A.; Rogers, J.H.; Campbell, C.T.; Daigle, S.R.; Deng, L.; Song, Y.; et al. DOT1L as a therapeutic target for the treatment of DNMT3A-mutant acute myeloid leukemia. Blood 2016, 128, 971–981. [Google Scholar] [CrossRef]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Basavapathruni, A.; Jin, L.; Boriack-Sjodin, P.A.; Allain, C.J.; Klaus, C.R.; Raimondi, A.; Scott, M.P.; et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 2013, 122, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Chory, E.J.; Wernimont, A.K.; Tempel, W.; Scopton, A.; Federation, A.; Marineau, J.J.; Qi, J.; Barsyte-Lovejoy, D.; Yi, J.; et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 2012, 3, 1288. [Google Scholar] [CrossRef]
- Konze, K.D.; Ma, A.; Li, F.; Barsyte-Lovejoy, D.; Parton, T.; Macnevin, C.J.; Liu, F.; Gao, C.; Huang, X.P.; Kuznetsova, E.; et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013, 8, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Yang, X.; Zhuang, L.; Jiang, X.; Chen, W.; Lee, P.L.; Karuturi, R.K.; Tan, P.B.; Liu, E.T.; Yu, Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007, 21, 1050–1063. [Google Scholar] [CrossRef]
- McCabe, M.T.; Ott, H.M.; Ganji, G.; Korenchuk, S.; Thompson, C.; Van Aller, G.S.; Liu, Y.; Graves, A.P.; Iii, A.D.P.; Diaz, E.; et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012, 492, 108–112. [Google Scholar] [CrossRef]
- Sarkozy, C.; Morschhauser, F.; Dubois, S.; Molina, T.; Michot, J.M.; Cullières-Dartigues, P.; Suttle, B.; Karlin, L.; Le Gouill, S.; Picquenot, J.M.; et al. A LYSA Phase Ib Study of Tazemetostat (EPZ-6438) plus R-CHOP in Patients with Newly Diagnosed Diffuse Large B-Cell Lymphoma (DLBCL) with Poor Prognosis Features. Clin. Cancer Res. 2020, 26, 3145–3153. [Google Scholar] [CrossRef]
- Qi, W.; Chan, H.; Teng, L.; Li, L.; Chuai, S.; Zhang, R.; Zeng, J.; Li, M.; Fan, H.; Lin, Y.; et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA 2012, 109, 21360–21365. [Google Scholar] [CrossRef]
- Knutson, S.K.; Wigle, T.J.; Warholic, N.M.; Sneeringer, C.J.; Allain, C.J.; Klaus, C.R.; Sacks, J.D.; Raimondi, A.; Majer, C.R.; Song, J.; et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012, 8, 890–896. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, X.; Horton, J.R.; Upadhyay, A.K.; Spannhoff, A.; Liu, J.; Snyder, J.P.; Bedford, M.T.; Cheng, X. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat. Struct. Mol. Biol. 2009, 16, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wang, Q.; Paulk, J.; Kubicek, S.; Kemp, M.M.; Adams, D.J.; Shamji, A.F.; Wagner, B.K.; Schreiber, S.L. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem. Biol. 2012, 7, 1152–1157. [Google Scholar] [CrossRef]
- Vedadi, M.; Barsyte-Lovejoy, D.; Liu, F.; Rival-Gervier, S.; Allali-Hassani, A.; Labrie, V.; Wigle, T.J.; Dimaggio, P.A.; Wasney, G.A.; Siarheyeva, A.; et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 2011, 7, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Greiner, D.; Bonaldi, T.; Eskeland, R.; Roemer, E.; Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat. Chem. Biol. 2005, 1, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.; Fung, T.K.; Zeisig, B.B.; Holmes, K.; Rane, J.K.; Mowen, K.A.; Finn, M.G.; Lenhard, B.; Chan, L.C.; So, C.W. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 2016, 29, 32–48. [Google Scholar] [CrossRef]
- Eram, M.S.; Shen, Y.; Szewczyk, M.; Wu, H.; Senisterra, G.; Li, F.; Butler, K.V.; Kaniskan, H.; Speed, B.A.; Dela Seña, C.; et al. A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases. ACS Chem. Biol. 2016, 11, 772–781. [Google Scholar] [CrossRef]
- Santer, F.R.; Höschele, P.P.; Oh, S.J.; Erb, H.H.; Bouchal, J.; Cavarretta, I.T.; Parson, W.; Meyers, D.J.; Cole, P.A.; Culig, Z. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol. Cancer Ther. 2011, 10, 1644–1655. [Google Scholar] [CrossRef]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef]
- Ullah, M.F. Sulforaphane (SFN): An Isothiocyanate in a Cancer Chemoprevention Paradigm. Medicines 2015, 2, 141–156. [Google Scholar] [CrossRef]
- von Tresckow, B.; Sayehli, C.; Aulitzky, W.E.; Goebeler, M.E.; Schwab, M.; Braz, E.; Krauss, B.; Krauss, R.; Hermann, F.; Bartz, R.; et al. Phase I study of domatinostat (4SC-202), a class I histone deacetylase inhibitor in patients with advanced hematological malignancies. Eur. J. Haematol. 2019, 102, 163–173. [Google Scholar] [CrossRef]
- Brunetto, A.T.; Ang, J.E.; Lal, R.; Olmos, D.; Molife, L.R.; Kristeleit, R.; Parker, A.; Casamayor, I.; Olaleye, M.; Mais, A.; et al. First-in-human, pharmacokinetic and pharmacodynamic phase I study of Resminostat, an oral histone deacetylase inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 2013, 19, 5494–5504. [Google Scholar] [CrossRef] [PubMed]
- Assouline, S.E.; Nielsen, T.H.; Yu, S.; Alcaide, M.; Chong, L.; MacDonald, D.; Tosikyan, A.; Kukreti, V.; Kezouh, A.; Petrogiannis-Haliotis, T.; et al. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood 2016, 128, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.A.; Kim, Y.H.; Kuzel, T.M.; Pacheco, T.R.; Foss, F.M.; Parker, S.; Frankel, S.R.; Chen, C.; Ricker, J.L.; Arduino, J.M.; et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 2007, 25, 3109–3115. [Google Scholar] [CrossRef] [PubMed]
- Piekarz, R.L.; Frye, R.; Prince, H.M.; Kirschbaum, M.H.; Zain, J.; Allen, S.L.; Jaffe, E.S.; Ling, A.; Turner, M.; Peer, C.J.; et al. Phase 2 trial of romidepsin in patients with peripheral T-cell lymphoma. Blood 2011, 117, 5827–5834. [Google Scholar] [CrossRef] [PubMed]
- Knipstein, J.; Gore, L. Entinostat for treatment of solid tumors and hematologic malignancies. Expert Opin. Investig. Drugs 2011, 20, 1455–1467. [Google Scholar] [CrossRef]
- Younes, A.; Oki, Y.; Bociek, R.G.; Kuruvilla, J.; Fanale, M.; Neelapu, S.; Copeland, A.; Buglio, D.; Galal, A.; Besterman, J.; et al. Mocetinostat for relapsed classical Hodgkin’s lymphoma: An open-label, single-arm, phase 2 trial. Lancet Oncol 2011, 12, 1222–1228. [Google Scholar] [CrossRef]
- Abaza, Y.M.; Kadia, T.M.; Jabbour, E.J.; Konopleva, M.Y.; Borthakur, G.; Ferrajoli, A.; Estrov, Z.; Wierda, W.G.; Alfonso, A.; Chong, T.H.; et al. Phase 1 dose escalation multicenter trial of pracinostat alone and in combination with azacitidine in patients with advanced hematologic malignancies. Cancer 2017, 123, 4851–4859. [Google Scholar] [CrossRef]
- Lee, H.Z.; Kwitkowski, V.E.; Del Valle, P.L.; Ricci, M.S.; Saber, H.; Habtemariam, B.A.; Bullock, J.; Bloomquist, E.; Li Shen, Y.; Chen, X.H.; et al. FDA Approval: Belinostat for the Treatment of Patients with Relapsed or Refractory Peripheral T-cell Lymphoma. Clin. Cancer Res. 2015, 21, 2666–2670. [Google Scholar] [CrossRef]
- von Tresckow, B.; Gundermann, S.; Eichenauer, D.A.; Aulitzky, W.E.; Göbeler, M.; Sayehli, C.; Bacchus, L.; Hauns, B.; Mais, A.; Hentsch, B.; et al. First-in-human study of 4SC-202, a novel oral HDAC inhibitor in advanced hematologic malignancies (TOPAS study). J. Clin. Oncol. 2014, 32, 8559. [Google Scholar] [CrossRef]
- Evens, A.M.; Balasubramanian, S.; Vose, J.M.; Harb, W.; Gordon, L.I.; Langdon, R.; Sprague, J.; Sirisawad, M.; Mani, C.; Yue, J.; et al. A Phase I/II Multicenter, Open-Label Study of the Oral Histone Deacetylase Inhibitor Abexinostat in Relapsed/Refractory Lymphoma. Clin. Cancer Res. 2016, 22, 1059–1066. [Google Scholar] [CrossRef]
- Kim, K.P.; Park, S.J.; Kim, J.E.; Hong, Y.S.; Lee, J.L.; Bae, K.S.; Cha, H.; Kwon, S.K.; Ro, S.; Cho, J.; et al. First-in-human study of the toxicity, pharmacokinetics, and pharmacodynamics of CG200745, a pan-HDAC inhibitor, in patients with refractory solid malignancies. Investig. New Drugs 2015, 33, 1048–1057. [Google Scholar] [CrossRef]
- Banerji, U.; van Doorn, L.; Papadatos-Pastos, D.; Kristeleit, R.; Debnam, P.; Tall, M.; Stewart, A.; Raynaud, F.; Garrett, M.D.; Toal, M.; et al. A phase I pharmacokinetic and pharmacodynamic study of CHR-3996, an oral class I selective histone deacetylase inhibitor in refractory solid tumors. Clin. Cancer Res. 2012, 18, 2687–2694. [Google Scholar] [CrossRef] [PubMed]
- Galloway, T.J.; Wirth, L.J.; Colevas, A.D.; Gilbert, J.; Bauman, J.E.; Saba, N.F.; Raben, D.; Mehra, R.; Ma, A.W.; Atoyan, R.; et al. A Phase I Study of CUDC-101, a Multitarget Inhibitor of HDACs, EGFR, and HER2, in Combination with Chemoradiation in Patients with Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2015, 21, 1566–1573. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Lai, C.J.; Bao, R.; Wang, D.G.; Wang, J.; Xu, G.X.; Atoyan, R.; Qu, H.; Yin, L.; Samson, M.; et al. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin. Cancer Res. 2012, 18, 4104–4113. [Google Scholar] [CrossRef]
- Furlan, A.; Monzani, V.; Reznikov, L.L.; Leoni, F.; Fossati, G.; Modena, D.; Mascagni, P.; Dinarello, C.A. Pharmacokinetics, safety and inducible cytokine responses during a phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat). Mol. Med. 2011, 17, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Galli, M.; Salmoiraghi, S.; Golay, J.; Gozzini, A.; Crippa, C.; Pescosta, N.; Rambaldi, A. A phase II multiple dose clinical trial of histone deacetylase inhibitor ITF2357 in patients with relapsed or progressive multiple myeloma. Ann. Hematol. 2010, 89, 185–190. [Google Scholar] [CrossRef]
- Zwergel, C.; Valente, S.; Jacob, C.; Mai, A. Emerging approaches for histone deacetylase inhibitor drug discovery. Expert Opin. Drug Discov. 2015, 10, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, B.; Baird, R.; Kristeleit, R.S.; Plummer, R.; Cowan, R.; Stewart, A.; Fourneau, N.; Hellemans, P.; Elsayed, Y.; McClue, S.; et al. A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clin. Cancer Res. 2013, 19, 4262–4272. [Google Scholar] [CrossRef]
- Shi, Y.; Dong, M.; Hong, X.; Zhang, W.; Feng, J.; Zhu, J.; Yu, L.; Ke, X.; Huang, H.; Shen, Z.; et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann. Oncol. 2015, 26, 1766–1771. [Google Scholar] [CrossRef]
- Santo, L.; Hideshima, T.; Kung, A.L.; Tseng, J.C.; Tamang, D.; Yang, M.; Jarpe, M.; van Duzer, J.H.; Mazitschek, R.; Ogier, W.C.; et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012, 119, 2579–2589. [Google Scholar] [CrossRef]
- Pauer, L.R.; Olivares, J.; Cunningham, C.; Williams, A.; Grove, W.; Kraker, A.; Olson, S.; Nemunaitis, J. Phase I study of oral CI-994 in combination with carboplatin and paclitaxel in the treatment of patients with advanced solid tumors. Cancer Investig. 2004, 22, 886–896. [Google Scholar] [CrossRef]
- Guzman, M.L.; Yang, N.; Sharma, K.K.; Balys, M.; Corbett, C.A.; Jordan, C.T.; Becker, M.W.; Steidl, U.; Abdel-Wahab, O.; Levine, R.L.; et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: A novel potential strategy in acute myelogenous leukemia. Mol. Cancer Ther. 2014, 13, 1979–1990. [Google Scholar] [CrossRef] [PubMed]
- Iannitti, T.; Palmieri, B. Clinical and experimental applications of sodium phenylbutyrate. Drugs R D 2011, 11, 227–249. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.; Valone, F.; Lipera, W.; Irwin, D.; Paroly, W.; Natale, R.; Sreedharan, S.; Keer, H.; Lum, B.; Scappaticci, F.; et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer 2004, 45, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Bilen, M.A.; Fu, S.; Falchook, G.S.; Ng, C.S.; Wheler, J.J.; Abdelrahim, M.; Erguvan-Dogan, B.; Hong, D.S.; Tsimberidou, A.M.; Kurzrock, R.; et al. Phase I trial of valproic acid and lenalidomide in patients with advanced cancer. Cancer Chemother. Pharmacol. 2015, 75, 869–874. [Google Scholar] [CrossRef]
- Zheng, Y.C.; Yu, B.; Jiang, G.Z.; Feng, X.J.; He, P.X.; Chu, X.Y.; Zhao, W.; Liu, H.M. Irreversible LSD1 Inhibitors: Application of Tranylcypromine and Its Derivatives in Cancer Treatment. Curr. Top. Med. Chem. 2016, 16, 2179–2188. [Google Scholar] [CrossRef]
- Maes, T.; Carceller, E.; Salas, J.; Ortega, A.; Buesa, C. Advances in the development of histone lysine demethylase inhibitors. Curr. Opin. Pharmacol. 2015, 23, 52–60. [Google Scholar] [CrossRef]
- Sugino, N.; Kawahara, M.; Tatsumi, G.; Kanai, A.; Matsui, H.; Yamamoto, R.; Nagai, Y.; Fujii, S.; Shimazu, Y.; Hishizawa, M.; et al. A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia 2017, 31, 2303–2314. [Google Scholar] [CrossRef]
- Heinemann, B.; Nielsen, J.M.; Hudlebusch, H.R.; Lees, M.J.; Larsen, D.V.; Boesen, T.; Labelle, M.; Gerlach, L.-O.; Birk, P.; Helin, K. Inhibition of demethylases by GSK-J1/J4. Nature 2014, 514, E1–E2. [Google Scholar] [CrossRef]
- Hancock, R.L.; Dunne, K.; Walport, L.J.; Flashman, E.; Kawamura, A. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics 2015, 7, 791–811. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef]
- Chaidos, A.; Caputo, V.; Gouvedenou, K.; Liu, B.; Marigo, I.; Chaudhry, M.S.; Rotolo, A.; Tough, D.F.; Smithers, N.N.; Bassil, A.K.; et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood 2014, 123, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Nepali, K.; Liou, J.-P. Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 2021, 28, 27. [Google Scholar] [CrossRef]
- Valdez, B.C.; Li, Y.; Murray, D.; Corn, P.; Champlin, R.E.; Andersson, B.S. 5-Aza-2’-deoxycytidine sensitizes busulfan-resistant myeloid leukemia cells by regulating expression of genes involved in cell cycle checkpoint and apoptosis. Leuk. Res. 2010, 34, 364–372. [Google Scholar] [CrossRef]
- Fu, S.; Hu, W.; Iyer, R.; Kavanagh, J.J.; Coleman, R.L.; Levenback, C.F.; Sood, A.K.; Wolf, J.K.; Gershenson, D.M.; Markman, M.; et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer 2011, 117, 1661–1669. [Google Scholar] [CrossRef]
- Singal, R.; Ramachandran, K.; Gordian, E.; Quintero, C.; Zhao, W.; Reis, I.M. Phase I/II study of azacitidine, docetaxel, and prednisone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel-based therapy. Clin. Genitourin. Cancer 2015, 13, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P.; Garcia-Manero, G.; Giles, F.J.; Mannari, R.; Thomas, D.; Faderl, S.; Bayar, E.; Lyons, J.; Rosenfeld, C.S.; Cortes, J.; et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 2004, 103, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Covre, A.; Coral, S.; Di Giacomo, A.M.; Taverna, P.; Azab, M.; Maio, M. Epigenetics meets immune checkpoints. Semin. Oncol. 2015, 42, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Maio, M.; Covre, A.; Fratta, E.; Di Giacomo, A.M.; Taverna, P.; Natali, P.G.; Coral, S.; Sigalotti, L. Molecular Pathways: At the Crossroads of Cancer Epigenetics and Immunotherapy. Clin. Cancer Res. 2015, 21, 4040–4047. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef]
- Dang, L.; Yen, K.; Attar, E.C. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann. Oncol. 2016, 27, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Damaschke, N.A.; Gawdzik, J.; Avilla, M.; Yang, B.; Svaren, J.; Roopra, A.; Luo, J.H.; Yu, Y.P.; Keles, S.; Jarrard, D.F. CTCF loss mediates unique DNA hypermethylation landscapes in human cancers. Clin. Epigenet. 2020, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.G.; Metierre, C.; Feng, Y.; Baidya, K.; Filippova, G.N.; Loukinov, D.I.; Lobanenkov, V.V.; Semaan, C.; Rasko, J.E. CTCF Expression is Essential for Somatic Cell Viability and Protection Against Cancer. Int. J. Mol. Sci. 2018, 19, 3832. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef]
- Campbell, P.; Thomas, C.M. Belinostat for the treatment of relapsed or refractory peripheral T-cell lymphoma. J. Oncol. Pharm. Pract. 2017, 23, 143–147. [Google Scholar] [CrossRef]
- Richon, V.M.; Sandhoff, T.W.; Rifkind, R.A.; Marks, P.A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 2000, 97, 10014–10019. [Google Scholar] [CrossRef] [PubMed]
- Xue, K.; Gu, J.J.; Zhang, Q.; Mavis, C.; Hernandez-Ilizaliturri, F.J.; Czuczman, M.S.; Guo, Y. Vorinostat, a histone deacetylase (HDAC) inhibitor, promotes cell cycle arrest and re-sensitizes rituximab- and chemo-resistant lymphoma cells to chemotherapy agents. J. Cancer Res. Clin. Oncol. 2016, 142, 379–387. [Google Scholar] [CrossRef]
- Garnock-Jones, K.P. Panobinostat: First global approval. Drugs 2015, 75, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Odenike, O. Emerging role of the histone deacetylase inhibitor romidepsin in hematologic malignancies. Expert Opin. Pharmacother. 2010, 11, 3073–3084. [Google Scholar] [CrossRef]
- Wang, Y.M.; Gu, M.L.; Meng, F.S.; Jiao, W.R.; Zhou, X.X.; Yao, H.P.; Ji, F. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int. J. Oncol. 2017, 51, 1860–1868. [Google Scholar] [CrossRef]
- Oike, T.; Komachi, M.; Ogiwara, H.; Amornwichet, N.; Saitoh, Y.; Torikai, K.; Kubo, N.; Nakano, T.; Kohno, T. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiother. Oncol. 2014, 111, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Heimbruch, K.E.; Fisher, J.B.; Stelloh, C.T.; Phillips, E.; Reimer, M.H., Jr.; Wargolet, A.J.; Meyer, A.E.; Pulakanti, K.; Viny, A.D.; Loppnow, J.J.; et al. DOT1L inhibitors block abnormal self-renewal induced by cohesin loss. Sci. Rep. 2021, 11, 7288. [Google Scholar] [CrossRef] [PubMed]
- Mould, D.P.; McGonagle, A.E.; Wiseman, D.H.; Williams, E.L.; Jordan, A.M. Reversible inhibitors of LSD1 as therapeutic agents in acute myeloid leukemia: Clinical significance and progress to date. Med. Res. Rev. 2015, 35, 586–618. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Yao, Y.; Zhou, C.; Chen, F.; Wu, F.; Wei, L.; Liu, W.; Dong, S.; Redell, M.; Mo, Q.; et al. Pharmacological inhibition of LSD1 for the treatment of MLL-rearranged leukemia. J. Hematol. Oncol. 2016, 9, 24. [Google Scholar] [CrossRef]
- Agboyibor, C.; Dong, J.; Effah, C.Y.; Drokow, E.K.; Pervaiz, W.; Liu, H.M. LSD1 as a Biomarker and the Outcome of Its Inhibitors in the Clinical Trial: The Therapy Opportunity in Tumor. J. Oncol. 2021, 2021, 5512524. [Google Scholar] [CrossRef] [PubMed]

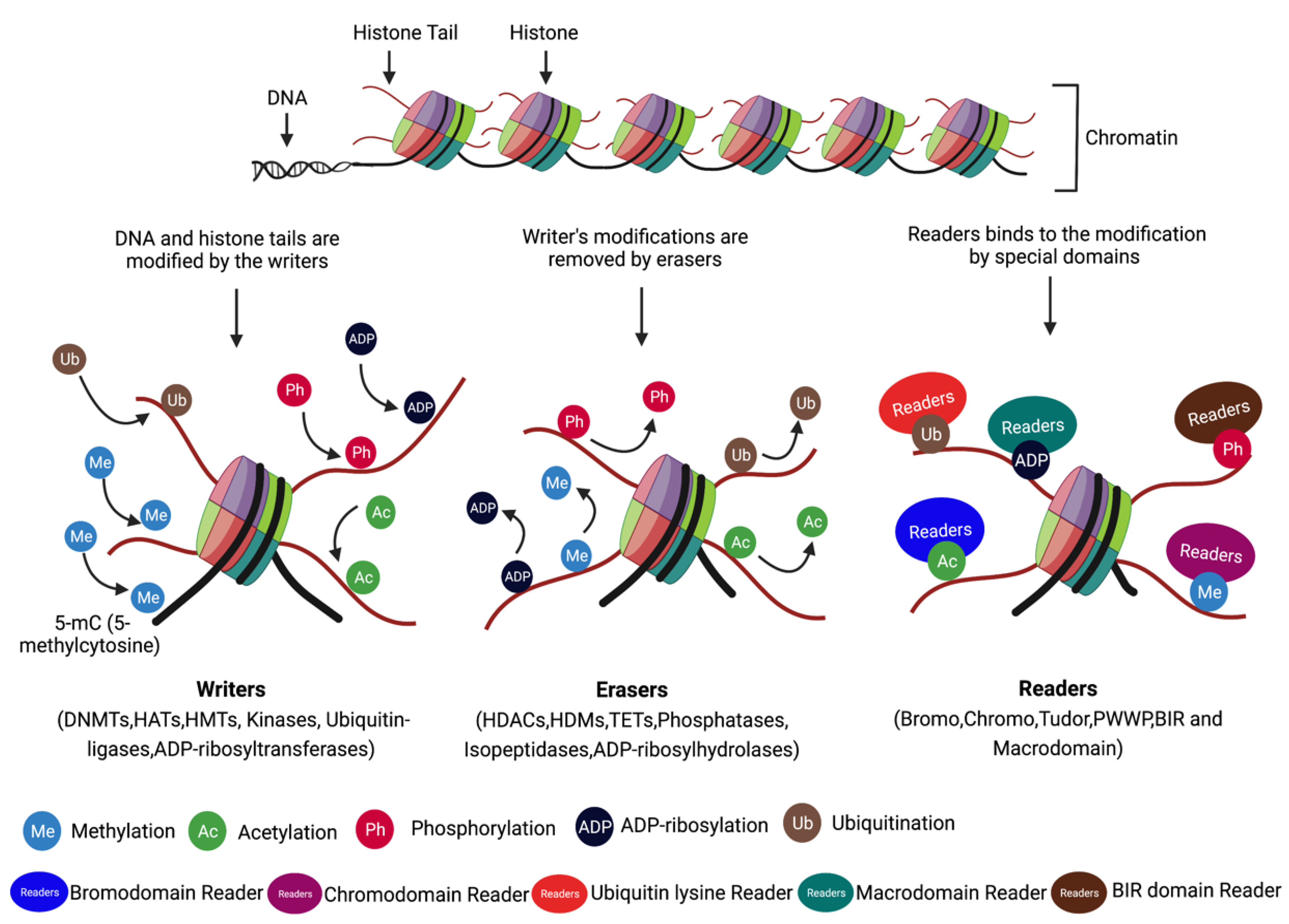
| Modification | Gene/Components | Enzyme/Action |
|---|---|---|
| Writers | DNMT1 | DNA methyltransferase |
| DNMT3a | DNA methyltransferase | |
| DNMT3b | DNA methyltransferase | |
| EZH1/2 | Histone methyltransferase | |
| DOT1L/KMT4 | Histone methyltransferase | |
| EP300 (P300/KAT3B) | Histone acetyltransferase | |
| CREBBP (CBP/KAT3A) | Histone acetyltransferase | |
| GNAT family (GCN5) | Histone acetyltransferase | |
| MYST family (TIP60/KAT5) | Histone acetyltransferase | |
| EHMT1/2 | Histone methyltransferase | |
| SUV39H1/2 | Histone methyltransferase | |
| PRMT1/2/4/5/6/7 | Histone methyltransferase | |
| KMT7 | Histone methyltransferase | |
| AuroraB (AURKB) | Kinases | |
| WSTF (BAZ1B) | Kinases | |
| BRCA1-BARD1 | Ubiquitin-ligases | |
| PARP1 | ADP-ribosyltransferases | |
| Erasers | TET1/TET2 | DNA demethylation |
| HDAC 1-3,8 (Class I) | Histone deacetylase | |
| HDAC 4,5,7,9 (Class IIa) | Histone deacetylase | |
| HDAC 6,10 (Class IIb) | Histone deacetylase | |
| HDAC Class III: SIRT 1-7 | Histone deacetylase | |
| HDAC 11 (Class IV) | Histone deacetylase | |
| KDM1A/LSD1, KDM1B/LSD2 | Histone demethylase | |
| KDM2A/2B, KDM3A/3B | Histone demethylase | |
| KDM4A/4B/4C/4D/4E | Histone demethylase | |
| KDM5A/5B/5C/5D | Histone demethylase | |
| KDM6A/6B/7/8 | Histone demethylase | |
| PP1 (PPP1CA)/PP2 (PPP2CA) | Phosphatases | |
| PPgamma (PPARG) | Phosphatases | |
| EYA1/3 | Phosphatases | |
| OTUB1/2, BRCC36, USP3/16/26/44 | Isopeptidases | |
| PARG, MDO1/2, TARG | ADP-ribosylhydrolases | |
| Readers | MORF, MRG15 (chromodomain) | Reads methylation |
| MBT, PHF1/19, TDRD7 (Tudor domain) | Reads methylation | |
| BRPF1, NSD1-3 (PWWP domain) | Reads methylation | |
| G9a/GLP (Ankyrin repeats) | Reads methylation | |
| BRD2/3/4/T (bromodomain) | Reads acetylation | |
| XRCC1, NBS1, BARD1 (BIR domain) | Reads Phosphorylation | |
| 14-3-3β/γ/η/ε/μ(14-3-3 proteins BRCT domain) | Reads Phosphorylation | |
| 53BP1 | Ubiquitin lysine reader | |
| RNF146 (Macrodomains) | Reads ADP-ribosylation | |
| APLF, CHFR (PBZ) | Reads ADP-ribosylation |
| Category | Epigenetic Regulation | Target | Cancer Types | Known Compound | Reference/https://clinicaltrials.gov Identifier |
|---|---|---|---|---|---|
| Writers | DNMT | DNMT1 | MDS, AML | 5-aza-2′-deoxycytidine (5-aza-CdR; decitabine, Dacogen®) | [109] |
| DNMT1 | MDS, AML | 5-azacytidine (5-aza-CR; Aza; Vidaza®) | [110] | ||
| DNMT1, DNMT3, | Hematological malignancies, and solid tumors | Zebularine (NSC309132; 4-deoxyuridine) | [111] | ||
| DNMT1 | Hematological malignancies and solid tumors | Guadecitabine (SGI-110) | NCT01261312, NCT01752933 | ||
| HMT | DOT1L | Hematological malignancies | EPZ00477 | [112] | |
| DOT1L | Hematological malignancies | Pinometostat (EPZ-5676) | [113] | ||
| DOT1L | Leukemia | SGC0946 | [114] | ||
| EZH1 | DLBCL | UNC1999 | [115] | ||
| EZH2 | AML | 3-Deazaneplanocin A (DZnep) | [116] | ||
| EZH2 | B-cell lymphoma | GSK126 | [117] | ||
| EZH2 | B-cell lymphoma | Tazemetostat (EPZ-6348) | [118] | ||
| EZH2 | DLBCL | EI1 | [119] | ||
| EZH2 | Non-Hodgkin lymphoma | EPZ005687 | [120] | ||
| EZH2 | Ovarian cancer | GSK343 | [117] | ||
| EZH2 | Breast, colon, prostate cancer | DZNep | [116] | ||
| G9a/EHMT2 | Leukemia, bladder cancer | BIX-01294 | [121] | ||
| G9a/EHMT2 | Pancreatic cancer | BRD4770 | [122] | ||
| G9a/EHMT2 | AML, breast cancer | UNC0638 | [123] | ||
| SUV39H1 | Lymphomas | Chaetocin | [124] | ||
| PRMT5 | Solid tumors, non-Hodgkin lymphoma | GSK3326595 | NCT02783300 | ||
| PRMT1 | AML | AMI-408 | [125] | ||
| PRMT1, 3, 4, 6, 8 | TBD | MS023 | [126] | ||
| HAT | p300 | Prostate cancer | C646 | [127] | |
| p300, CBP | MM, breast cancer, pancreatic cancer | Curcumin | [128] | ||
| Erasers | HDAC | Class I, II, IV | Leukemia, colorectal cancer, prostate cancer, and other solid tumors | Sulforaphane (SFN) | [129] |
| Class I | Leukemia, colorectal cancer | Domatinostat (4SC-202) | [130] | ||
| Class I, II, IV | Leukemia, colorectal cancer, head and neck cancer, hepatocellular carcinoma | Resminostat (4SC-201, RAS2410) | [131] | ||
| Class I, II, IV | CTCL, Hodgkin’s lymphoma, breast cancer, head and neck cancer, prostate cancer, colorectal cancer, thyroid cancer | Panobinostat (LBH589) | [132] | ||
| Class I, II, IV | CTCL, leukemia, prostate cancer, bladder cancer, breast cancer | Vorinostat (SAHA, Zolinza®) | [133] | ||
| Class I, II, IV | CTCL | Romidepsin (depsipeptide, FK228) | [134] | ||
| Class I (HDAC1, 9, 11) | Hodgkin lymphoma, kidney cancer, breast cancer | Entinostat (MS-275, SNDX-275) | [135] | ||
| Class I, IV | Follicular lymphoma, Hodgkin’s lymphoma and AML, CLL, MDS, solid tumors | Mocetinostat (MGCD0103) | [136] | ||
| Class I, II, IV | MDS, AML | Pracinostat (SB939) | [137] | ||
| Class I, II, IV | Leukemia, colorectal cancer, lung cancer, pancreatic cancer | Belinostat (PXD101) | [138] | ||
| HDAC-LSD1 | Hematological malignancies | HDAC-LSD1 4SC-202 | [139] | ||
| Class I, II | Clinical trials: Hodgkin lymphoma, non-Hodgkin lymphoma, CLL | Abexinostat | [140] | ||
| Class I, II, IV | Clinical trials: solid tumors | CG200745 | [141] | ||
| Class I | Clinical trials: solid tumors | CHR-3996 | [142] | ||
| Class I, II | Clinical trials: SCC | CUDC-101 | [143] | ||
| Class I, II | Clinical trials: MM, lymphoma, solid tumors | CUDC-907 | [144] | ||
| Class I, II | Clinical trials: CLL, MM, Hodgkin lymphoma | Givinostat | [145,146] | ||
| Class I, II (HDAC1, 2, 6) | Clinical trials: solid tumors | MPT0E028 | [147] | ||
| Class I, II | Clinical trials: solid tumors, lymphoma, CTCL | Quisinostat | [148] | ||
| Class I, II (HDAC1, 2, 3, 10) | Clinical trials: breast cancer, NSCLC | Chidamide | [149] | ||
| Class II (HDAC6) | Clinical trials: MM, lymphoma | Ricolinostat | [150] | ||
| Class I | Clinical trials: MM, lung cancer, pancreatic cancer | Tacedinaline | [151] | ||
| Class I (HDAC1, 2) | Approved: CTCL, PTCL | Romidepsin | [134] | ||
| Class I, II | Clinical trials: AML, MM | AR-42 | [152] | ||
| Class I, II | Clinical trials: solid tumors, hematological malignancies | Phenylbutyrate | [153] | ||
| Class I, II | Clinical trials: NSCLC, MM, CLL | Pivanex | [154] | ||
| Class I, II | Clinical trials: solid tumors, hematological malignancies | Valproic acid | [155] | ||
| HDM | LSD1 | AML, small cell lung cancer | GSK2879552 | [156] | |
| LSD1 | AML | GSK354, GSK690 | [157] | ||
| LSD1 | MDS | NCD25, NCD38 | [158] | ||
| LSD1 | Acute leukemia | ORY-1001 | [156] | ||
| LSD1 | MDS, AML | Tranylcypromine | [156] | ||
| HDAC-LSD1 | Hematological malignancies | 4SC-202 | [139] | ||
| JmjC domain proteins | TBD | GSK-J1, GSK-J4 | [159] | ||
| KDM5B | Hematological malignancies and solid tumors | EPT-103182 | [160] | ||
| Readers | BET | BET proteins | Prostate cancer, AML with mixed lineage leukemia translocations, MM, NUT midline carcinoma | JQ1 | [161] |
| BET proteins | Clinical trials: Hematological malignancies, NUT midline carcinoma, solid tumors | I-BET762 | NCT01943851, NCT01587703, [162] | ||
| BRD2, 3, 4 | Clinical trials: AML | OTX015 | NCT01713582 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, A.; Jha, S. Epimutations and Their Effect on Chromatin Organization: Exciting Avenues for Cancer Treatment. Cancers 2023, 15, 215. https://doi.org/10.3390/cancers15010215
Mohammad A, Jha S. Epimutations and Their Effect on Chromatin Organization: Exciting Avenues for Cancer Treatment. Cancers. 2023; 15(1):215. https://doi.org/10.3390/cancers15010215
Chicago/Turabian StyleMohammad, Asad, and Sudhakar Jha. 2023. "Epimutations and Their Effect on Chromatin Organization: Exciting Avenues for Cancer Treatment" Cancers 15, no. 1: 215. https://doi.org/10.3390/cancers15010215
APA StyleMohammad, A., & Jha, S. (2023). Epimutations and Their Effect on Chromatin Organization: Exciting Avenues for Cancer Treatment. Cancers, 15(1), 215. https://doi.org/10.3390/cancers15010215







