The Structural Dynamics, Complexity of Interactions, and Functions in Cancer of Multi-SAM Containing Proteins
Abstract
Simple Summary
Abstract
1. Introduction
2. Domain Architectures of the MSCPs
3. Structural Dynamics of the SAM Domains in MSCPs
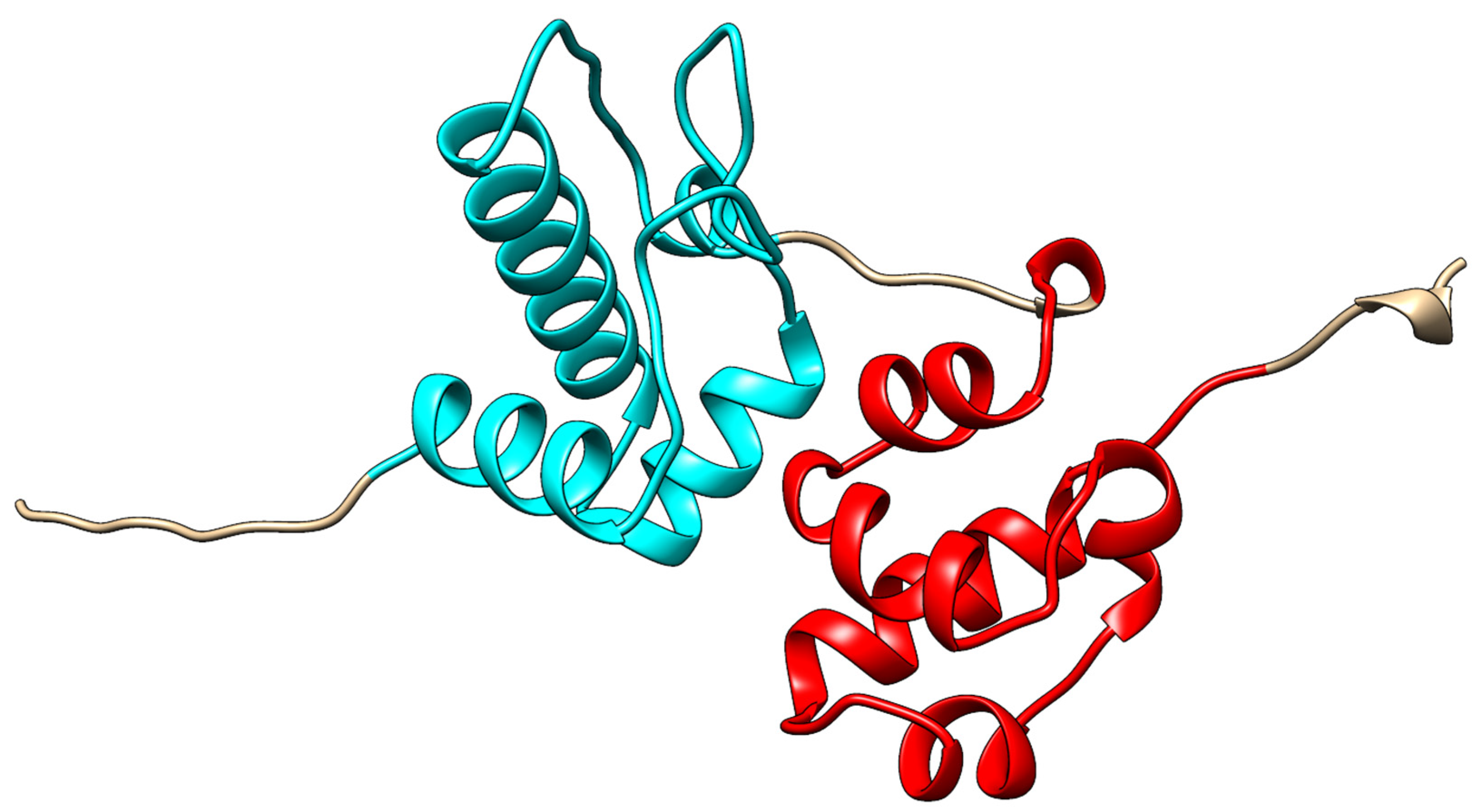
3.1. Structure and Interaction Arrangements of Singular SAM Domains
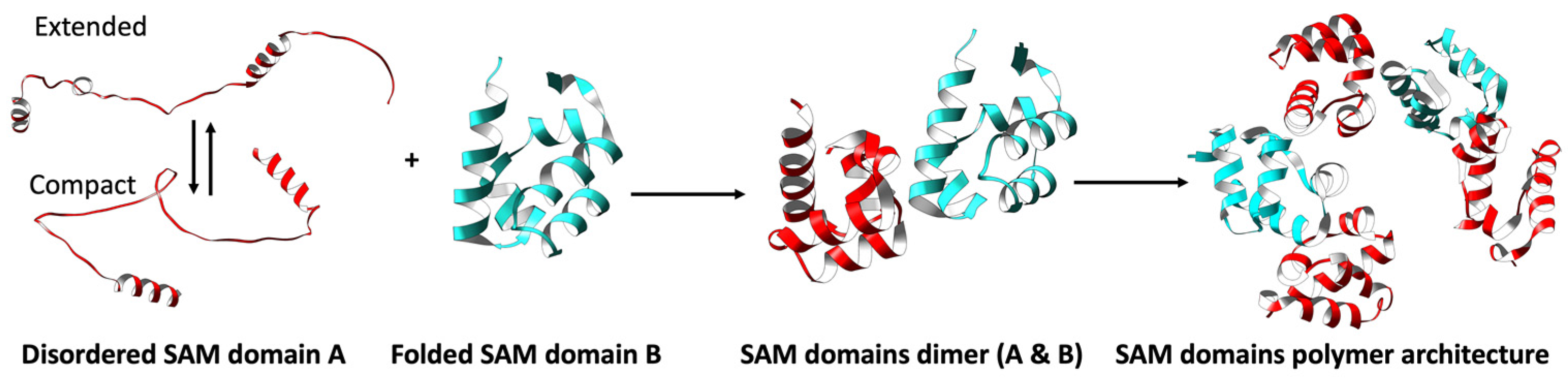
3.2. Disordered and Unique Scaffolding in Multi-SAM Domains
3.2.1. Intrinsic Disorder in Multi-SAMs and Protein Function
3.2.2. Multi-SAM Domains Increase the Complexity of SAM-Mediated Oligomers
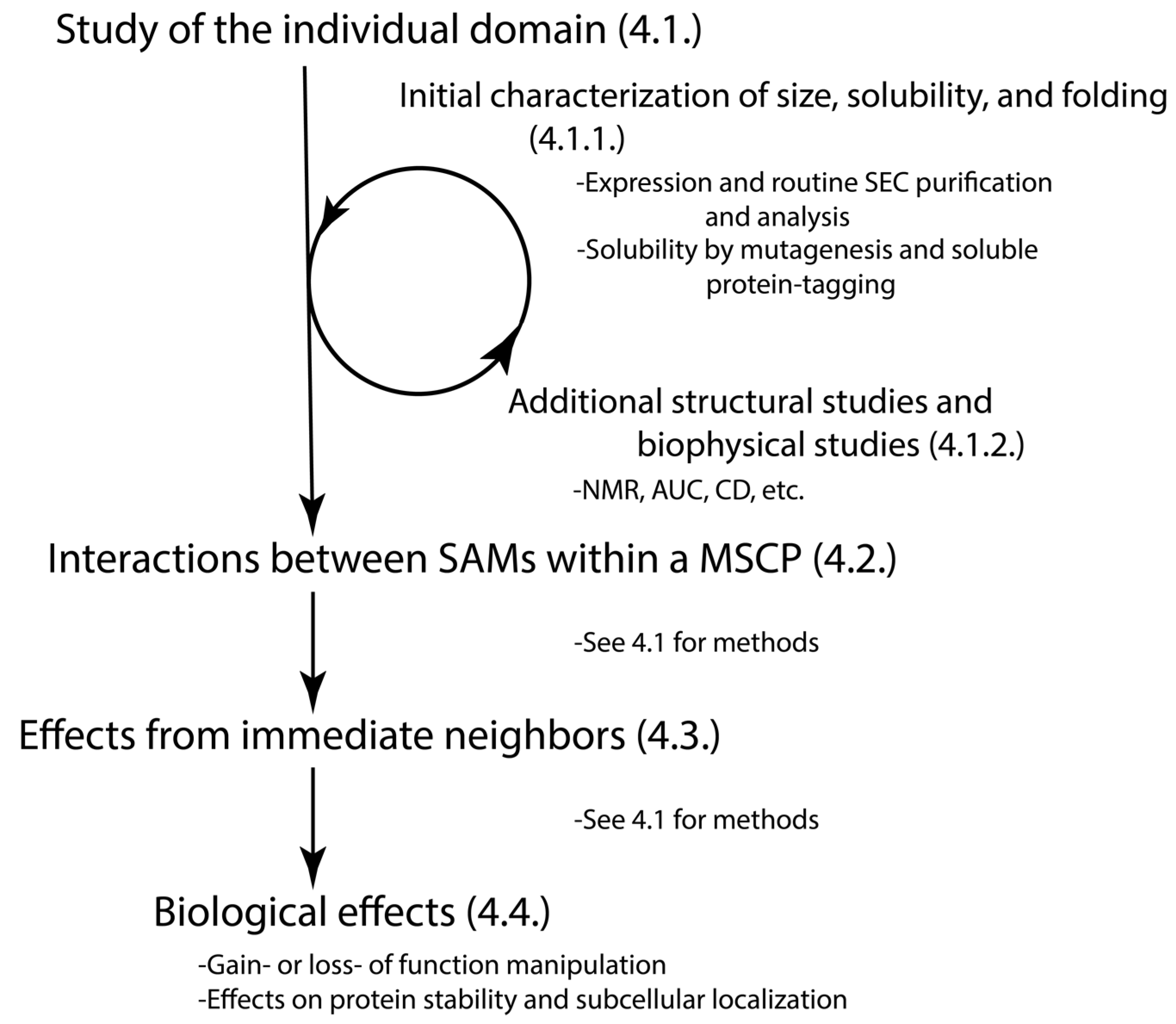
4. Methods to Study the SAM Domains from MSCPs
4.1. Does the Individual SAM Domain Exist as a Well-Folded or Disordered Monomer, a Dimer, or an Oligomer in Solution?
4.1.1. Initial Assessment on the Size, Solubility, and the Status of Folded vs. Disorder of the Individual SAM Domain
4.1.2. Additional Structural Studies of Individual SAM Domain
4.2. How Do the Multiple SAM Domains within the Protein Interact with Each Other?
4.3. How Do the SAM Domains Exist in the Context of Their Immediate Neighbors and Full Proteins?
4.4. How Do the SAM Domains Regulate the Protein Function?
5. MSCPs’ Major Functions, Their Roles in Cancer, and the Structural Regulation of Their SAMs
5.1. Overview
5.2. SASH1
5.3. ANKS1 Family
5.4. Liprins
5.5. CASKINs
5.6. SARM1
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 26 April 2023).
- Rana, J.S.; Khan, S.S.; Lloyd-Jones, D.M.; Sidney, S. Changes in Mortality in Top 10 Causes of Death from 2011 to 2018. J. Gen. Intern. Med. 2021, 36, 2517–2518. [Google Scholar] [CrossRef]
- Ahmad, F.B.; Anderson, R.N. The Leading Causes of Death in the US for 2020. J. Am. Med. Assoc. 2021, 325, 1829–1830. [Google Scholar] [CrossRef]
- Knight, M.J.; Leettola, C.; Gingery, M.; Li, H.; Bowie, J.U. A Human Sterile Alpha Motif Domain Polymerizome. Protein Sci. 2011, 20, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- DaRosa, P.A.; Ovchinnikov, S.; Xu, W.; Klevit, R.E. Structural Insights into SAM Domain-Mediated Tankyrase Oligomerization. Protein Sci. 2016, 25, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, L.; Templeton, C.M.; Ranes, M.; Paracuellos, P.; Cronin, N.; Beuron, F.; Morris, E.; Guettler, S. Tankyrase Requires SAM Domain-Dependent Polymerization to Support Wnt-β-Catenin Signaling. Mol. Cell 2016, 63, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Kwan, J.J.; Siu, R.; Gao, X.; Zoidl, G.; Demeler, B.; Saridakis, V.; Donaldson, L.W. A New Mode of SAM Domain Mediated Oligomerization Observed in the CASKIN2 Neuronal Scaffolding Protein. Cell Commun. Signal. 2016, 14, 17. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Marasco, D.; Pirone, L.; Pedone, E.M.; Pellecchia, M.; Leone, M. Solution Structure of the First Sam Domain of Odin and Binding Studies with the EphA2 Receptor. Biochemistry 2012, 51, 2136–2145. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Marasco, D.; Pirone, L.; Scognamiglio, P.L.; Pedone, E.M.; Pellecchia, M.; Leone, M. Heterotypic Sam-Sam Association between Odin-Sam1 and Arap3-Sam: Binding Affinity and Structural Insights. ChemBioChem 2013, 14, 100–106. [Google Scholar] [CrossRef]
- Barrera, F.N.; Poveda, J.A.; González-Ros, J.M.; Neira, J.L. Binding of the C-Terminal Sterile α Motif (SAM) Domain of Human P73 to Lipid Membranes. J. Biol. Chem. 2003, 278, 46878–46885. [Google Scholar] [CrossRef]
- Bhunia, A.; Domadia, P.N.; Mohanram, H.; Bhattacharjya, S. NMR Structural Studies of the Ste11 SAM Domain in the Dodecyl Phosphocholine Micelle. Proteins Struct. Funct. Bioinform. 2009, 74, 328–343. [Google Scholar] [CrossRef]
- Green, J.B.; Gardner, C.D.; Wharton, R.P.; Aggarwal, A.K. RNA Recognition via the SAM Domain of Smaug and Brain Tumor (Brat), in Turn Represses the Translation. Mol. Cell 2003, 11, 1537–1548. [Google Scholar] [CrossRef]
- Pillay, N.; Mariotti, L.; Zaleska, M.; Inian, O.; Jessop, M.; Hibbs, S.; Desfosses, A.; Hopkins, P.C.R.; Templeton, C.M.; Beuron, F.; et al. Structural Basis of Tankyrase Activation by Polymerization. Nature 2022, 612, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Zheng, S.; Spangler, S.A.; Yu, C.; Hoogenraad, C.C.; Zhang, M. Liprin-Mediated Large Signaling Complex Organization Revealed by the Liprin-α/CASK and Liprin-α/Liprin-β Complex Structures. Mol. Cell 2011, 43, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Luo, L.; Liang, M.; Zhang, W.; Zhang, T.; Yu, C.; Wei, Z. Structural Basis of Liprin-α-Promoted LAR-RPTP Clustering for Modulation of Phosphatase Activity. Nat. Commun. 2020, 11, 169. [Google Scholar] [CrossRef]
- Sporny, M.; Guez-Haddad, J.; Lebendiker, M.; Ulisse, V.; Volf, A.; Mim, C.; Isupov, M.N.; Opatowsky, Y. Structural Evidence for an Octameric Ring Arrangement of SARM1. J. Mol. Biol. 2019, 431, 3591–3605. [Google Scholar] [CrossRef]
- Clements, C.M.; Vögeli, B.; Shellman, Y.G.; Henen, M.A. SAM1 Domain of SASH1 Harbors Distinctive Structural Heterogeneity. J. Struct. Biol. 2022, 214, 107914. [Google Scholar] [CrossRef] [PubMed]
- Kurabi, A.; Brener, S.; Mobli, M.; Kwan, J.J.; Donaldson, L.W. A Nuclear Localization Signal at the SAM-SAM Domain Interface of AIDA-1 Suggests a Requirement for Domain Uncoupling Prior to Nuclear Import. J. Mol. Biol. 2009, 392, 1168–1177. [Google Scholar] [CrossRef]
- Stafford, R.L.; Hinde, E.; Knight, M.J.; Pennella, M.A.; Ear, J.; Digman, M.A.; Gratton, E.; Bowie, J.U. Tandem SAM Domain Structure of Human Caskin1: A Presynaptic, Self-Assembling Scaffold for CASK. Structure 2011, 19, 1826–1836. [Google Scholar] [CrossRef]
- Vincenzi, M.; Mercurio, F.A.; Leone, M. Sam Domains in Multiple Diseases. Curr. Med. Chem. 2020, 27, 450–476. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Bowie, J.U. The Many Faces of SAM. Sci. STKE 2005, 2005, re7. [Google Scholar] [CrossRef]
- Kukuk, L.; Dingley, A.J.; Granzin, J.; Nagel-Steger, L.; Thiagarajan-Rosenkranz, P.; Ciupka, D.; Hänel, K.; Batra-Safferling, R.; Pacheco, V.; Stoldt, M.; et al. Structure of the SLy1 SAM Homodimer Reveals a New Interface for SAM Domain Self-Association. Nat. Sci. Rep. 2019, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, F.; Cortés, B.I.; Findlay, G.M.; Cancino, G.I. LAR Receptor Tyrosine Phosphatase Family in Healthy and Diseased Brain. Front. Cell Dev. Biol. 2021, 9, 3475. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.A.; Bowie, J.U. SAM Domains: Uniform Structure, Diversity of Function. Trends Biochem. Sci. 2003, 28, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Zeller, C.; Hinzmann, B.; Seitz, S.; Prokoph, H.; Burkhard-Goettges, E.; Fischer, J.; Jandrig, B.; Schwarz, L.E.; Rosenthal, A.; Scherneck, S. SASH1: A Candidate Tumor Suppressor Gene on Chromosome 6q24.3 Is Downregulated in Breast Cancer. Oncogene 2003, 22, 2972–2983. [Google Scholar] [CrossRef] [PubMed]
- Clements, C.M.; Vogeli, B.; Shellman, Y.G.; Henen, M.A. Solution NMR Backbone Assignment of the SASH1 SLy Proteins Associated Disordered Region (SPIDER). Biomol. NMR Assign. 2023, 17, 151–157. [Google Scholar] [CrossRef]
- Sporny, M.; Guez-Haddad, J.; Khazma, T.; Yaron, A.; Dessau, M.; Shkolnisky, Y.; Mim, C.; Isupov, M.N.; Zalk, R.; Hons, M.; et al. Structural Basis for Sarm1 Inhibition and Activation under Energetic Stress. Elife 2020, 9, e22153. [Google Scholar] [CrossRef]
- Serra-Pagès, C.; Medley, Q.G.; Tang, M.; Hart, A.; Streuli, M. Liprins, a Family of LAR Transmembrane Protein-Tyrosine Phosphatase-Interacting Proteins. J. Biol. Chem. 1998, 273, 15611–15620. [Google Scholar] [CrossRef]
- Stafford, R.L.; Tang, M.Y.; Sawaya, M.R.; Phillips, M.L.; Bowie, J.U. Crystal Structure of the Central Coiled-Coil Domain from Human Liprin-Β2. Biochemistry 2011, 50, 3807–3815. [Google Scholar] [CrossRef]
- Wakita, M.; Yamagata, A.; Shiroshima, T.; Izumi, H.; Maeda, A.; Sendo, M.; Imai, A.; Kubota, K.; Goto-Ito, S.; Sato, Y.; et al. Structural Insights into Selective Interaction between Type IIa Receptor Protein Tyrosine Phosphatases and Liprin-α. Nat. Commun. 2020, 11, 649. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why Are “Natively Unfolded” Proteins Unstructured Under Physiologic Conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The Importance of Intrinsic Disorder for Protein Phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.O.; Yu, L.; Campuzano, I.; Grant, S.G.N.; Choudhary, J.S. Phosphoproteomic Analysis of the Mouse Brain Cytosol Reveals a Predominance of Protein Phosphorylation in Regions of Intrinsic Sequence Disorder. Mol. Cell. Proteom. 2008, 7, 1331–1348. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Madan Babu, M. The Rules of Disorder or Why Disorder Rules. Prog. Biophys. Mol. Biol. 2009, 99, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Fuxreiter, M. Fuzzy Complexes: Polymorphism and Structural Disorder in Protein–Protein Interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef]
- Xie, X.; Liang, M.; Yu, C.; Wei, Z. Liprin-α-Mediated Assemblies and Their Roles in Synapse Formation. Front. Cell Dev. Biol. 2021, 9, 595. [Google Scholar] [CrossRef]
- Ogawa, T.; Hirokawa, N. Multiple Analyses of Protein Dynamics in Solution. Biophys. Rev. 2018, 10, 299–306. [Google Scholar] [CrossRef]
- Fekete, S.; Beck, A.; Veuthey, J.L.; Guillarme, D. Theory and Practice of Size Exclusion Chromatography for the Analysis of Protein Aggregates. J. Pharm. Biomed. Anal. 2014, 101, 161–173. [Google Scholar] [CrossRef]
- Leettola, C.N.; Knight, M.J.; Cascio, D.; Hoffman, S.; Bowie, J.U. Characterization of the SAM Domain of the PKD-Related Protein ANKS6 and Its Interaction with ANKS3. BMC Struct. Biol. 2014, 14, 17. [Google Scholar] [CrossRef]
- Marion, D. An Introduction to Biological NMR Spectroscopy. Mol. Cell. Proteom. 2013, 12, 3006–3025. [Google Scholar] [CrossRef]
- Cole, J.L.; Lary, J.W.; Moody, T.P.; Laue, T.M. Analytical Ultracentrifugation: Sedimentation Velocity and Sedimentation Equilibrium. Methods Cell Biol. 2008, 84, 143–179. [Google Scholar]
- Wang, Y.; Shang, Y.; Li, J.; Chen, W.; Li, G.; Wan, J.; Liu, W.; Zhang, M. Specific Eph Receptor-Cytoplasmic Effector Signaling Mediated by SAM-SAM Domain Interactions. Elife 2018, 7, e35677. [Google Scholar] [CrossRef]
- Henen, M.A.; Myers, W.; Schmitt, L.R.; Wade, K.J.; Born, A.; Nichols, P.J.; Vögeli, B. The Disordered Spindly C-Terminus Interacts with RZZ Subunits ROD-1 and ZWL-1 in the Kinetochore through the Same Sites in C. Elegans. J. Mol. Biol. 2021, 433, 166812. [Google Scholar] [CrossRef]
- Wider, G.; Wüthrich, K. NMR Spectroscopy of Large Molecules and Multimolecular Assemblies in Solution. Curr. Opin. Struct. Biol 1999, 9, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to Study Proteins by Circular Dichroism. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhou, Y.; Zhang, J.; Zhang, H.X.; Jia, R. Molecular Dynamics Simulation Investigation of the Binding and Interaction of the EphA6-Odin Protein Complex. J. Phys. Chem. B 2022, 126, 4914–4924. [Google Scholar] [CrossRef] [PubMed]
- Issaian, A.; Schmitt, L.; Born, A.; Nichols, P.J.; Sikela, J.; Hansen, K.; Vögeli, B.; Henen, M.A. Solution NMR Backbone Assignment Reveals Interaction-Free Tumbling of Human Lineage-Specific Olduvai Protein Domains. Biomol. NMR Assign. 2019, 13, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Stanley, C.B.; Heller, W.T.; Friedman, P.A.; Bu, Z. Dynamic Structure of the Full-Length Scaffolding Protein NHERF1 Influences Signaling Complex Assembly. J. Biol. Chem. 2019, 294, 11297–11310. [Google Scholar] [CrossRef]
- Gerdts, J.; Summers, D.W.; Sasaki, Y.; DiAntonio, A.; Milbrandt, J. Sarm1-Mediated Axon Degeneration Requires Both SAM and TIR Interactions. J. Neurosci. 2013, 33, 13569–13580. [Google Scholar] [CrossRef]
- Chen, E.G.; Chen, Y.; Dong, L.L.; Zhang, J.S. Effects of SASH1 on Lung Cancer Cell Proliferation, Apoptosis, and Invasion in Vitro. Tumor Biol. 2012, 33, 1393–1401. [Google Scholar] [CrossRef]
- Jaufmann, J.; Franke, F.C.; Sperlich, A.; Blumendeller, C.; Kloos, I.; Schneider, B.; Sasaki, D.; Janssen, K.P.; Beer-Hammer, S. The Emerging and Diverse Roles of the SLy/SASH1-Protein Family in Health and Disease—Overview of Three Multifunctional Proteins. FASEB J. 2021, 35, e21470. [Google Scholar] [CrossRef]
- Martini, M.; Gnann, A.; Scheikl, D.; Holzmann, B.; Janssen, K.P. The Candidate Tumor Suppressor SASH1 Interacts with the Actin Cytoskeleton and Stimulates Cell-Matrix Adhesion. Int. J. Biochem. Cell Biol. 2011, 43, 1630–1640. [Google Scholar] [CrossRef]
- Astro, V.; Chiaretti, S.; Magistrati, E.; Fivaz, M.; De Curtis, I. Liprin-A1, ERC1 and LL5 Define Polarized and Dynamic Structures That Are Implicated in Cell Migration. J. Cell Sci. 2014, 127, 3862–3876. [Google Scholar] [CrossRef]
- Pehkonen, H.; Lento, M.; Von Nandelstadh, P.; Filippou, A.; Grénman, R.; Lehti, K.; Monni, O. Liprin-A1 Modulates Cancer Cell Signaling by Transmembrane Protein CD82 in Adhesive Membrane Domains Linked to Cytoskeleton. Cell Commun. Signal. 2018, 16, 41. [Google Scholar] [CrossRef]
- Pehkonen, H.; de Curtis, I.; Monni, O. Liprins in Oncogenic Signaling and Cancer Cell Adhesion. Oncogene 2021, 40, 6406–6416. [Google Scholar] [CrossRef]
- Pandey, A.; Blagoev, B.; Kratchmarova, I.; Fernandez, M.; Nielsen, M.; Zakarias Kristiansen, T.; Ohara, O.; Podtelejnikov, A.V.; Roche, S.; Lodish, H.F.; et al. Cloning of a Novel Phosphotyrosine Binding Domain Containing Molecule, Odin, Involved in Signaling by Receptor Tyrosine Kinases. Oncogene 2002, 21, 8029–8036. [Google Scholar] [CrossRef]
- Emaduddin, M.; Edelmann, M.J.; Kessler, B.M.; Feller, S.M. Odin (ANKS1A) Is a Src Family Kinase Target in Colorectal Cancer Cells. Cell Commun. Signal. 2008, 6, 7. [Google Scholar] [CrossRef]
- Tong, J.; Sydorskyy, Y.; St-Germain, J.R.; Taylor, P.; Tsao, M.S.; Moran, M.F. Odin (ANKS1A) Modulates EGF Receptor Recycling and Stability. PLoS ONE 2013, 8, e64817. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, H.; Kim, Y.; Yoo, S.; Park, E.; Park, S. The SAM Domains of Anks Family Proteins Are Critically Involved in Modulating the Degradation of EphA Receptors. Mol. Cell. Biol. 2010, 30, 1582–1592. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Noh, H.; Mun, J.; Gu, C.; Sever, S.; Park, S. Anks1a Regulates COPII-Mediated Anterograde Transport of Receptor Tyrosine Kinases Critical for Tumorigenesis. Nat. Commun. 2016, 7, 12799. [Google Scholar] [CrossRef] [PubMed]
- Dauphinee, S.M.; Clayton, A.; Hussainkhel, A.; Yang, C.; Park, Y.-J.; Fuller, M.E.; Blonder, J.; Veenstra, T.D.; Karsan, A. SASH1 Is a Scaffold Molecule in Endothelial TLR4 Signaling. J. Immunol. 2013, 191, 892–901. [Google Scholar] [CrossRef]
- Mukherjee, P.; Winkler, C.W.; Taylor, K.G.; Woods, T.A.; Nair, V.; Khan, B.A.; Peterson, K.E. SARM1, Not MyD88, Mediates TLR7/TLR9-Induced Apoptosis in Neurons. J. Immunol. 2015, 195, 4913–4921. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Kumari, N.; Mukherjee, P. The Curious Case of SARM1: Dr. Jekyll and Mr. Hyde in Cell Death and Immunity? FEBS J. 2023, 290, 340–358. [Google Scholar] [CrossRef] [PubMed]
- Stafford, R.L.; Ear, J.; Knight, M.J.; Bowie, J.U. The Molecular Basis of the Caskin1 and Mint1 Interaction with CASK. J. Mol. Biol. 2011, 412, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pazyra-Murphy, M.F.; Avizonis, D.; Russo, M.T.; Tang, S.; Chen, C.Y.; Hsueh, Y.P.; Bergholz, J.S.; Jiang, T.; Zhao, J.J.; et al. Sarm1 Activation Produces CADPR to Increase Intra-Axonal Ca++ and Promote Axon Degeneration in PIPN. J. Cell Biol. 2022, 221, e202106080. [Google Scholar] [CrossRef]
- Younis, R.M.; Taylor, R.M.; Beardsley, P.M.; McClay, J.L. The ANKS1B Gene and Its Associated Phenotypes: Focus on CNS Drug Response. Pharmacogenomics 2019, 20, 669–684. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, H.; Yao, Q.; Yan, Y.; Wu, R.; Liu, M. Clinical Significance of SASH1 Expression in Glioma. Dis. Markers 2015, 2015, 383046. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, J.; Xu, J.; Wang, H.; Sang, Q.; Xing, Q.; He, L. Effects of SASH1 on Melanoma Cell Proliferation and Apoptosis in Vitro. Mol. Med. Rep. 2012, 6, 1243–1248. [Google Scholar] [CrossRef]
- Franke, F.C.; Müller, J.; Abal, M.; Medina, E.D.; Nitsche, U.; Weidmann, H.; Chardonnet, S.; Ninio, E.; Janssen, K.P. The Tumor Suppressor SASH1 Interacts With the Signal Adaptor CRKL to Inhibit Epithelial-Mesenchymal Transition and Metastasis in Colorectal Cancer. Cell. Mol. Gastroenterol. Hepatol. 2018, 7, 33–53. [Google Scholar] [CrossRef]
- Coulombe, P.; Paliouras, G.N.; Clayton, A.; Hussainkhel, A.; Fuller, M.; Jovanovic, V.; Dauphinee, S.; Umlandt, P.; Xiang, P.; Kyle, A.H.; et al. Endothelial Sash1 Is Required for Lung Maturation through Nitric Oxide Signaling. Cell Rep. 2019, 27, 1769–1780. [Google Scholar] [CrossRef]
- Zhou, D.; Wei, Z.; Deng, S.; Wang, T.; Zai, M.; Wang, H.; Guo, L.; Zhang, J.; Zhong, H.; He, L.; et al. SASH1 Regulates Melanocyte Transepithelial Migration through a Novel Gαs-SASH1-IQGAP1-E-Cadherin Dependent Pathway. Cell. Signal. 2013, 25, 1526–1538. [Google Scholar] [CrossRef]
- Zhou, D.; Wei, Z.; Kuang, Z.; Luo, H.; Ma, J.; Zeng, X.; Wang, K.; Liu, B.; Gong, F.; Wang, J.; et al. A Novel P53/POMC/Gαs/SASH1 Autoregulatory Feedback Loop Activates Mutated SASH1 to Cause Pathologic Hyperpigmentation. J. Cell. Mol. Med. 2017, 21, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Courcet, J.B.; Elalaoui, S.C.; Duplomb, L.; Tajir, M.; Rivière, J.B.; Thevenon, J.; Gigot, N.; Marle, N.; Aral, B.; Duffourd, Y.; et al. Autosomal-Recessive SASH1 Variants Associated with a New Genodermatosis with Pigmentation Defects, Palmoplantar Keratoderma and Skin Carcinoma. Eur. J. Hum. Genet. 2015, 23, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Shellman, Y.G.; Lambert, K.A.; Brauweiler, A.; Fain, P.; Spritz, R.A.; Martini, M.; Janssen, K.P.; Box, N.F.; Terzian, T.; Rewers, M.; et al. SASH1 Is Involved in an Autosomal Dominant Lentiginous Phenotype. J. Investig. Dermatol. 2015, 135, 3192–3194. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, J.; Li, X.; Wang, Z.; Lei, D.; Wang, G.; Li, J.; Zhang, S.; Li, Z.; Li, M. A Novel de Novo Mutation of the SASH1 Gene in a Chinese Family with Multiple Lentigines. Acta Derm.-Venereol. 2017, 97, 530–531. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.L.; Wang, H.J.; Lin, Z.M.; Yang, Y. Novel Mutations in SASH1 Associated with Dyschromatosis Universalis Hereditaria. Indian J. Dermatol. Venereol. Leprol. 2019, 85, 440. [Google Scholar]
- Cui, H.; Guo, S.; He, H.; Guo, H.; Zhang, Y.; Wang, B. SASH1 Promotes Melanin Synthesis and Migration via Suppression of TGF-Β1 Secretion in Melanocytes Resulting in Pathologic Hyperpigmentation. Int. J. Biol. Sci. 2020, 16, 1264–1273. [Google Scholar] [CrossRef]
- Araki, A.; Okamura, K.; Saito, T. Five Novel Mutations in SASH1 Contribute to Lentiginous Phenotypes in Japanese Families. Pigment. Cell Melanoma Res. 2021, 34, 174–178. [Google Scholar] [CrossRef]
- Guo, K.; Liu, J.W.; Zhang, R.; Wang, R.; Ma, D.L.; Zhang, X. Genetic and Phenotypic Heterogeneity of Multiple Lentigines and Precise Diagnosis in Four Chinese Families with Multiple Lentigines. Pigment. Cell Melanoma Res. 2023, 36, 288–298. [Google Scholar] [CrossRef]
- Rimkus, C.; Martini, M.; Friederichs, J.; Rosenberg, R.; Doll, D.; Siewert, J.R.; Holzmann, B.; Janssen, K.P. Prognostic Significance of Downregulated Expression of the Candidate Tumour Suppressor Gene SASH1 in Colon Cancer. Br. J. Cancer 2006, 95, 1419. [Google Scholar] [CrossRef]
- He, P.; Zhang, H.X.; Sun, C.Y.; Chen, C.Y.; Jiang, H.Q. Overexpression of SASH1 Inhibits the Proliferation, Invasion, and EMT in Hepatocarcinoma Cells. Oncol. Res. 2016, 24, 25. [Google Scholar] [CrossRef]
- Sun, D.; Zhou, R.; Liu, H.; Sun, W.; Dong, A.; Zhang, H. SASH1 Inhibits Proliferation and Invasion of Thyroid Cancer Cells through PI3K/Akt Signaling Pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 12276. [Google Scholar]
- Yang, L.; Liu, M.; Gu, Z.; Chen, J.; Yan, Y.; Li, J. Overexpression of SASH1 Related to the Decreased Invasion Ability of Human Glioma U251 Cells. Tumor Biol. 2012, 33, 2255–2263. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lu, C.; Stewart, D.J.; Gu, J.; Huang, M.; Chang, D.W.; Lippman, S.M.; Wu, X. Systematic Evaluation of Apoptotic Pathway Gene Polymorphisms and Lung Cancer Risk. Carcinogenesis 2012, 33, 1699. [Google Scholar] [CrossRef]
- Meng, Q.; Zheng, M.; Liu, H.; Song, C.; Zhang, W.; Yan, J.; Qin, L.; Liu, X. SASH1 Regulates Proliferation, Apoptosis, and Invasion of Osteosarcoma Cell. Mol. Cell. Biochem. 2013, 373, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Burgess, J.T.; Bolderson, E.; Adams, M.N.; Baird, A.M.; Zhang, S.D.; Gately, K.A.; Umezawa, K.; O’Byrne, K.J.; Richard, D.J. Activation and Cleavage of SASH1 by Caspase-3 Mediates an Apoptotic Response. Cell Death Dis. 2016, 7, e2469. [Google Scholar] [CrossRef] [PubMed]
- Stubblefield, K.; Chean, J.; Nguyen, T.; Chen, C.-J.; Shively, J.E. The Adapter SASH1 Acts through NOTCH1 and Its Inhibitor DLK1 in a 3D Model of Lumenogenesis Involving CEACAM1. Exp. Cell Res. 2017, 359, 384. [Google Scholar] [CrossRef]
- Zong, W.; Yu, C.; Wang, P.; Dong, L. Overexpression of SASH1 Inhibits TGF-Β1-Induced EMT in Gastric Cancer Cells. Oncol. Res. 2016, 24, 17. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Arora, S.; Scott, A.M.; Janes, P.W. Eph Receptors in Cancer. Biomedicines 2023, 11, 315. [Google Scholar] [CrossRef]
- Jacob, A.L.; Jordan, B.A.; Weinberg, R.J. Organization of Amyloid-b Protein Precursor Intracellular Domain-Associated Protein-1 in the Rat Brain. J. Comp. Neurol. 2010, 518, 3221–3236. [Google Scholar] [CrossRef]
- Marcello, E.; Di Luca, M.; Gardoni, F. Synapse-to-Nucleus Communication: From Developmental Disorders to Alzheimer’s Disease. Curr. Opin. Neurobiol. 2018, 48, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Serie, D.J.; Bot, B.M.; Joseph, R.W.; Cheville, J.C.; Parker, A.S. ANKS1B Is a Smoking-Related Molecular Alteration in Clear Cell Renal Cell Carcinoma. BMC Urol. 2014, 14, 14. [Google Scholar] [CrossRef]
- Zeng, K.; He, B.; Yang, B.B.; Xu, T.; Chen, X.; Xu, M.; Liu, X.; Sun, H.; Pan, Y.; Wang, S. The Pro-Metastasis Effect of CircANKS1B in Breast Cancer. Mol. Cancer 2018, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, R.; Yang, L.; Wang, D.; Zhou, X.; Sun, Y. CircANKS1B Regulates FOXM1 Expression and Promotes Cell Migration and Invasion by Functioning as a Sponge of the MiR-149 in Colorectal Cancer. OncoTargets Ther. 2019, 12, 4065–4073. [Google Scholar] [CrossRef]
- Tao, L.J.; Pan, X.Y.; Wang, J.W.; Zhang, L.; Tao, L.S.; Liang, C.Z. Circular RNA CircANKS1B Acts as a Sponge for MiR-152-3p and Promotes Prostate Cancer Progression by Upregulating TGF-α Expression. Prostate 2021, 81, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xu, H. Regulation of Transforming Growth Factor-Beta1 by CircANKS1B/MiR-515-5p Affects the Metastatic Potential and Cisplatin Resistance in Oral Squamous Cell Carcinoma. Bioengineered 2021, 12, 12420–12430. [Google Scholar] [CrossRef] [PubMed]
- Han, K.A.; Jeon, S.; Um, J.W.; Ko, J. Emergent Synapse Organizers: LAR-RPTPs and Their Companions. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2016; Volume 324, pp. 39–65. ISBN 9780128048078. [Google Scholar]
- Nachat, R.; Cipolat, S.; Sevilla, L.M.; Chhatriwala, M.; Groot, K.R.; Watt, F.M. KazrinE Is a Desmosome-Associated Liprin That Colocalises with Acetylated Microtubules. J. Cell Sci. 2009, 122, 4035–4041. [Google Scholar] [CrossRef] [PubMed]
- Järvinen, A.K.; Autio, R.; Kilpinen, S.; Saarela, M.; Leivo, I.; Grénman, R.; Mäkitie, A.A.; Monni, O. High-Resolution Copy Number and Gene Expression Microarray Analyses of Head and Neck Squamous Cell Carcinoma Cell Lines of Tongue and Larynx. Genes Chromosom. Cancer 2008, 47, 500–509. [Google Scholar] [CrossRef]
- Carneiro, A.; Isinger, A.; Karlsson, A.; Johansson, J.; Jönsson, G.; Bendahl, P.O.; Falkenback, D.; Halvarsson, B.; Nilbert, M. Prognostic Impact of Array-Based Genomic Profiles in Esophageal Squamous Cell Cancer. BMC Cancer 2008, 8, 98. [Google Scholar] [CrossRef]
- Ramos-García, P.; Ruiz-Ávila, I.; Gil-Montoya, J.A.; Ayén, Á.; González-Ruiz, L.; Navarro-Triviño, F.J.; González-Moles, M.Á. Relevance of Chromosomal Band 11q13 in Oral Carcinogenesis: An Update of Current Knowledge. Oral Oncol. 2017, 72, 7–16. [Google Scholar] [CrossRef]
- Barros-Filho, M.C.; Reis-Rosa, L.A.; Hatakeyama, M.; Marchi, F.A.; Chulam, T.; Scapulatempo-Neto, C.; Nicolau, U.R.; Carvalho, A.L.; Pinto, C.A.L.; Drigo, S.A.; et al. Oncogenic Drivers in 11q13 Associated with Prognosis and Response to Therapy in Advanced Oropharyngeal Carcinomas. Oral Oncol. 2018, 83, 81–90. [Google Scholar] [CrossRef]
- Xu, C.; Liu, Y.; Wang, P.; Fan, W.; Rue, T.C.; Upton, M.P.; Houck, J.R.; Lohavanichbutr, P.; Doody, D.R.; Futran, N.D.; et al. Integrative Analysis of DNA Copy Number and Gene Expression in Metastatic Oral Squamous Cell Carcinoma Identifies Genes Associated with Poor Survival. Mol. Cancer 2010, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gollin, S.M.; Raja, S.; Godfrey, T.E. High-Resolution Mapping of the 11q13 Amplicon and Identification of a Gene, TAOS1, That Is Amplified and Overexpressed in Oral Cancer Cells. Proc. Natl. Acad. Sci. USA 2002, 99, 11369. [Google Scholar] [CrossRef] [PubMed]
- Ormandy, C.J.; Musgrove, E.A.; Hui, R.; Daly, R.J.; Sutherland, R.L. Cyclin D1, EMS1 and 11q13 Amplification in Breast Cancer. Breast Cancer Res. Treat. 2003, 78, 323–335. [Google Scholar] [CrossRef]
- Song, J.; Wu, S.; Xia, X.; Wang, Y.; Fan, Y.; Yang, Z. Cell Adhesion-Related Gene Somatic Mutations Are Enriched in Aggressive Papillary Thyroid Microcarcinomas 06 Biological Sciences 0604 Genetics. J. Transl. Med. 2018, 16, 269. [Google Scholar] [CrossRef]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Sugawara, E.; Hatano, S.; Asaka, R.; Okumura, S.; Nakagawa, K.; Mano, H.; Ishikawa, Y. Pulmonary Inflammatory Myofibroblastic Tumor Expressing a Novel Fusion, PPFIBP1-ALK: Reappraisal of Anti-ALK Immunohistochemistry as a Tool for Novel ALK Fusion Identification. Clin. Cancer Res. 2011, 17, 3341–3348. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; He, J.; Yelensky, R.; Esteve-Puig, R.; Botton, T.; Yeh, I.; Lipson, D.; Otto, G.; Brennan, K.; Murali, R.; et al. Kinase Fusions Are Frequent in Spitz Tumours and Spitzoid Melanomas. Nat. Commun. 2014, 5, 3116. [Google Scholar] [CrossRef]
- Yeh, I.; Botton, T.; Talevich, E.; Shain, A.H.; Sparatta, A.J.; De La Fouchardiere, A.; Mully, T.W.; North, J.P.; Garrido, M.C.; Gagnon, A.; et al. Activating MET Kinase Rearrangements in Melanoma and Spitz Tumours. Nat. Commun. 2015, 6, 7174. [Google Scholar] [CrossRef]
- de Curtis, I. Function of Liprins in Cell Motility. Exp. Cell Res. 2011, 317, 1–8. [Google Scholar] [CrossRef]
- Astro, V.; Asperti, C.; Cangi, M.G.; Doglioni, C.; De Curtis, I. Liprin-A1 Regulates Breast Cancer Cell Invasion by Affecting Cell Motility, Invadopodia and Extracellular Matrix Degradation. Oncogene 2010, 30, 1841–1849. [Google Scholar] [CrossRef]
- Asperti, C.; Pettinato, E.; de Curtis, I. Liprin-Alpha1 Affects the Distribution of Low-Affinity Beta1 Integrins and Stabilizes Their Permanence at the Cell Surface. Exp. Cell Res. 2010, 316, 915–926. [Google Scholar] [CrossRef]
- Robertson, J.; Jacquemet, G.; Byron, A.; Jones, M.C.; Warwood, S.; Selley, J.N.; Knight, D.; Humphries, J.D.; Humphries, M.J. Defining the Phospho-Adhesome through the Phosphoproteomic Analysis of Integrin Signalling. Nat. Commun. 2015, 6, 6265. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Ishizaki, T.; Okawa, K.; Watanabe, S.; Arakawa, T.; Watanabe, N.; Narumiya, S. Liprin-α Controls Stress Fiber Formation by Binding to MDia and Regulating Its Membrane Localization. J. Cell Sci. 2012, 125, 108–120. [Google Scholar] [CrossRef]
- Chiaretti, S.; Astro, V.; Chiricozzi, E.; de Curtis, I. Effects of the Scaffold Proteins Liprin-A1, Β1 and Β2 on Invasion by Breast Cancer Cells. Biol. Cell 2016, 108, 65–75. [Google Scholar] [CrossRef]
- von Thun, A.; Birtwistle, M.; Kalna, G.; Grindlay, J.; Strachan, D.; Kolch, W.; von Kriegsheim, A.; Norman, J.C. ERK2 Drives Tumour Cell Migration in Threedimensional Microenvironments by Suppressing Expression of Rab17 and Liprin-Β2. J. Cell Sci. 2012, 125, 1465–1477. [Google Scholar] [CrossRef]
- Astro, V.; Tonoli, D.; Chiaretti, S.; Badanai, S.; Sala, K.; Zerial, M.; De Curtis, I. Liprin-A1 and ERC1 Control Cell Edge Dynamics by Promoting Focal Adhesion Turnover. Sci. Rep. 2016, 6, 33653. [Google Scholar] [CrossRef] [PubMed]
- Sala, K.; Raimondi, A.; Tonoli, D.; Tacchetti, C.; De Curtis, I. Identification of a Membrane-Less Compartment Regulating Invadosome Function and Motility. Sci. Rep. 2018, 8, 1164. [Google Scholar] [CrossRef]
- Pehkonen, H.; Von Nandelstadh, P.; Karhemo, P.R.; Lepikhova, T.; Grenman, R.; Lehti, K.; Monni, O. Liprin-A1 Is a Regulator of Vimentin Intermediate Filament Network in the Cancer Cell Adhesion Machinery. Sci. Rep. 2016, 6, 24486. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.C.; Unoki, M.; Ythier, D.; Duperray, A.; Varticovski, L.; Kumamoto, K.; Pedeux, R.; Harris, C.C. Inhibitor of Growth 4 Suppresses Cell Spreading and Cell Migration by Interacting with a Novel Binding Partner, Liprin Alpha1. Cancer Res. 2007, 67, 2552. [Google Scholar] [CrossRef]
- Buraschi, S.; Neill, T.; Xu, S.Q.; Palladino, C.; Belfiore, A.; Iozzo, R.V.; Morrione, A. Progranulin/EphA2 Axis: A Novel Oncogenic Mechanism in Bladder Cancer. Matrix Biol. 2020, 93, 10–24. [Google Scholar] [CrossRef]
- Tan, K.D.; Zhu, Y.; Tan, H.K.; Rajasegaran, V.; Aggarwal, A.; Wu, J.; Wu, H.Y.; Hwang, J.; Lim, D.T.H.; Chee Soo, K.; et al. Amplification and Overexpression of PPFIA1, a Putative 11q13 Invasion Suppressor Gene, in Head and Neck Squamous Cell Carcinoma. Genes Chromosom. Cancer 2008, 47, 353–362. [Google Scholar] [CrossRef]
- Weng, Y.L.; Liu, N.; DiAntonio, A.; Broihier, H.T. The Cytoplasmic Adaptor Protein Caskin Mediates Lar Signal Transduction during Drosophila Motor Axon Guidance. J. Neurosci. 2011, 31, 4421–4433. [Google Scholar] [CrossRef]
- Xiang, S.; Li, J.; Shen, J.; Zhao, Y.; Wu, X.; Li, M.; Yang, X.; Kaboli, P.J.; Du, F.; Zheng, Y.; et al. Identification of Prognostic Genes in the Tumor Microenvironment of Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 653836. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, X.; Cai, M.; Huang, P.; Guan, Z. Novel Necroptosis-Related Gene Signature for Predicting the Prognosis of Pancreatic Adenocarcinoma. Aging 2022, 14, 869–891. [Google Scholar] [CrossRef] [PubMed]
- Morale, M.G.; Tamura, R.E.; Cintra, R.; Araújo, N.M.; Villa, L.L. TLR4 and SARM1 Modulate Survival and Chemoresistance in an HPV-Positive Cervical Cancer Cell Line. Sci. Rep. 2022, 12, 6714. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, H.Y.; Gu, W.; Shi, Y.; Kobe, B.; Ve, T. SARM1-Dependent Axon Degeneration: Nucleotide Signaling, Neurodegenerative Disorders, Toxicity, and Therapeutic Opportunities. Neuroscientist 2023. [Google Scholar] [CrossRef] [PubMed]
- Carty, M.; Bowie, A.G. SARM: From Immune Regulator to Cell Executioner. Biochem. Pharmacol. 2019, 161, 52–62. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clements, C.M.; Henen, M.A.; Vögeli, B.; Shellman, Y.G. The Structural Dynamics, Complexity of Interactions, and Functions in Cancer of Multi-SAM Containing Proteins. Cancers 2023, 15, 3019. https://doi.org/10.3390/cancers15113019
Clements CM, Henen MA, Vögeli B, Shellman YG. The Structural Dynamics, Complexity of Interactions, and Functions in Cancer of Multi-SAM Containing Proteins. Cancers. 2023; 15(11):3019. https://doi.org/10.3390/cancers15113019
Chicago/Turabian StyleClements, Christopher M., Morkos A. Henen, Beat Vögeli, and Yiqun G. Shellman. 2023. "The Structural Dynamics, Complexity of Interactions, and Functions in Cancer of Multi-SAM Containing Proteins" Cancers 15, no. 11: 3019. https://doi.org/10.3390/cancers15113019
APA StyleClements, C. M., Henen, M. A., Vögeli, B., & Shellman, Y. G. (2023). The Structural Dynamics, Complexity of Interactions, and Functions in Cancer of Multi-SAM Containing Proteins. Cancers, 15(11), 3019. https://doi.org/10.3390/cancers15113019






