Modern Risk Stratification of Acute Myeloid Leukemia in 2023: Integrating Established and Emerging Prognostic Factors
Abstract
Simple Summary
Abstract
1. Introduction
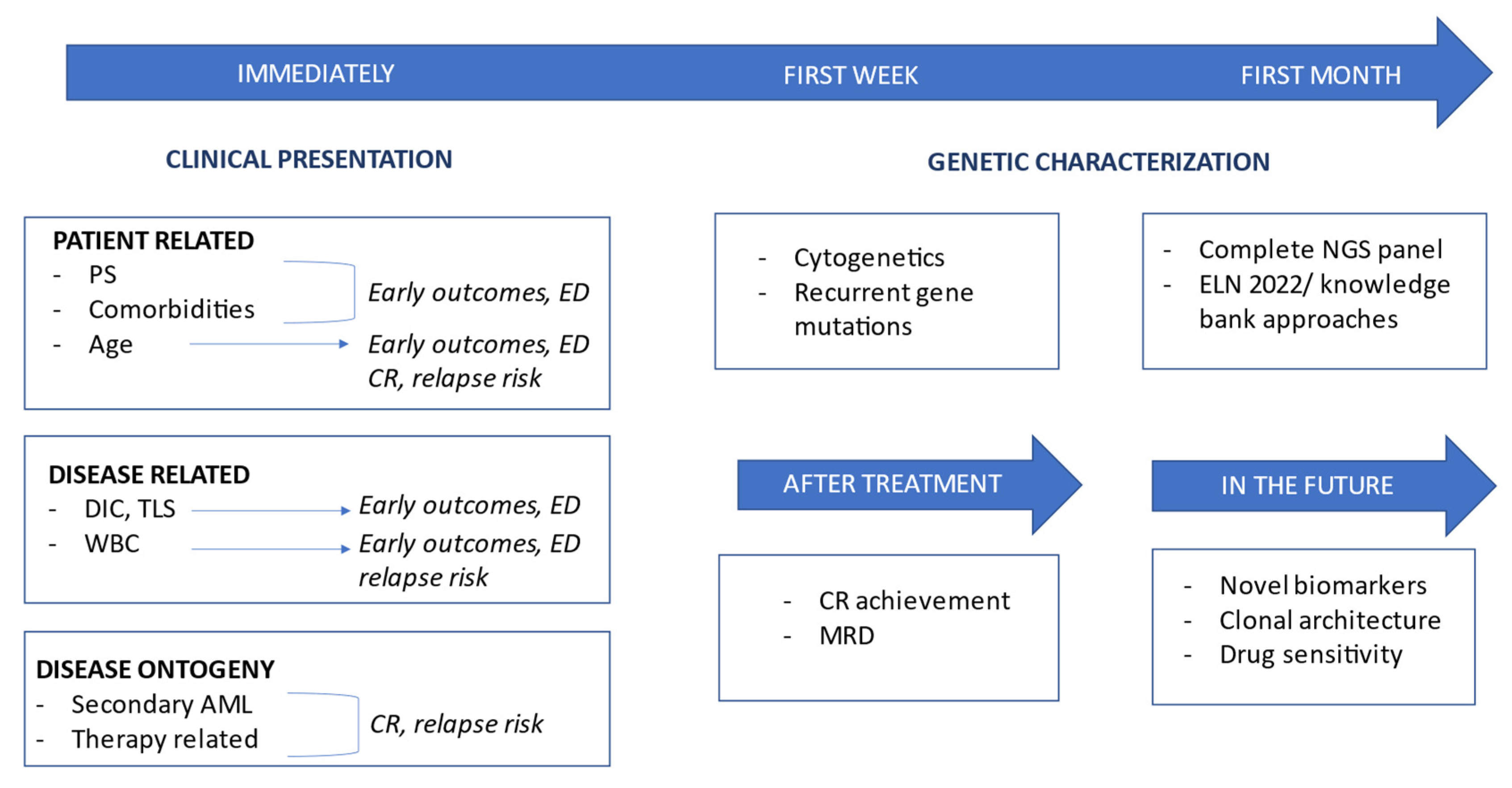
2. Clinical Risk Profile
2.1. Patient-Related Risk Factors: Age, Performance Status, and Frailty
2.2. Disease-Related Risk Factors: Disease Presentation, Extramedullary Disease, and Inaugural Complications
2.3. Disease Ontogeny
3. Genetic Risk Profile
3.1. Cytogenetics
3.1.1. Favorable Risk Recurrent Rearrangements
3.1.2. Intermediate and Adverse Risk Recurrent Rearrangements
3.1.3. Aneuploidies
3.2. Gene Mutations
3.2.1. FLT3
3.2.2. NPM1
3.2.3. CEBPA
3.2.4. TP53
3.2.5. RUNX1, ASXL1, and Other Myelodysplasia-Related Gene Mutations
3.2.6. Other Genes
4. Measurable Residual Disease
4.1. MRD in Less-Intensively Treated Patients
4.2. MRD and HCT
5. Current Risk Stratification Algorithms
- Patients with in-frame bZIP CEBPA mutations are now considered favorable-risk irrespective of CEBPA biallelic or monoallelic mutational status.
- NPM1-mutated patients with adverse cytogenetics are considered at adverse risk.
- Hyperdiploid karyotypes with three or more trisomies without structural abnormalities are excluded from the group of CK.
- In addition to RUNX1 and ASXL1, other MDS-related gene mutations (BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2) are added as poor-risk prognostic markers in the absence of favorable risk genetics.
- New high-risk rearrangements are included, namely t(3q26.2;v)/MECOM and t(8;16)(p11;p13)/KAT6A::CREBB.
- At least a 10% VAF is required to classify patients as TP53-mutated.
6. Emerging Biological Risk Factors
6.1. RNA
6.2. Methylation
6.3. Leukemia-Stem Cells
6.4. Proteomics
6.5. BH3 Profiling
7. Prognostic Impact of Clonal Architecture in AMLs
8. Global Risk Assessment in AML
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Urbino, I.; Secreto, C.; Olivi, M.; Apolito, V.; D’Ardia, S.; Frairia, C.; Giai, V.; Aydin, S.; Freilone, R.; Dellacasa, C.; et al. Evolving Therapeutic Approaches for Older Patients with Acute Myeloid Leukemia in 2021. Cancers 2021, 13, 5075. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Wei, A.H. How I Treat Acute Myeloid Leukemia in the Era of New Drugs. Blood 2020, 135, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Cerrano, M.; Esteve, J. Prognostic Factors in AML. In Acute Myeloid Leukemia; Röllig, C., Ossenkoppele, G.J., Eds.; Hematologic Malignancies; Springer International Publishing: Cham, Switzerland, 2021; pp. 127–175. ISBN 978-3-030-72676-8. [Google Scholar]
- Elgarten, C.W.; Aplenc, R. Pediatric acute myeloid leukemia: Updates on biology, risk stratification, and therapy. Curr. Opin. Pediatr. 2020, 32, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Ravandi, F.; Kadia, T.M.; DiNardo, C.D.; Short, N.J.; Borthakur, G.; Jabbour, E.; Kantarjian, H.M. De Novo Acute Myeloid Leukemia: A Population-Based Study of Outcome in the United States Based on the Surveillance, Epidemiology, and End Results (SEER) Database, 1980 to 2017. Cancer 2021, 127, 2049–2061. [Google Scholar] [CrossRef]
- Lazarevic, V.; Hörstedt, A.-S.; Johansson, B.; Antunovic, P.; Billström, R.; Derolf, Å.; Hulegårdh, E.; Lehmann, S.; Möllgård, L.; Nilsson, C.; et al. Incidence and Prognostic Significance of Karyotypic Subgroups in Older Patients with Acute Myeloid Leukemia: The Swedish Population-Based Experience. Blood Cancer J. 2014, 4, e188. [Google Scholar] [CrossRef]
- Appelbaum, F.R.; Gundacker, H.; Head, D.R.; Slovak, M.L.; Willman, C.L.; Godwin, J.E.; Anderson, J.E.; Petersdorf, S.H. Age and Acute Myeloid Leukemia. Blood 2006, 107, 3481–3485. [Google Scholar] [CrossRef]
- Juliusson, G.; Antunovic, P.; Derolf, A.; Lehmann, S.; Möllgård, L.; Stockelberg, D.; Tidefelt, U.; Wahlin, A.; Höglund, M. Age and Acute Myeloid Leukemia: Real World Data on Decision to Treat and Outcomes from the Swedish Acute Leukemia Registry. Blood 2009, 113, 4179–4187. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, R.; Maurillo, L.; Del Principe, M.I.; Paterno, G.; Walter, R.B.; Venditti, A.; Buccisano, F. Time for Dynamic Assessment of Fitness in Acute Myeloid Leukemia. Cancers 2022, 15, 136. [Google Scholar] [CrossRef]
- Ferrara, F.; Barosi, G.; Venditti, A.; Angelucci, E.; Gobbi, M.; Pane, F.; Tosi, P.; Zinzani, P.; Tura, S. Consensus-Based Definition of Unfitness to Intensive and Non-Intensive Chemotherapy in Acute Myeloid Leukemia: A Project of SIE, SIES and GITMO Group on a New Tool for Therapy Decision Making. Leukemia 2013, 27, 997–999. [Google Scholar] [CrossRef]
- Graf, I.; Greiner, G.; Marculescu, R.; Gleixner, K.V.; Herndlhofer, S.; Stefanzl, G.; Knoebl, P.; Jäger, U.; Hauswirth, A.; Schwarzinger, I.; et al. N-Terminal pro-Brain Natriuretic Peptide Is a Prognostic Marker for Response to Intensive Chemotherapy, Early Death, and Overall Survival in Acute Myeloid Leukemia. Am. J. Hematol. 2023, 98, 290–299. [Google Scholar] [CrossRef]
- Min, G.-J.; Cho, B.-S.; Park, S.-S.; Park, S.; Jeon, Y.-W.; Shin, S.-H.; Yahng, S.-A.; Yoon, J.-H.; Lee, S.-E.; Eom, K.-S.; et al. Geriatric Assessment Predicts Nonfatal Toxicities and Survival for Intensively Treated Older Adults with AML. Blood 2022, 139, 1646–1658. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Passera, R.; Cerrano, M.; Giai, V.; D’Ardia, S.; Iovino, G.; Dellacasa, C.M.; Audisio, E.; Busca, A. Combining the HCT-CI, G8, and AML-Score for Fitness Evaluation of Elderly Patients with Acute Myeloid Leukemia: A Single Center Analysis. Cancers 2023, 15, 1002. [Google Scholar] [CrossRef] [PubMed]
- Ganzel, C.; Becker, J.; Mintz, P.D.; Lazarus, H.M.; Rowe, J.M. Hyperleukocytosis, Leukostasis and Leukapheresis: Practice Management. Blood Rev. 2012, 26, 117–122. [Google Scholar] [CrossRef]
- Cerrano, M.; Seegers, V.; Raffoux, E.; Rabian, F.; Sébert, M.; Itzykson, R.; Lemiale, V.; Adès, L.; Boissel, N.; Dombret, H.; et al. Predictors and Outcomes Associated with Hydroxyurea Sensitivity in Acute Myeloid Leukemia Patients with High Hyperleukocytosis. Leuk Lymphoma 2020, 61, 737–740. [Google Scholar] [CrossRef]
- Frairia, C.; Nicolino, B.; Secreto, C.; Messa, E.; Arrigo, G.; Busca, A.; Cerrano, M.; D’Ardìa, S.; Dellacasa, C.; Evangelista, A.; et al. Validation of National Early Warning Score and Quick Sequential (Sepsis-Related) Organ Failure Assessment in Acute Myeloid Leukaemia Patients Treated with Intensive Chemotherapy. Eur. J. Haematol. 2023, 110, 696–705. [Google Scholar] [CrossRef]
- Cerrano, M.; Chevret, S.; Raffoux, E.; Rabian, F.; Sébert, M.; Valade, S.; Itzykson, R.; Lemiale, V.; Adès, L.; Boissel, N.; et al. Benefits of Dexamethasone on Early Outcomes in Patients with Acute Myeloid Leukemia with Hyperleukocytosis: A Propensity Score Matched Analysis. Ann. Hematol. 2023, 102, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Canaani, J.; Labopin, M.; Socié, G.; Nihtinen, A.; Huynh, A.; Cornelissen, J.; Deconinck, E.; Gedde-Dahl, T.; Forcade, E.; Chevallier, P.; et al. Long Term Impact of Hyperleukocytosis in Newly Diagnosed Acute Myeloid Leukemia Patients Undergoing Allogeneic Stem Cell Transplantation: An Analysis from the Acute Leukemia Working Party of the EBMT. Am. J. Hematol. 2017, 92, 653–659. [Google Scholar] [CrossRef]
- Tien, F.-M.; Hou, H.-A.; Tsai, C.-H.; Tang, J.-L.; Chen, C.-Y.; Kuo, Y.-Y.; Li, C.-C.; Lin, C.-T.; Yao, M.; Huang, S.-Y.; et al. Hyperleukocytosis Is Associated with Distinct Genetic Alterations and Is an Independent Poor-Risk Factor in de novo Acute Myeloid Leukemia Patients. Eur. J. Haematol. 2018, 101, 86–94. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Ganzel, C.; Manola, J.; Douer, D.; Rowe, J.M.; Fernandez, H.F.; Paietta, E.M.; Litzow, M.R.; Lee, J.-W.; Luger, S.M.; Lazarus, H.M.; et al. Extramedullary Disease in Adult Acute Myeloid Leukemia Is Common but Lacks Independent Significance: Analysis of Patients in ECOG-ACRIN Cancer Research Group Trials, 1980–2008. J. Clin. Oncol. 2016, 34, 3544–3553. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Granfeldt Østgård, L.S.; Medeiros, B.C.; Sengeløv, H.; Nørgaard, M.; Andersen, M.K.; Dufva, I.H.; Friis, L.S.; Kjeldsen, E.; Marcher, C.W.; Preiss, B.; et al. Epidemiology and Clinical Significance of Secondary and Therapy-Related Acute Myeloid Leukemia: A National Population-Based Cohort Study. J. Clin. Oncol. 2015, 33, 3641–3649. [Google Scholar] [CrossRef] [PubMed]
- Schmaelter, A.-K.; Labopin, M.; Socié, G.; Itälä-Remes, M.; Blaise, D.; Yakoub-Agha, I.; Forcade, E.; Cornelissen, J.; Ganser, A.; Beelen, D.; et al. Inferior Outcome of Allogeneic Stem Cell Transplantation for Secondary Acute Myeloid Leukemia in First Complete Remission as Compared to de Novo Acute Myeloid Leukemia. Blood Cancer J. 2020, 10, 26. [Google Scholar] [CrossRef]
- Chanswangphuwana, C.; Polprasert, C.; Owattanapanich, W.; Kungwankiattichai, S.; Tantiworawit, A.; Rattanathammethee, T.; Limvorapitak, W.; Saengboon, S.; Niparuck, P.; Puavilai, T.; et al. Characteristics and Outcomes of Secondary Acute Myeloid Leukemia and Acute Myeloid Leukemia with Myelodysplasia-Related Changes: Multicenter Study from the Thai Acute Leukemia Study Group. Clin. Lymphoma Myeloma Leuk. 2022, 22, e1075–e1083. [Google Scholar] [CrossRef] [PubMed]
- Higgins, A.; Shah, M.V. Genetic and Genomic Landscape of Secondary and Therapy-Related Acute Myeloid Leukemia. Genes 2020, 11, 749. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute Myeloid Leukemia Ontogeny Is Defined by Distinct Somatic Mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Gurnari, C.; Fabiani, E.; Falconi, G.; Travaglini, S.; Ottone, T.; Cristiano, A.; Voso, M.T. From Clonal Hematopoiesis to Therapy-Related Myeloid Neoplasms: The Silent Way of Cancer Progression. Biology 2021, 10, 128. [Google Scholar] [CrossRef]
- Gurnari, C.; Pagliuca, S.; Prata, P.H.; Galimard, J.-E.; Catto, L.F.B.; Larcher, L.; Sebert, M.; Allain, V.; Patel, B.J.; Durmaz, A.; et al. Clinical and Molecular Determinants of Clonal Evolution in Aplastic Anemia and Paroxysmal Nocturnal Hemoglobinuria. J. Clin. Oncol. 2023, 41, 132–142. [Google Scholar] [CrossRef]
- Schoch, C.; Kern, W.; Schnittger, S.; Hiddemann, W.; Haferlach, T. Karyotype Is an Independent Prognostic Parameter in Therapy-Related Acute Myeloid Leukemia (t-AML): An Analysis of 93 Patients with t-AML in Comparison to 1091 Patients with de Novo AML. Leukemia 2004, 18, 120–125. [Google Scholar] [CrossRef]
- Ossenkoppele, G.; Montesinos, P. Challenges in the Diagnosis and Treatment of Secondary Acute Myeloid Leukemia. Crit. Rev. Oncol. Hematol. 2019, 138, 6–13. [Google Scholar] [CrossRef]
- Rogers, H.J.; Wang, X.; Xie, Y.; Davis, A.R.; Thakral, B.; Wang, S.A.; Borthakur, G.; Cantu, M.D.; Margolskee, E.M.; Philip, J.K.S.; et al. Comparison of Therapy-Related and de Novo Core Binding Factor Acute Myeloid Leukemia: A Bone Marrow Pathology Group Study. Am. J. Hematol. 2020, 95, 799–808. [Google Scholar] [CrossRef]
- Nilsson, C.; Linde, F.; Hulegårdh, E.; Garelius, H.; Lazarevic, V.; Antunovic, P.; Cammenga, J.; Deneberg, S.; Eriksson, A.; Jädersten, M.; et al. Characterization of Therapy-Related Acute Myeloid Leukemia: Increasing Incidence and Prognostic Implications. Haematologica 2023, 108, 1015–1025. [Google Scholar] [CrossRef]
- Braun, T.; Cereja, S.; Chevret, S.; Raffoux, E.; Beaumont, M.; Detourmignies, L.; Pigneux, A.; Thomas, X.; Bordessoule, D.; Guerci, A.; et al. Evolving Characteristics and Outcome of Secondary Acute Promyelocytic Leukemia (APL): A Prospective Analysis by the French-Belgian-Swiss APL Group. Cancer 2015, 121, 2393–2399. [Google Scholar] [CrossRef]
- Heuser, M. Therapy-Related Myeloid Neoplasms: Does Knowing the Origin Help to Guide Treatment? Hematol. Am. Soc. Hematol. Educ. Program 2016, 2016, 24–32. [Google Scholar] [CrossRef]
- Rücker, F.G.; Schlenk, R.F.; Bullinger, L.; Kayser, S.; Teleanu, V.; Kett, H.; Habdank, M.; Kugler, C.-M.; Holzmann, K.; Gaidzik, V.I.; et al. TP53 Alterations in Acute Myeloid Leukemia with Complex Karyotype Correlate with Specific Copy Number Alterations, Monosomal Karyotype, and Dismal Outcome. Blood 2012, 119, 2114–2121. [Google Scholar] [CrossRef]
- Hochman, M.J.; Othus, M.; Hasserjian, R.P.; Ambinder, A.J.; Brunner, A.M.; Percival, M.-E.M.; Hourigan, C.S.; Swords, R.; DeZern, A.E.; Estey, E.H.; et al. Prognostic Impact of Secondary Versus De Novo Ontogeny in Acute Myeloid Leukemia (AML) Is Predominantly Accounted for By the European Leukemianet (ELN) 2022 Risk Classification. Blood 2022, 140, 1430–1432. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.-M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating Morphologic, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 Recommendations from an International Expert Panel on Behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Boddu, P.; Kantarjian, H.M.; Garcia-Manero, G.; Ravandi, F.; Verstovsek, S.; Jabbour, E.; Borthakur, G.; Konopleva, M.; Bhalla, K.N.; Daver, N.; et al. Treated Secondary Acute Myeloid Leukemia: A Distinct High-Risk Subset of AML with Adverse Prognosis. Blood Adv. 2017, 1, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K. On behalf of the National Cancer Research Institute Adult Leukaemia Working Group Refinement of Cytogenetic Classification in Acute Myeloid Leukemia: Determination of Prognostic Significance of Rare Recurring Chromosomal Abnormalities among 5876 Younger Adult Patients Treated in the United Kingdom Medical Research Council Trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef]
- Slovak, M.L.; Kopecky, K.J.; Cassileth, P.A.; Harrington, D.H.; Theil, K.S.; Mohamed, A.; Paietta, E.; Willman, C.L.; Head, D.R.; Rowe, J.M.; et al. Karyotypic Analysis Predicts Outcome of Preremission and Postremission Therapy in Adult Acute Myeloid Leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 2000, 96, 4075–4083. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Mrózek, K.; Dodge, R.K.; Carroll, A.J.; Edwards, C.G.; Arthur, D.C.; Pettenati, M.J.; Patil, S.R.; Rao, K.W.; Watson, M.S.; et al. Pretreatment Cytogenetic Abnormalities Are Predictive of Induction Success, Cumulative Incidence of Relapse, and Overall Survival in Adult Patients with de Novo Acute Myeloid Leukemia: Results from Cancer and Leukemia Group B (CALGB 8461)Presented in Part at the 43rd Annual Meeting of the American Society of Hematology, Orlando, FL, December 10, 2001, and Published in Abstract Form.59. Blood 2002, 100, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.A.; Fenaux, P.; Tallman, M.S.; Estey, E.H.; Löwenberg, B.; Naoe, T.; Lengfelder, E.; Döhner, H.; Burnett, A.K.; Chen, S.-J.; et al. Management of Acute Promyelocytic Leukemia: Updated Recommendations from an Expert Panel of the European LeukemiaNet. Blood 2019, 133, 1630–1643. [Google Scholar] [CrossRef]
- Kuykendall, A.; Duployez, N.; Boissel, N.; Lancet, J.E.; Welch, J.S. Acute Myeloid Leukemia: The Good, the Bad, and the Ugly. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 555–573. [Google Scholar] [CrossRef]
- Grimwade, D.; Mrózek, K. Diagnostic and Prognostic Value of Cytogenetics in Acute Myeloid Leukemia. Hematol./Oncol. Clin. N. Am. 2011, 25, 1135–1161. [Google Scholar] [CrossRef]
- Jourdan, E.; Boissel, N.; Chevret, S.; Delabesse, E.; Renneville, A.; Cornillet, P.; Blanchet, O.; Cayuela, J.-M.; Recher, C.; Raffoux, E.; et al. Prospective Evaluation of Gene Mutations and Minimal Residual Disease in Patients with Core Binding Factor Acute Myeloid Leukemia. Blood 2013, 121, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Opatz, S.; Bamopoulos, S.A.; Metzeler, K.H.; Herold, T.; Ksienzyk, B.; Bräundl, K.; Tschuri, S.; Vosberg, S.; Konstandin, N.P.; Wang, C.; et al. The Clinical Mutatome of Core Binding Factor Leukemia. Leukemia 2020, 34, 1553–1562. [Google Scholar] [CrossRef]
- Cher, C.Y.; Leung, G.M.K.; Au, C.H.; Chan, T.L.; Ma, E.S.K.; Sim, J.P.Y.; Gill, H.; Lie, A.K.W.; Liang, R.; Wong, K.F.; et al. Next-Generation Sequencing with a Myeloid Gene Panel in Core-Binding Factor AML Showed KIT Activation Loop and TET2 Mutations Predictive of Outcome. Blood Cancer J. 2016, 6, e442. [Google Scholar] [CrossRef]
- Han, S.Y.; Mrózek, K.; Voutsinas, J.; Wu, Q.; Morgan, E.A.; Vestergaard, H.; Ohgami, R.; Kluin, P.M.; Kristensen, T.K.; Pullarkat, S.; et al. Secondary Cytogenetic Abnormalities in Core-Binding Factor AML Harboring Inv(16) vs t(8;21). Blood Adv. 2021, 5, 2481–2489. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Benner, A.; Krauter, J.; Büchner, T.; Sauerland, C.; Ehninger, G.; Schaich, M.; Mohr, B.; Niederwieser, D.; Krahl, R.; et al. Individual Patient Data-Based Meta-Analysis of Patients Aged 16 to 60 Years with Core Binding Factor Acute Myeloid Leukemia: A Survey of the German Acute Myeloid Leukemia Intergroup. J. Clin. Oncol. 2004, 22, 3741–3750. [Google Scholar] [CrossRef]
- Herold, T.; Rothenberg-Thurley, M.; Grunwald, V.V.; Janke, H.; Goerlich, D.; Sauerland, M.C.; Konstandin, N.P.; Dufour, A.; Schneider, S.; Neusser, M.; et al. Validation and Refinement of the Revised 2017 European LeukemiaNet Genetic Risk Stratification of Acute Myeloid Leukemia. Leukemia 2020, 34, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Boddu, P.; Gurguis, C.; Sanford, D.; Cortes, J.; Akosile, M.; Ravandi, F.; Garcia-Manero, G.; Patel, K.P.; Kadia, T.; Brandt, M.; et al. Response Kinetics and Factors Predicting Survival in Core-Binding Factor Leukemia. Leukemia 2018, 32, 2698–2701. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Kawashima, N.; Atsuta, Y.; Sugiura, I.; Sawa, M.; Dobashi, N.; Yokoyama, H.; Doki, N.; Tomita, A.; Kiguchi, T.; et al. Prospective Evaluation of Prognostic Impact of KIT Mutations on Acute Myeloid Leukemia with RUNX1-RUNX1T1 and CBFB-MYH11. Blood Adv. 2020, 4, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Paschka, P.; Du, J.; Schlenk, R.F.; Gaidzik, V.I.; Bullinger, L.; Corbacioglu, A.; Späth, D.; Kayser, S.; Schlegelberger, B.; Krauter, J.; et al. Secondary Genetic Lesions in Acute Myeloid Leukemia with Inv(16) or t(16;16): A Study of the German-Austrian AML Study Group (AMLSG). Blood 2013, 121, 170–177. [Google Scholar] [CrossRef]
- Christen, F.; Hoyer, K.; Yoshida, K.; Hou, H.-A.; Waldhueter, N.; Heuser, M.; Hills, R.K.; Chan, W.; Hablesreiter, R.; Blau, O.; et al. Genomic Landscape and Clonal Evolution of Acute Myeloid Leukemia with t(8;21): An International Study on 331 Patients. Blood 2019, 133, 1140–1151. [Google Scholar] [CrossRef]
- Tazi, Y.; Arango-Ossa, J.E.; Zhou, Y.; Bernard, E.; Thomas, I.; Gilkes, A.; Freeman, S.; Pradat, Y.; Johnson, S.J.; Hills, R.; et al. Unified Classification and Risk-Stratification in Acute Myeloid Leukemia. Nat. Commun. 2022, 13, 4622. [Google Scholar] [CrossRef]
- Boissel, N.; Leroy, H.; Brethon, B.; Philippe, N.; de Botton, S.; Auvrignon, A.; Raffoux, E.; Leblanc, T.; Thomas, X.; Hermine, O.; et al. Incidence and Prognostic Impact of C-Kit, FLT3, and Ras Gene Mutations in Core Binding Factor Acute Myeloid Leukemia (CBF-AML). Leukemia 2006, 20, 965–970. [Google Scholar] [CrossRef]
- Paschka, P.; Marcucci, G.; Ruppert, A.S.; Mrózek, K.; Chen, H.; Kittles, R.A.; Vukosavljevic, T.; Perrotti, D.; Vardiman, J.W.; Carroll, A.J.; et al. Adverse Prognostic Significance of KIT Mutations in Adult Acute Myeloid Leukemia with Inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2006, 24, 3904–3911. [Google Scholar] [CrossRef]
- Rücker, F.G.; Agrawal, M.; Corbacioglu, A.; Weber, D.; Kapp-Schwoerer, S.; Gaidzik, V.I.; Jahn, N.; Schroeder, T.; Wattad, M.; Lübbert, M.; et al. Measurable Residual Disease Monitoring in Acute Myeloid Leukemia with t(8;21)(Q22;Q22.1): Results from the AML Study Group. Blood 2019, 134, 1608–1618. [Google Scholar] [CrossRef]
- Chen, W.; Xie, H.; Wang, H.; Chen, L.; Sun, Y.; Chen, Z.; Li, Q. Prognostic Significance of KIT Mutations in Core-Binding Factor Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0146614. [Google Scholar] [CrossRef]
- Klein, K.; Kaspers, G.; Harrison, C.J.; Beverloo, H.B.; Reedijk, A.; Bongers, M.; Cloos, J.; Pession, A.; Reinhardt, D.; Zimmerman, M.; et al. Clinical Impact of Additional Cytogenetic Aberrations, CKIT and RAS Mutations, and Treatment Elements in Pediatric t(8;21)-AML: Results From an International Retrospective Study by the International Berlin-Frankfurt-Münster Study Group. J. Clin. Oncol. 2015, 33, 4247–4258. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Duployez, N.; Fasan, A.; Decool, G.; Marceau-Renaut, A.; Meggendorfer, M.; Jourdan, E.; Petit, A.; Lapillonne, H.; Micol, J.-B.; et al. Clonal Interference of Signaling Mutations Worsens Prognosis in Core-Binding Factor Acute Myeloid Leukemia. Blood 2018, 132, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Kramer, M.; Martínez-Cuadrón, D.; Grenet, J.; Metzeler, K.H.; Sustkova, Z.; Luskin, M.R.; Brunner, A.M.; Elliott, M.A.; Gil, C.; et al. Characteristics and Outcome of Patients with Core-Binding Factor Acute Myeloid Leukemia and FLT3-ITD: Results from an International Collaborative Study. Haematologica 2022, 107, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.P.S.; Jones, D.; Qiao, W.; Cortes, J.E.; Ravandi, F.; Estey, E.E.; Verma, D.; Kantarjian, H.; Borthakur, G. Prognostic Value of FLT3 Mutations among Different Cytogenetic Subgroups in Acute Myeloid Leukemia. Cancer 2011, 117, 2145–2155. [Google Scholar] [CrossRef]
- Meyer, C.; Burmeister, T.; Gröger, D.; Tsaur, G.; Fechina, L.; Renneville, A.; Sutton, R.; Venn, N.C.; Emerenciano, M.; Pombo-de-Oliveira, M.S.; et al. The MLL Recombinome of Acute Leukemias in 2017. Leukemia 2018, 32, 273–284. [Google Scholar] [CrossRef]
- Tarlock, K.; Alonzo, T.A.; Moraleda, P.P.; Gerbing, R.B.; Raimondi, S.C.; Hirsch, B.A.; Ravindranath, Y.; Lange, B.; Woods, W.G.; Gamis, A.S.; et al. Acute Myeloid Leukaemia (AML) with t(6;9)(P23;Q34) Is Associated with Poor Outcome in Childhood AML Regardless of FLT3-ITD Status: A Report from the Children’s Oncology Group. Br. J. Haematol. 2014, 166, 254–259. [Google Scholar] [CrossRef]
- Díaz-Beyá, M.; Labopin, M.; Maertens, J.; Alijurf, M.; Passweg, J.; Dietrich, B.; Schouten, H.; Socié, G.; Schaap, N.; Schwerdtfeger, R.; et al. Allogeneic Stem Cell Transplantation in AML with t(6;9)(P23;Q34);DEK-NUP214 Shows a Favourable Outcome When Performed in First Complete Remission. Br. J. Haematol. 2020, 189, 920–925. [Google Scholar] [CrossRef]
- Lugthart, S.; Gröschel, S.; Beverloo, H.B.; Kayser, S.; Valk, P.J.M.; van Zelderen-Bhola, S.L.; Jan Ossenkoppele, G.; Vellenga, E.; van den Berg-de Ruiter, E.; Schanz, U.; et al. Clinical, Molecular, and Prognostic Significance of WHO Type Inv(3)(Q21q26.2)/t(3;3)(Q21;Q26.2) and Various Other 3q Abnormalities in Acute Myeloid Leukemia. J. Clin. Oncol. 2010, 28, 3890–3898. [Google Scholar] [CrossRef]
- Richard-Carpentier, G.; Rausch, C.R.; Sasaki, K.; Hammond, D.; Morita, K.; Takahashi, K.; Tang, G.; Kanagal-Shamanna, R.; Bhalla, K.; Dinardo, C.D.; et al. Characteristics and Clinical Outcomes of Patients with Acute Myeloid Leukemia with Inv(3)(Q21q26.2) or t(3;3)(Q21;Q26.2). Haematologica 2023. [Google Scholar] [CrossRef]
- Ottema, S.; Mulet-Lazaro, R.; Beverloo, H.B.; Erpelinck, C.; van Herk, S.; van der Helm, R.; Havermans, M.; Grob, T.; Valk, P.J.M.; Bindels, E.; et al. Atypical 3q26/MECOM Rearrangements Genocopy Inv(3)/t(3;3) in Acute Myeloid Leukemia. Blood 2020, 136, 224–234. [Google Scholar] [CrossRef]
- Neuendorff, N.R.; Burmeister, T.; Dörken, B.; Westermann, J. BCR-ABL-Positive Acute Myeloid Leukemia: A New Entity? Analysis of Clinical and Molecular Features. Ann. Hematol. 2016, 95, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Neuendorff, N.R.; Hemmati, P.; Arnold, R.; Ihlow, J.; Dörken, B.; Müller-Tidow, C.; Westermann, J. BCR-ABL+ Acute Myeloid Leukemia: Are We Always Dealing with a High-Risk Disease? Blood Adv. 2018, 2, 1409–1411. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, V.L.; Labopin, M.; Depei, W.; Yakoub-Agha, I.; Huynh, A.; Ljungman, P.; Schaap, N.; Cornelissen, J.J.; Maillard, N.; Pioltelli, P.; et al. Relatively Favorable Outcome after Allogeneic Stem Cell Transplantation for BCR-ABL1-Positive AML: A Survey from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation (EBMT). Am. J. Hematol. 2018, 93, 31–39. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhao, D.; Eladl, E.; Capo-Chichi, J.-M.; Kim, D.D.H.; Chang, H. Molecular Genetic Characterization of Philadelphia Chromosome-Positive Acute Myeloid Leukemia. Leuk. Res. 2023, 124, 107002. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Hills, R.K.; Langova, R.; Kramer, M.; Guijarro, F.; Sustkova, Z.; Estey, E.H.; Shaw, C.M.; Ráčil, Z.; Mayer, J.; et al. Characteristics and Outcome of Patients with Acute Myeloid Leukaemia and t(8;16)(P11;P13): Results from an International Collaborative Study*. Br. J. Haematol. 2021, 192, 832–842. [Google Scholar] [CrossRef]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Solé, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised International Prognostic Scoring System for Myelodysplastic Syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef]
- Creutzig, U.; Zimmermann, M.; Reinhardt, D.; Rasche, M.; von Neuhoff, C.; Alpermann, T.; Dworzak, M.; Perglerová, K.; Zemanova, Z.; Tchinda, J.; et al. Changes in Cytogenetics and Molecular Genetics in Acute Myeloid Leukemia from Childhood to Adult Age Groups. Cancer 2016, 122, 3821–3830. [Google Scholar] [CrossRef]
- Stölzel, F.; Mohr, B.; Kramer, M.; Oelschlägel, U.; Bochtler, T.; Berdel, W.E.; Kaufmann, M.; Baldus, C.D.; Schäfer-Eckart, K.; Stuhlmann, R.; et al. Karyotype Complexity and Prognosis in Acute Myeloid Leukemia. Blood Cancer J. 2016, 6, e386. [Google Scholar] [CrossRef]
- Haferlach, C.; Alpermann, T.; Schnittger, S.; Kern, W.; Chromik, J.; Schmid, C.; Pielken, H.J.; Kreuzer, K.-A.; Höffkes, H.-G.; Haferlach, T. Prognostic Value of Monosomal Karyotype in Comparison to Complex Aberrant Karyotype in Acute Myeloid Leukemia: A Study on 824 Cases with Aberrant Karyotype. Blood 2012, 119, 2122–2125. [Google Scholar] [CrossRef]
- Schoch, C.; Kern, W.; Kohlmann, A.; Hiddemann, W.; Schnittger, S.; Haferlach, T. Acute Myeloid Leukemia with a Complex Aberrant Karyotype Is a Distinct Biological Entity Characterized by Genomic Imbalances and a Specific Gene Expression Profile. Genes Chromosomes Cancer 2005, 43, 227–238. [Google Scholar] [CrossRef]
- Mrózek, K.; Eisfeld, A.-K.; Kohlschmidt, J.; Carroll, A.J.; Walker, C.J.; Nicolet, D.; Blachly, J.S.; Bill, M.; Papaioannou, D.; Wang, E.S.; et al. Complex Karyotype in de Novo Acute Myeloid Leukemia: Typical and Atypical Subtypes Differ Molecularly and Clinically. Leukemia 2019, 33, 1620–1634. [Google Scholar] [CrossRef]
- Breems, D.A.; Van Putten, W.L.J.; De Greef, G.E.; Van Zelderen-Bhola, S.L.; Gerssen-Schoorl, K.B.J.; Mellink, C.H.M.; Nieuwint, A.; Jotterand, M.; Hagemeijer, A.; Beverloo, H.B.; et al. Monosomal Karyotype in Acute Myeloid Leukemia: A Better Indicator of Poor Prognosis than a Complex Karyotype. J. Clin. Oncol. 2008, 26, 4791–4797. [Google Scholar] [CrossRef]
- Medeiros, B.C.; Othus, M.; Fang, M.; Roulston, D.; Appelbaum, F.R. Prognostic Impact of Monosomal Karyotype in Young Adult and Elderly Acute Myeloid Leukemia: The Southwest Oncology Group (SWOG) Experience. Blood 2010, 116, 2224–2228. [Google Scholar] [CrossRef]
- Lazarevic, V.; Rosso, A.; Juliusson, G.; Antunovic, P.; Rangert-Derolf, Å.; Lehmann, S.; Möllgård, L.; Uggla, B.; Wennström, L.; Wahlin, A.; et al. Prognostic Significance of High Hyperdiploid and Triploid/Tetraploid Adult Acute Myeloid Leukemia. Am. J. Hematol. 2015, 90, 800–805. [Google Scholar] [CrossRef]
- Kayser, S.; Martínez-Cuadrón, D.; Hanoun, M.; Stölzel, F.; Gil, C.; Reinhardt, H.C.; Aguiar, E.; Schäfer-Eckart, K.; Burgues, J.M.B.; Steffen, B.; et al. Characteristics and Outcome of Patients with Acute Myeloid Leukemia and Trisomy 4. Haematologica 2023, 108, 34–41. [Google Scholar] [CrossRef]
- Chilton, L.; Hills, R.K.; Harrison, C.J.; Burnett, A.K.; Grimwade, D.; Moorman, A.V. Hyperdiploidy with 49-65 Chromosomes Represents a Heterogeneous Cytogenetic Subgroup of Acute Myeloid Leukemia with Differential Outcome. Leukemia 2014, 28, 321–328. [Google Scholar] [CrossRef]
- Grimwade, D.; Ivey, A.; Huntly, B.J.P. Molecular Landscape of Acute Myeloid Leukemia in Younger Adults and Its Clinical Relevance. Blood 2016, 127, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Whitman, S.P.; Maharry, K.; Radmacher, M.D.; Becker, H.; Mrózek, K.; Margeson, D.; Holland, K.B.; Wu, Y.-Z.; Schwind, S.; Metzeler, K.H.; et al. FLT3 Internal Tandem Duplication Associates with Adverse Outcome and Gene- and MicroRNA-Expression Signatures in Patients 60 Years of Age or Older with Primary Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. Blood 2010, 116, 3622–3626. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.-K.; Kohlschmidt, J.; Mrózek, K.; Blachly, J.S.; Walker, C.J.; Nicolet, D.; Orwick, S.; Maharry, S.E.; Carroll, A.J.; Stone, R.M.; et al. Mutation Patterns Identify Adult Patients with de Novo Acute Myeloid Leukemia Aged 60 Years or Older Who Respond Favorably to Standard Chemotherapy: An Analysis of Alliance Studies. Leukemia 2018, 32, 1338–1348. [Google Scholar] [CrossRef]
- Liu, S.-B.; Dong, H.-J.; Bao, X.-B.; Qiu, Q.-C.; Li, H.-Z.; Shen, H.-J.; Ding, Z.-X.; Wang, C.; Chu, X.-L.; Yu, J.-Q.; et al. Impact of FLT3-ITD Length on Prognosis of Acute Myeloid Leukemia. Haematologica 2019, 104, e9–e12. [Google Scholar] [CrossRef] [PubMed]
- Bacher, U.; Haferlach, C.; Kern, W.; Haferlach, T.; Schnittger, S. Prognostic Relevance of FLT3-TKD Mutations in AML: The Combination Matters—An Analysis of 3082 Patients. Blood 2008, 111, 2527–2537. [Google Scholar] [CrossRef] [PubMed]
- Mead, A.J.; Linch, D.C.; Hills, R.K.; Wheatley, K.; Burnett, A.K.; Gale, R.E. FLT3 Tyrosine Kinase Domain Mutations Are Biologically Distinct from and Have a Significantly More Favorable Prognosis than FLT3 Internal Tandem Duplications in Patients with Acute Myeloid Leukemia. Blood 2007, 110, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Boddu, P.; Kantarjian, H.; Borthakur, G.; Kadia, T.; Daver, N.; Pierce, S.; Andreeff, M.; Ravandi, F.; Cortes, J.; Kornblau, S.M. Co-Occurrence of FLT3-TKD and NPM1 Mutations Defines a Highly Favorable Prognostic AML Group. Blood Adv. 2017, 1, 1546–1550. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic Nucleophosmin in Acute Myelogenous Leukemia with a Normal Karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Thiede, C.; Koch, S.; Creutzig, E.; Steudel, C.; Illmer, T.; Schaich, M.; Ehninger, G. Prevalence and Prognostic Impact of NPM1 Mutations in 1485 Adult Patients with Acute Myeloid Leukemia (AML). Blood 2006, 107, 4011–4020. [Google Scholar] [CrossRef]
- Schneider, F.; Hoster, E.; Unterhalt, M.; Schneider, S.; Dufour, A.; Benthaus, T.; Mellert, G.; Zellmeier, E.; Kakadia, P.M.; Bohlander, S.K.; et al. The FLT3ITD MRNA Level Has a High Prognostic Impact in NPM1 Mutated, but Not in NPM1 Unmutated, AML with a Normal Karyotype. Blood 2012, 119, 4383–4386. [Google Scholar] [CrossRef]
- Patel, S.S.; Kuo, F.C.; Gibson, C.J.; Steensma, D.P.; Soiffer, R.J.; Alyea, E.P.; Chen, Y.-B.A.; Fathi, A.T.; Graubert, T.A.; Brunner, A.M.; et al. High NPM1 Mutant Allele Burden at Diagnosis Predicts Unfavorable Outcomes in de Novo AML. Blood 2018, 131, 2816–2825. [Google Scholar] [CrossRef]
- Wang, Y.; Quesada, A.E.; Zuo, Z.; Medeiros, L.J.; Yin, C.C.; Li, S.; Xu, J.; Borthakur, G.; Li, Y.; Yang, C.; et al. The Impact of Mutation of Myelodysplasia-Related Genes in De Novo Acute Myeloid Leukemia Carrying NPM1 Mutation. Cancers 2023, 15, 198. [Google Scholar] [CrossRef]
- Haferlach, C.; Mecucci, C.; Schnittger, S.; Kohlmann, A.; Mancini, M.; Cuneo, A.; Testoni, N.; Rege-Cambrin, G.; Santucci, A.; Vignetti, M.; et al. AML with Mutated NPM1 Carrying a Normal or Aberrant Karyotype Show Overlapping Biologic, Pathologic, Immunophenotypic, and Prognostic Features. Blood 2009, 114, 3024–3032. [Google Scholar] [CrossRef]
- Angenendt, L.; Röllig, C.; Montesinos, P.; Martínez-Cuadrón, D.; Barragan, E.; García, R.; Botella, C.; Martínez, P.; Ravandi, F.; Kadia, T.; et al. Chromosomal Abnormalities and Prognosis in NPM1-Mutated Acute Myeloid Leukemia: A Pooled Analysis of Individual Patient Data From Nine International Cohorts. J. Clin. Oncol. 2019, 37, 2632–2642. [Google Scholar] [CrossRef]
- Othman, J.; Meggendorfer, M.; Tiacci, E.; Thiede, C.; Schlenk, R.; Dillon, R.; Stasik, S.; Venanzi, A.; Bertoli, S.; Delabesse, E.; et al. Overlapping Features of Therapy-Related and de Novo NPM1-Mutated AML. Blood 2023, 141, 1846–1857. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Döhner, K.; Krauter, J.; Fröhling, S.; Corbacioglu, A.; Bullinger, L.; Habdank, M.; Späth, D.; Morgan, M.; Benner, A.; et al. Mutations and Treatment Outcome in Cytogenetically Normal Acute Myeloid Leukemia. N. Engl. J. Med. 2008, 358, 1909–1918. [Google Scholar] [CrossRef]
- Fröhling, S.; Schlenk, R.F.; Stolze, I.; Bihlmayr, J.; Benner, A.; Kreitmeier, S.; Tobis, K.; Döhner, H.; Döhner, K. CEBPA Mutations in Younger Adults with Acute Myeloid Leukemia and Normal Cytogenetics: Prognostic Relevance and Analysis of Cooperating Mutations. J. Clin. Oncol. 2004, 22, 624–633. [Google Scholar] [CrossRef]
- Renneville, A.; Boissel, N.; Gachard, N.; Naguib, D.; Bastard, C.; de Botton, S.; Nibourel, O.; Pautas, C.; Reman, O.; Thomas, X.; et al. The Favorable Impact of CEBPA Mutations in Patients with Acute Myeloid Leukemia Is Only Observed in the Absence of Associated Cytogenetic Abnormalities and FLT3 Internal Duplication. Blood 2009, 113, 5090–5093. [Google Scholar] [CrossRef] [PubMed]
- Taskesen, E.; Bullinger, L.; Corbacioglu, A.; Sanders, M.A.; Erpelinck, C.A.J.; Wouters, B.J.; van der Poel-van de Luytgaarde, S.C.; Damm, F.; Krauter, J.; Ganser, A.; et al. Prognostic Impact, Concurrent Genetic Mutations, and Gene Expression Features of AML with CEBPA Mutations in a Cohort of 1182 Cytogenetically Normal AML Patients: Further Evidence for CEBPA Double Mutant AML as a Distinctive Disease Entity. Blood 2011, 117, 2469–2475. [Google Scholar] [CrossRef] [PubMed]
- Taube, F.; Georgi, J.A.; Kramer, M.; Stasik, S.; Middeke, J.M.; Röllig, C.; Krug, U.; Krämer, A.; Scholl, S.; Hochhaus, A.; et al. CEBPA Mutations in 4708 Patients with Acute Myeloid Leukemia: Differential Impact of BZIP and TAD Mutations on Outcome. Blood 2022, 139, 87–103. [Google Scholar] [CrossRef]
- Wakita, S.; Sakaguchi, M.; Oh, I.; Kako, S.; Toya, T.; Najima, Y.; Doki, N.; Kanda, J.; Kuroda, J.; Mori, S.; et al. Prognostic Impact of CEBPA BZIP Domain Mutation in Acute Myeloid Leukemia. Blood Adv. 2022, 6, 238–247. [Google Scholar] [CrossRef]
- Konstandin, N.P.; Pastore, F.; Herold, T.; Dufour, A.; Rothenberg-Thurley, M.; Hinrichsen, T.; Ksienzyk, B.; Tschuri, S.; Schneider, S.; Hoster, E.; et al. Genetic Heterogeneity of Cytogenetically Normal AML with Mutations of CEBPA. Blood Adv. 2018, 2, 2724–2731. [Google Scholar] [CrossRef] [PubMed]
- Tien, F.-M.; Hou, H.-A.; Tang, J.-L.; Kuo, Y.-Y.; Chen, C.-Y.; Tsai, C.-H.; Yao, M.; Lin, C.-T.; Li, C.-C.; Huang, S.-Y.; et al. Concomitant WT1 Mutations Predict Poor Prognosis in Acute Myeloid Leukemia Patients with Double Mutant CEBPA. Haematologica 2018, 103, e510–e513. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, O.K.; Siddon, A.; Madanat, Y.F.; Gagan, J.; Arber, D.A.; Dal Cin, P.; Narayanan, D.; Ouseph, M.M.; Kurzer, J.H.; Hasserjian, R.P. TP53 Mutation Defines a Unique Subgroup within Complex Karyotype de Novo and Therapy-Related MDS/AML. Blood Adv. 2022, 6, 2847–2853. [Google Scholar] [CrossRef]
- Grob, T.; Al Hinai, A.S.A.; Sanders, M.A.; Kavelaars, F.G.; Rijken, M.; Gradowska, P.L.; Biemond, B.J.; Breems, D.A.; Maertens, J.; van Marwijk Kooy, M.; et al. Molecular Characterization of Mutant TP53 Acute Myeloid Leukemia and High-Risk Myelodysplastic Syndrome. Blood 2022, 139, 2347–2354. [Google Scholar] [CrossRef]
- Dutta, S.; Moritz, J.; Pregartner, G.; Thallinger, G.G.; Brandstätter, I.; Lind, K.; Rezania, S.; Lyssy, F.; Reinisch, A.; Zebisch, A.; et al. Comparison of Acute Myeloid Leukemia and Myelodysplastic Syndromes with TP53 Aberrations. Ann. Hematol. 2022, 101, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 Allelic State for Genome Stability, Clinical Presentation and Outcomes in Myelodysplastic Syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef] [PubMed]
- Fenwarth, L.; Vasseur, L.; Duployez, N.; Gardin, C.; Terré, C.; Lambert, J.; de Botton, S.; Celli-Lebras, K.; Turlure, P.; Cluzeau, T.; et al. Prognostic Impact of Monoallelic Versus Biallelic TP53 Alterations in Intensively-Treated Adults AML Patients: A Retrospective Study from the ALFA Group. Blood 2022, 140, 737–738. [Google Scholar] [CrossRef]
- Bahaj, W.; Kewan, T.; Gurnari, C.; Durmaz, A.; Ponvilawan, B.; Pandit, I.; Kubota, Y.; Ogbue, O.; Aly, M.; Madanat, Y.F.; et al. A Clinically Practicable Approach to Predict TP53 Allelic Configurations in Myeloid Neoplasia. Blood 2022, 140, 2075–2077. [Google Scholar] [CrossRef]
- Short, N.J.; Montalban-Bravo, G.; Hwang, H.; Ning, J.; Franquiz, M.J.; Kanagal-Shamanna, R.; Patel, K.P.; DiNardo, C.D.; Ravandi, F.; Garcia-Manero, G.; et al. Prognostic and Therapeutic Impacts of Mutant TP53 Variant Allelic Frequency in Newly Diagnosed Acute Myeloid Leukemia. Blood Adv. 2020, 4, 5681–5689. [Google Scholar] [CrossRef]
- Kihara, R.; Nagata, Y.; Kiyoi, H.; Kato, T.; Yamamoto, E.; Suzuki, K.; Chen, F.; Asou, N.; Ohtake, S.; Miyawaki, S.; et al. Comprehensive Analysis of Genetic Alterations and Their Prognostic Impacts in Adult Acute Myeloid Leukemia Patients. Leukemia 2014, 28, 1586–1595. [Google Scholar] [CrossRef]
- Gaidzik, V.I.; Teleanu, V.; Papaemmanuil, E.; Weber, D.; Paschka, P.; Hahn, J.; Wallrabenstein, T.; Kolbinger, B.; Köhne, C.H.; Horst, H.A.; et al. RUNX1 Mutations in Acute Myeloid Leukemia Are Associated with Distinct Clinico-Pathologic and Genetic Features. Leukemia 2016, 30, 2160–2168. [Google Scholar] [CrossRef]
- Greif, P.A.; Konstandin, N.P.; Metzeler, K.H.; Herold, T.; Pasalic, Z.; Ksienzyk, B.; Dufour, A.; Schneider, F.; Schneider, S.; Kakadia, P.M.; et al. RUNX1 Mutations in Cytogenetically Normal Acute Myeloid Leukemia Are Associated with a Poor Prognosis and Up-Regulation of Lymphoid Genes. Haematologica 2012, 97, 1909–1915. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Herold, T.; Rothenberg-Thurley, M.; Amler, S.; Sauerland, M.C.; Görlich, D.; Schneider, S.; Konstandin, N.P.; Dufour, A.; Bräundl, K.; et al. Spectrum and Prognostic Relevance of Driver Gene Mutations in Acute Myeloid Leukemia. Blood 2016, 128, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Eder, C.; Jeromin, S.; Alpermann, T.; Fasan, A.; Grossmann, V.; Kohlmann, A.; Illig, T.; Klopp, N.; Wichmann, H.-E.; et al. ASXL1 Exon 12 Mutations Are Frequent in AML with Intermediate Risk Karyotype and Are Independently Associated with an Adverse Outcome. Leukemia 2013, 27, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Pratcorona, M.; Abbas, S.; Sanders, M.A.; Koenders, J.E.; Kavelaars, F.G.; Erpelinck-Verschueren, C.A.J.; Zeilemakers, A.; Löwenberg, B.; Valk, P.J.M. Acquired Mutations in ASXL1 in Acute Myeloid Leukemia: Prevalence and Prognostic Value. Haematologica 2012, 97, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Chiche, E.; Rahmé, R.; Bertoli, S.; Dumas, P.-Y.; Micol, J.-B.; Hicheri, Y.; Pasquier, F.; Peterlin, P.; Chevallier, P.; Thomas, X.; et al. Real-Life Experience with CPX-351 and Impact on the Outcome of High-Risk AML Patients: A Multicentric French Cohort. Blood Adv. 2021, 5, 176–184. [Google Scholar] [CrossRef]
- Othman, J.; Wilhelm-Benartzi, C.S.; Dillon, R.; Knapper, S.; Freeman, S.D.; Batten, L.M.; Canham, J.; Hinson, E.L.; Wych, J.; Betteridge, S.; et al. A Randomised Comparison of CPX-351 and FLAG-Ida in Adverse Karyotype AML and High-Risk MDS: The UK NCRI AML19 Trial. Blood Adv. 2023. [Google Scholar] [CrossRef]
- Kayser, S.; Levis, M.J. The Clinical Impact of the Molecular Landscape of Acute Myeloid Leukemia. Haematologica 2023, 108, 308–320. [Google Scholar] [CrossRef]
- Bullinger, L.; Döhner, K.; Döhner, H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. 2017, 35, 934–946. [Google Scholar] [CrossRef]
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3A Mutations in Acute Myeloid Leukemia. N. Engl. J. Med. 2010, 363, 2424–2433. [Google Scholar] [CrossRef]
- Thol, F.; Damm, F.; Lüdeking, A.; Winschel, C.; Wagner, K.; Morgan, M.; Yun, H.; Göhring, G.; Schlegelberger, B.; Hoelzer, D.; et al. Incidence and Prognostic Influence of DNMT3A Mutations in Acute Myeloid Leukemia. J. Clin. Oncol. 2011, 29, 2889–2896. [Google Scholar] [CrossRef]
- Ahn, J.-S.; Kim, H.-J.; Kim, Y.-K.; Lee, S.-S.; Jung, S.-H.; Yang, D.-H.; Lee, J.-J.; Kim, N.Y.; Choi, S.H.; Jung, C.W.; et al. DNMT3A R882 Mutation with FLT3-ITD Positivity Is an Extremely Poor Prognostic Factor in Patients with Normal-Karyotype Acute Myeloid Leukemia after Allogeneic Hematopoietic Cell Transplantation. Biol. Blood Marrow Transplant. 2016, 22, 61–70. [Google Scholar] [CrossRef]
- Vetro, C.; Haferlach, T.; Meggendorfer, M.; Stengel, A.; Jeromin, S.; Kern, W.; Haferlach, C. Cytogenetic and Molecular Genetic Characterization of KMT2A-PTD Positive Acute Myeloid Leukemia in Comparison to KMT2A-Rearranged Acute Myeloid Leukemia. Cancer Genet. 2020, 240, 15–22. [Google Scholar] [CrossRef]
- Hinai, A.S.A.A.; Pratcorona, M.; Grob, T.; Kavelaars, F.G.; Bussaglia, E.; Sanders, M.A.; Nomdedeu, J.; Valk, P.J.M. The Landscape of KMT2A-PTD AML: Concurrent Mutations, Gene Expression Signatures, and Clinical Outcome. Hemasphere 2019, 3, e181. [Google Scholar] [CrossRef] [PubMed]
- Paschka, P.; Marcucci, G.; Ruppert, A.S.; Whitman, S.P.; Mrózek, K.; Maharry, K.; Langer, C.; Baldus, C.D.; Zhao, W.; Powell, B.L.; et al. Wilms’ Tumor 1 Gene Mutations Independently Predict Poor Outcome in Adults with Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008, 26, 4595–4602. [Google Scholar] [CrossRef] [PubMed]
- Gaidzik, V.I.; Schlenk, R.F.; Moschny, S.; Becker, A.; Bullinger, L.; Corbacioglu, A.; Krauter, J.; Schlegelberger, B.; Ganser, A.; Döhner, H.; et al. Prognostic Impact of WT1 Mutations in Cytogenetically Normal Acute Myeloid Leukemia: A Study of the German-Austrian AML Study Group. Blood 2009, 113, 4505–4511. [Google Scholar] [CrossRef]
- Sargas, C.; Ayala, R.; Larráyoz, M.J.; Chillón, M.C.; Carrillo-Cruz, E.; Bilbao-Sieyro, C.; Prados de la Torre, E.; Martínez-Cuadrón, D.; Rodríguez-Veiga, R.; Boluda, B.; et al. Molecular Landscape and Validation of New Genomic Classification in 2668 Adult AML Patients: Real Life Data from the PETHEMA Registry. Cancers 2023, 15, 438. [Google Scholar] [CrossRef] [PubMed]
- Alfayez, M.; Issa, G.C.; Patel, K.P.; Wang, F.; Wang, X.; Short, N.J.; Cortes, J.E.; Kadia, T.; Ravandi, F.; Pierce, S.; et al. The Clinical Impact of PTPN11 Mutations in Adults with Acute Myeloid Leukemia. Leukemia 2021, 35, 691–700. [Google Scholar] [CrossRef]
- Stasik, S.; Eckardt, J.-N.; Kramer, M.; Röllig, C.; Krämer, A.; Scholl, S.; Hochhaus, A.; Crysandt, M.; Brümmendorf, T.H.; Naumann, R.; et al. Impact of PTPN11 Mutations on Clinical Outcome Analyzed in 1529 Patients with Acute Myeloid Leukemia. Blood Adv. 2021, 5, 3279–3289. [Google Scholar] [CrossRef]
- Fobare, S.; Kohlschmidt, J.; Ozer, H.G.; Mrózek, K.; Nicolet, D.; Mims, A.S.; Garzon, R.; Blachly, J.S.; Orwick, S.; Carroll, A.J.; et al. Molecular, Clinical, and Prognostic Implications of PTPN11 Mutations in Acute Myeloid Leukemia. Blood Adv. 2022, 6, 1371–1380. [Google Scholar] [CrossRef]
- Haferlach, T.; Kohlmann, A.; Klein, H.-U.; Ruckert, C.; Dugas, M.; Williams, P.M.; Kern, W.; Schnittger, S.; Bacher, U.; Löffler, H.; et al. AML with Translocation t(8;16)(P11;P13) Demonstrates Unique Cytomorphological, Cytogenetic, Molecular and Prognostic Features. Leukemia 2009, 23, 934–943. [Google Scholar] [CrossRef]
- Lamble, A.J.; Hagiwara, K.; Gerbing, R.B.; Smith, J.L.; Kolekar, P.; Ries, R.E.; Kolb, E.A.; Alonzo, T.A.; Ma, X.; Meshinchi, S. CREBBP Alterations Are Associated with a Poor Prognosis in de Novo AML. Blood 2023, 141, 2156–2159. [Google Scholar] [CrossRef] [PubMed]
- Sébert, M.; Passet, M.; Raimbault, A.; Rahmé, R.; Raffoux, E.; Sicre de Fontbrune, F.; Cerrano, M.; Quentin, S.; Vasquez, N.; Da Costa, M.; et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood 2019, 134, 1441–1444. [Google Scholar] [CrossRef]
- Duployez, N.; Largeaud, L.; Duchmann, M.; Kim, R.; Rieunier, J.; Lambert, J.; Bidet, A.; Larcher, L.; Lemoine, J.; Delhommeau, F.; et al. Prognostic Impact of DDX41 Germline Mutations in Intensively Treated Acute Myeloid Leukemia Patients: An ALFA-FILO Study. Blood 2022, 140, 756–768. [Google Scholar] [CrossRef]
- Bassan, R.; Brüggemann, M.; Radcliffe, H.-S.; Hartfield, E.; Kreuzbauer, G.; Wetten, S. A Systematic Literature Review and Meta-Analysis of Minimal Residual Disease as a Prognostic Indicator in Adult B-Cell Acute Lymphoblastic Leukemia. Haematologica 2019, 104, 2028–2039. [Google Scholar] [CrossRef]
- Blachly, J.S.; Walter, R.B.; Hourigan, C.S. The Present and Future of Measurable Residual Disease Testing in Acute Myeloid Leukemia. Haematologica 2022, 107, 2810–2822. [Google Scholar] [CrossRef]
- Schuurhuis, G.J.; Heuser, M.; Freeman, S.; Béné, M.-C.; Buccisano, F.; Cloos, J.; Grimwade, D.; Haferlach, T.; Hills, R.K.; Hourigan, C.S.; et al. Minimal/Measurable Residual Disease in AML: A Consensus Document from the European LeukemiaNet MRD Working Party. Blood 2018, 131, 1275–1291. [Google Scholar] [CrossRef]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.-C.; et al. 2021 Update on MRD in Acute Myeloid Leukemia: A Consensus Document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef]
- Bernardi, M.; Ferrara, F.; Carrabba, M.G.; Mastaglio, S.; Lorentino, F.; Vago, L.; Ciceri, F. MRD in Venetoclax-Based Treatment for AML: Does It Really Matter? Front. Oncol. 2022, 12, 890871. [Google Scholar] [CrossRef]
- Pratz, K.W.; Jonas, B.A.; Pullarkat, V.; Recher, C.; Schuh, A.C.; Thirman, M.J.; Garcia, J.S.; DiNardo, C.D.; Vorobyev, V.; Fracchiolla, N.S.; et al. Measurable Residual Disease Response and Prognosis in Treatment-Naïve Acute Myeloid Leukemia with Venetoclax and Azacitidine. J. Clin. Oncol. 2022, 40, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A.; DiNardo, C.D.; Wang, S.A.; Jorgensen, J.; Kadia, T.M.; Daver, N.G.; Short, N.J.; Yilmaz, M.; Pemmaraju, N.; Borthakur, G.; et al. Prognostic Value of Measurable Residual Disease after Venetoclax and Decitabine in Acute Myeloid Leukemia. Blood Adv. 2021, 5, 1876–1883. [Google Scholar] [CrossRef]
- Walter, R.B.; Buckley, S.A.; Pagel, J.M.; Wood, B.L.; Storer, B.E.; Sandmaier, B.M.; Fang, M.; Gyurkocza, B.; Delaney, C.; Radich, J.P.; et al. Significance of Minimal Residual Disease before Myeloablative Allogeneic Hematopoietic Cell Transplantation for AML in First and Second Complete Remission. Blood 2013, 122, 1813–1821. [Google Scholar] [CrossRef]
- Anthias, C.; Dignan, F.L.; Morilla, R.; Morilla, A.; Ethell, M.E.; Potter, M.N.; Shaw, B.E. Pre-Transplant MRD Predicts Outcome Following Reduced-Intensity and Myeloablative Allogeneic Hemopoietic SCT in AML. Bone Marrow Transpl. 2014, 49, 679–683. [Google Scholar] [CrossRef]
- Buckley, S.A.; Wood, B.L.; Othus, M.; Hourigan, C.S.; Ustun, C.; Linden, M.A.; DeFor, T.E.; Malagola, M.; Anthias, C.; Valkova, V.; et al. Minimal Residual Disease Prior to Allogeneic Hematopoietic Cell Transplantation in Acute Myeloid Leukemia: A Meta-Analysis. Haematologica 2017, 102, 865–873. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Altman, J.K.; Assi, R.; Bixby, D.; Fathi, A.T.; Foran, J.M.; Gojo, I.; Hall, A.C.; Jonas, B.A.; Kishtagari, A.; et al. Acute Myeloid Leukemia, Version 3.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2023, 21, 503–513. [Google Scholar] [CrossRef]
- Döhner, K.; Thiede, C.; Jahn, N.; Panina, E.; Gambietz, A.; Larson, R.A.; Prior, T.W.; Marcucci, G.; Jones, D.; Krauter, J.; et al. Impact of NPM1/FLT3-ITD Genotypes Defined by the 2017 European LeukemiaNet in Patients with Acute Myeloid Leukemia. Blood 2020, 135, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lachowiez, C.A.; Long, N.; Saultz, J.; Gandhi, A.; Newell, L.F.; Hayes-Lattin, B.; Maziarz, R.T.; Leonard, J.; Bottomly, D.; McWeeney, S.; et al. Comparison and Validation of the 2022 European LeukemiaNet Guidelines in Acute Myeloid Leukemia. Blood Adv. 2023, 7, 1899–1909. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.-Y.; Tsai, X.C.-H.; Lin, C.-C.; Tien, F.-M.; Kuo, Y.-Y.; Lee, W.-H.; Peng, Y.-L.; Liu, M.-C.; Tseng, M.-H.; Hsu, C.-A.; et al. Validation of the Prognostic Significance of the 2022 European LeukemiaNet Risk Stratification System in Intensive Chemotherapy Treated Aged 18 to 65 Years Patients with de Novo Acute Myeloid Leukemia. Am. J. Hematol. 2023, 98, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Rothenberg-Thurley, M.; Dufour, A.; Schneider, S.; Gittinger, H.; Sauerland, C.; Görlich, D.; Krug, U.; Berdel, W.E.; Woermann, B.J.; et al. Validation and Refinement of the 2022 European LeukemiaNet Genetic Risk Stratification of Acute Myeloid Leukemia. Leukemia 2023, 37, 1234–1244. [Google Scholar] [CrossRef]
- Mrózek, K.; Kohlschmidt, J.; Blachly, J.S.; Nicolet, D.; Carroll, A.J.; Archer, K.J.; Mims, A.S.; Larkin, K.T.; Orwick, S.; Oakes, C.C.; et al. Outcome Prediction by the 2022 European LeukemiaNet Genetic-Risk Classification for Adults with Acute Myeloid Leukemia: An Alliance Study. Leukemia 2023, 37, 788–798. [Google Scholar] [CrossRef]
- Döhner, H.; Pratz, K.W.; DiNardo, C.D.; Jonas, B.A.; Pullarkat, V.A.; Thirman, M.J.; Recher, C.; Schuh, A.C.; Babu, S.; Dail, M.; et al. ELN Risk Stratification Is Not Predictive of Outcomes for Treatment-Naïve Patients with Acute Myeloid Leukemia Treated with Venetoclax and Azacitidine. Blood 2022, 140, 1441–1444. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of MicroRNAs, CircRNAs and Long Noncoding RNAs in Acute Myeloid Leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef]
- Li, J.; Sun, C.-K. Long Noncoding RNA SNHG5 Is Up-Regulated and Serves as a Potential Prognostic Biomarker in Acute Myeloid Leukemia. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3342–3347. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Volinia, S.; Papaioannou, D.; Nicolet, D.; Kohlschmidt, J.; Yan, P.S.; Mrózek, K.; Bucci, D.; Carroll, A.J.; Baer, M.R.; et al. Expression and Prognostic Impact of LncRNAs in Acute Myeloid Leukemia. Proc. Natl. Acad. Sci. USA 2014, 111, 18679–18684. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.; Thoms, J.A.I.; Palu, C.; Herold, T.; Shah, A.; Olivier, J.; Boelen, L.; Huang, Y.; Chacon, D.; Brown, A.; et al. A Four-Gene LincRNA Expression Signature Predicts Risk in Multiple Cohorts of Acute Myeloid Leukemia Patients. Leukemia 2018, 32, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Farrar, J.E.; Smith, J.L.; Othus, M.; Huang, B.J.; Wang, Y.-C.; Ries, R.; Hylkema, T.; Pogosova-Agadjanyan, E.L.; Challa, S.; Leonti, A.; et al. Long Noncoding RNA Expression Independently Predicts Outcome in Pediatric Acute Myeloid Leukemia. J. Clin. Oncol. 2023, 41, 2949–2962. [Google Scholar] [CrossRef]
- Marcucci, G.; Radmacher, M.D.; Maharry, K.; Mrózek, K.; Ruppert, A.S.; Paschka, P.; Vukosavljevic, T.; Whitman, S.P.; Baldus, C.D.; Langer, C.; et al. MicroRNA Expression in Cytogenetically Normal Acute Myeloid Leukemia. N. Engl. J. Med. 2008, 358, 1919–1928. [Google Scholar] [CrossRef]
- Marcucci, G.; Maharry, K.S.; Metzeler, K.H.; Volinia, S.; Wu, Y.-Z.; Mrózek, K.; Nicolet, D.; Kohlschmidt, J.; Whitman, S.P.; Mendler, J.H.; et al. Clinical Role of MicroRNAs in Cytogenetically Normal Acute Myeloid Leukemia: MiR-155 Upregulation Independently Identifies High-Risk Patients. J. Clin. Oncol. 2013, 31, 2086–2093. [Google Scholar] [CrossRef]
- Díaz-Beyá, M.; Brunet, S.; Nomdedéu, J.; Tejero, R.; Díaz, T.; Pratcorona, M.; Tormo, M.; Ribera, J.M.; Escoda, L.; Duarte, R.; et al. MicroRNA Expression at Diagnosis Adds Relevant Prognostic Information to Molecular Categorization in Patients with Intermediate-Risk Cytogenetic Acute Myeloid Leukemia. Leukemia 2014, 28, 804–812. [Google Scholar] [CrossRef]
- L′Abbate, A.; Tolomeo, D.; Cifola, I.; Severgnini, M.; Turchiano, A.; Augello, B.; Squeo, G.; D′Addabbo, P.; Traversa, D.; Daniele, G.; et al. MYC-Containing Amplicons in Acute Myeloid Leukemia: Genomic Structures, Evolution, and Transcriptional Consequences. Leukemia 2018, 32, 2152–2166. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Lugthart, S.; Li, Y.; Erpelinck-Verschueren, C.; Deng, X.; Christos, P.J.; Schifano, E.; Booth, J.; van Putten, W.; Skrabanek, L.; et al. DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer Cell 2010, 17, 13–27. [Google Scholar] [CrossRef]
- Bullinger, L.; Ehrich, M.; Döhner, K.; Schlenk, R.F.; Döhner, H.; Nelson, M.R.; van den Boom, D. Quantitative DNA Methylation Predicts Survival in Adult Acute Myeloid Leukemia. Blood 2010, 115, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Yan, P.; Maharry, K.; Frankhouser, D.; Nicolet, D.; Metzeler, K.H.; Kohlschmidt, J.; Mrózek, K.; Wu, Y.-Z.; Bucci, D.; et al. Epigenetics Meets Genetics in Acute Myeloid Leukemia: Clinical Impact of a Novel Seven-Gene Score. J. Clin. Oncol. 2014, 32, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Deneberg, S.; Guardiola, P.; Lennartsson, A.; Qu, Y.; Gaidzik, V.; Blanchet, O.; Karimi, M.; Bengtzén, S.; Nahi, H.; Uggla, B.; et al. Prognostic DNA Methylation Patterns in Cytogenetically Normal Acute Myeloid Leukemia Are Predefined by Stem Cell Chromatin Marks. Blood 2011, 118, 5573–5582. [Google Scholar] [CrossRef] [PubMed]
- Jost, E.; Lin, Q.; Weidner, C.I.; Wilop, S.; Hoffmann, M.; Walenda, T.; Schemionek, M.; Herrmann, O.; Zenke, M.; Brümmendorf, T.H.; et al. Epimutations Mimic Genomic Mutations of DNMT3A in Acute Myeloid Leukemia. Leukemia 2014, 28, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Luskin, M.R.; Gimotty, P.A.; Smith, C.; Loren, A.W.; Figueroa, M.E.; Harrison, J.; Sun, Z.; Tallman, M.S.; Paietta, E.M.; Litzow, M.R.; et al. A Clinical Measure of DNA Methylation Predicts Outcome in de Novo Acute Myeloid Leukemia. JCI Insight 2016, 1, e87323. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Luskin, M.R.; Carroll, M.; Smith, C.; Harrison, J.; Pierce, S.; Kornblau, S.; Konopleva, M.; Kadia, T.; Kantarjian, H.; et al. Validation of a Clinical Assay of Multi-Locus DNA Methylation for Prognosis of Newly Diagnosed AML. Am. J. Hematol. 2017, 92, E14–E15. [Google Scholar] [CrossRef] [PubMed]
- Metzeler, K.H.; Hummel, M.; Bloomfield, C.D.; Spiekermann, K.; Braess, J.; Sauerland, M.-C.; Heinecke, A.; Radmacher, M.; Marcucci, G.; Whitman, S.P.; et al. An 86-Probe-Set Gene-Expression Signature Predicts Survival in Cytogenetically Normal Acute Myeloid Leukemia. Blood 2008, 112, 4193–4201. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.H.; Simonds, E.F.; Bendall, S.C.; Davis, K.L.; Amir, E.D.; Tadmor, M.D.; Litvin, O.; Fienberg, H.G.; Jager, A.; Zunder, E.R.; et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells That Correlate with Prognosis. Cell 2015, 162, 184–197. [Google Scholar] [CrossRef]
- Eppert, K.; Takenaka, K.; Lechman, E.R.; Waldron, L.; Nilsson, B.; van Galen, P.; Metzeler, K.H.; Poeppl, A.; Ling, V.; Beyene, J.; et al. Stem Cell Gene Expression Programs Influence Clinical Outcome in Human Leukemia. Nat. Med. 2011, 17, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.W.K.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; et al. A 17-Gene Stemness Score for Rapid Determination of Risk in Acute Leukaemia. Nature 2016, 540, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Duployez, N.; Marceau-Renaut, A.; Villenet, C.; Petit, A.; Rousseau, A.; Ng, S.W.K.; Paquet, A.; Gonzales, F.; Barthélémy, A.; Leprêtre, F.; et al. The Stem Cell-Associated Gene Expression Signature Allows Risk Stratification in Pediatric Acute Myeloid Leukemia. Leukemia 2019, 33, 348–357. [Google Scholar] [CrossRef]
- Bill, M.; Nicolet, D.; Kohlschmidt, J.; Walker, C.J.; Mrózek, K.; Eisfeld, A.-K.; Papaioannou, D.; Rong-Mullins, X.; Brannan, Z.; Kolitz, J.E.; et al. Mutations Associated with a 17-Gene Leukemia Stem Cell Score and Its Prognostic Relevance in the Context of the European LeukemiaNet Classification for Acute Myeloid Leukemia. Haematologica 2019, 105, 721–729. [Google Scholar] [CrossRef]
- Vasseur, L.; Fenwarth, L.; Lambert, J.; de Botton, S.; Figeac, M.; Villenet, C.; Heiblig, M.; Dumas, P.; Récher, C.; Berthon, C.; et al. LSC17 Score Complements Genetics and Measurable Residual Disease in Acute Myeloid Leukemia: An ALFA Study. Blood Adv. 2023, 7, 1219–1224. [Google Scholar] [CrossRef]
- Legrand, O.; Simonin, G.; Perrot, J.Y.; Zittoun, R.; Marie, J.P. Pgp and MRP Activities Using Calcein-AM Are Prognostic Factors in Adult Acute Myeloid Leukemia Patients. Blood 1998, 91, 4480–4488. [Google Scholar] [CrossRef] [PubMed]
- Laupeze, B.; Amiot, L.; Drenou, B.; Bernard, M.; Branger, B.; Grosset, J.M.; Lamy, T.; Fauchet, R.; Fardel, O. High Multidrug Resistance Protein Activity in Acute Myeloid Leukaemias Is Associated with Poor Response to Chemotherapy and Reduced Patient Survival. Br. J. Haematol. 2002, 116, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Ong, Y.L.; McMullin, M.F.; Bailie, K.E.; Lappin, T.R.; Jones, F.G.; Irvine, A.E. High Bax Expression Is a Good Prognostic Indicator in Acute Myeloid Leukaemia. Br. J. Haematol. 2000, 111, 182–189. [Google Scholar] [CrossRef]
- Del Poeta, G.; Venditti, A.; Del Principe, M.I.; Maurillo, L.; Buccisano, F.; Tamburini, A.; Cox, M.C.; Franchi, A.; Bruno, A.; Mazzone, C.; et al. Amount of Spontaneous Apoptosis Detected by Bax/Bcl-2 Ratio Predicts Outcome in Acute Myeloid Leukemia (AML). Blood 2003, 101, 2125–2131. [Google Scholar] [CrossRef] [PubMed]
- Venditti, A.; Poeta, G.D.; Maurillo, L.; Buccisano, F.; Principe, M.D.; Mazzone, C.; Tamburini, A.; Cox, C.; Panetta, P.; Neri, B.; et al. Combined Analysis of Bcl-2 and MDR1 Proteins in 256 Cases of Acute Myeloid Leukemia. Haematologica 2004, 89, 934–939. [Google Scholar]
- Carter, B.Z.; Qiu, Y.; Huang, X.; Diao, L.; Zhang, N.; Coombes, K.R.; Mak, D.H.; Konopleva, M.; Cortes, J.; Kantarjian, H.M.; et al. Survivin Is Highly Expressed in CD34+38− Leukemic Stem/Progenitor Cells and Predicts Poor Clinical Outcomes in AML. Blood 2012, 120, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Kornblau, S.M.; Tibes, R.; Qiu, Y.H.; Chen, W.; Kantarjian, H.M.; Andreeff, M.; Coombes, K.R.; Mills, G.B. Functional Proteomic Profiling of AML Predicts Response and Survival. Blood 2009, 113, 154–164. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Wolf, S.; Buettner, F.; Alexe, G.; Häupl, B.; Comoglio, F.; Schneider, C.; Doebele, C.; Fuhrmann, D.C.; Wagner, S.; et al. The Proteogenomic Subtypes of Acute Myeloid Leukemia. Cancer Cell 2022, 40, 301–317.e12. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.-T.; Ryan, J.; Carrasco, R.; Neuberg, D.; Rossi, D.J.; Stone, R.M.; Deangelo, D.J.; Frattini, M.G.; Letai, A. Relative Mitochondrial Priming of Myeloblasts and Normal HSCs Determines Chemotherapeutic Success in AML. Cell 2012, 151, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Dal Bello, R.; Pacchiardi, K.; Chauvel, C.; Adès, L.; Braun, T.; Pasanisi, J.; Fournier, E.; Berthon, C.; Clappier, E.; Raffoux, E.; et al. Relative Mitochondrial Priming Predicts Survival in Older AML Patients Treated Intensively. HemaSphere 2023, 7, e819. [Google Scholar] [CrossRef] [PubMed]
- Benard, B.A.; Leak, L.B.; Azizi, A.; Thomas, D.; Gentles, A.J.; Majeti, R. Clonal Architecture Predicts Clinical Outcomes and Drug Sensitivity in Acute Myeloid Leukemia. Nat. Commun. 2021, 12, 7244. [Google Scholar] [CrossRef]
- Miles, L.A.; Bowman, R.L.; Merlinsky, T.R.; Csete, I.S.; Ooi, A.T.; Durruthy-Durruthy, R.; Bowman, M.; Famulare, C.; Patel, M.A.; Mendez, P.; et al. Single-Cell Mutation Analysis of Clonal Evolution in Myeloid Malignancies. Nature 2020, 587, 477–482. [Google Scholar] [CrossRef]
- Duchmann, M.; Micol, J.-B.; Duployez, N.; Raffoux, E.; Thomas, X.; Marolleau, J.-P.; Braun, T.; Adès, L.; Chantepie, S.; Lemasle, E.; et al. Prognostic Significance of Concurrent Gene Mutations in Intensively Treated Patients with IDH-Mutated AML: An ALFA Study. Blood 2021, 137, 2827–2837. [Google Scholar] [CrossRef]
- Bochtler, T.; Stölzel, F.; Heilig, C.E.; Kunz, C.; Mohr, B.; Jauch, A.; Janssen, J.W.G.; Kramer, M.; Benner, A.; Bornhäuser, M.; et al. Clonal Heterogeneity as Detected by Metaphase Karyotyping Is an Indicator of Poor Prognosis in Acute Myeloid Leukemia. J. Clin. Oncol. 2013, 31, 3898–3905. [Google Scholar] [CrossRef]
- Li, S.; Garrett-Bakelman, F.E.; Chung, S.S.; Sanders, M.A.; Hricik, T.; Rapaport, F.; Patel, J.; Dillon, R.; Vijay, P.; Brown, A.L.; et al. Distinct Evolution and Dynamics of Epigenetic and Genetic Heterogeneity in Acute Myeloid Leukemia. Nat. Med. 2016, 22, 792–799. [Google Scholar] [CrossRef]
- Cerrano, M.; Duchmann, M.; Kim, R.; Vasseur, L.; Hirsch, P.; Thomas, X.; Quentin, S.; Pasanisi, J.; Passet, M.; Rabian, F.; et al. Clonal Dominance Is an Adverse Prognostic Factor in Acute Myeloid Leukemia Treated with Intensive Chemotherapy. Leukemia 2021, 35, 712–723. [Google Scholar] [CrossRef]
- Morita, K.; Wang, F.; Jahn, K.; Hu, T.; Tanaka, T.; Sasaki, Y.; Kuipers, J.; Loghavi, S.; Wang, S.A.; Yan, Y.; et al. Clonal Evolution of Acute Myeloid Leukemia Revealed by High-Throughput Single-Cell Genomics. Nat. Commun. 2020, 11, 5327. [Google Scholar] [CrossRef]
- Duchmann, M.; Laplane, L.; Itzykson, R. Clonal Architecture and Evolutionary Dynamics in Acute Myeloid Leukemias. Cancers 2021, 13, 4887. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Meira, A.; Buck, G.; Clark, S.-A.; Povinelli, B.J.; Alcolea, V.; Louka, E.; McGowan, S.; Hamblin, A.; Sousos, N.; Barkas, N.; et al. Unravelling Intratumoral Heterogeneity through High-Sensitivity Single-Cell Mutational Analysis and Parallel RNA Sequencing. Mol. Cell. 2019, 73, 1292–1305.e8. [Google Scholar] [CrossRef]
- Duchmann, M.; Joudinaud, R.; Boudry, A.; Pasanisi, J.; Di Feo, G.; Kim, R.; Bucci, M.; Chauvel, C.; Chat, L.; Larcher, L.; et al. Hematopoietic Differentiation at Single-Cell Resolution in NPM1-Mutated AML. Blood Cancer J. 2022, 12, 136. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, J.J.; Blaise, D. Hematopoietic Stem Cell Transplantation for Patients with AML in First Complete Remission. Blood 2016, 127, 62–70. [Google Scholar] [CrossRef]
- Pastore, F.; Dufour, A.; Benthaus, T.; Metzeler, K.H.; Maharry, K.S.; Schneider, S.; Ksienzyk, B.; Mellert, G.; Zellmeier, E.; Kakadia, P.M.; et al. Combined Molecular and Clinical Prognostic Index for Relapse and Survival in Cytogenetically Normal Acute Myeloid Leukemia. J. Clin. Oncol. 2014, 32, 1586–1594. [Google Scholar] [CrossRef]
- Peterlin, P.; Renneville, A.; Abdelali, R.B.; Nibourel, O.; Thomas, X.; Pautas, C.; de Botton, S.; Raffoux, E.; Cayuela, J.-M.; Boissel, N.; et al. Impact of Additional Genetic Alterations on the Outcome of Patients with NPM1-Mutated Cytogenetically Normal Acute Myeloid Leukemia. Haematologica 2015, 100, e196–e199. [Google Scholar] [CrossRef]
- Bezerra, M.F.; Lima, A.S.; Piqué-Borràs, M.-R.; Silveira, D.R.; Coelho-Silva, J.L.; Pereira-Martins, D.A.; Weinhäuser, I.; Franca-Neto, P.L.; Quek, L.; Corby, A.; et al. Co-Occurrence of DNMT3A, NPM1, FLT3 Mutations Identifies a Subset of Acute Myeloid Leukemia with Adverse Prognosis. Blood 2020, 135, 870–875. [Google Scholar] [CrossRef]
- Hernández Sánchez, A.; Villaverde Ramiro, A.; Sträng, E.; Gastone, C.; Heckman, C.A.; Versluis, J.; Abáigar, M.; Sobas, M.A.; Azibeiro Melchor, R.; Benner, A.; et al. Machine Learning Allows the Identification of New Co-Mutational Patterns with Prognostic Implications in NPM1 Mutated AML—Results of the European Harmony Alliance. Blood 2022, 140, 739–742. [Google Scholar] [CrossRef]
- Gerstung, M.; Papaemmanuil, E.; Martincorena, I.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Heuser, M.; Thol, F.; Bolli, N.; Ganly, P.; et al. Precision Oncology for Acute Myeloid Leukemia Using a Knowledge Bank Approach. Nat. Genet. 2017, 49, 332–340. [Google Scholar] [CrossRef]
- Huet, S.; Paubelle, E.; Lours, C.; Grange, B.; Courtois, L.; Chabane, K.; Charlot, C.; Mosnier, I.; Simonet, T.; Hayette, S.; et al. Validation of the Prognostic Value of the Knowledge Bank Approach to Determine AML Prognosis in Real Life. Blood 2018, 132, 865–867. [Google Scholar] [CrossRef]
- Bill, M.; Mrózek, K.; Giacopelli, B.; Kohlschmidt, J.; Nicolet, D.; Papaioannou, D.; Eisfeld, A.-K.; Kolitz, J.E.; Powell, B.L.; Carroll, A.J.; et al. Precision Oncology in AML: Validation of the Prognostic Value of the Knowledge Bank Approach and Suggestions for Improvement. J. Hematol. Oncol. 2021, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Fenwarth, L.; Thomas, X.; de Botton, S.; Duployez, N.; Bourhis, J.-H.; Lesieur, A.; Fortin, G.; Meslin, P.-A.; Yakoub-Agha, I.; Sujobert, P.; et al. A Personalized Approach to Guide Allogeneic Stem Cell Transplantation in Younger Adults with Acute Myeloid Leukemia. Blood 2021, 137, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Fournier, E.; Berthon, C.; Röllig, C.; Braun, T.; Marceau-Renaut, A.; Pautas, C.; Nibourel, O.; Lemasle, E.; Micol, J.-B.; et al. Genetic Identification of Patients with AML Older than 60 Years Achieving Long-Term Survival with Intensive Chemotherapy. Blood 2021, 138, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, S.; Borthakur, G.; Daver, N.; DiNardo, C.D.; Pemmaraju, N.; Short, N.J.; Abbas, H.A.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F.; et al. Validation of ALFA 1200 Score in Patients with AML >60 Years Treated with Double Nucleoside-Based Low-Intensity Therapy. Blood Adv. 2022, 6, 5546–5549. [Google Scholar] [CrossRef]
- Cerrano, M.; Itzykson, R. New Treatment Options for Acute Myeloid Leukemia in 2019. Curr. Oncol. Rep. 2019, 21, 16. [Google Scholar] [CrossRef]
- Calleja, A.; Loschi, M.; Bailly, L.; Morisot, A.; Marceau, A.; Mannone, L.; Robert, G.; Auberger, P.; Preudhomme, C.; Raynaud, S.; et al. Real-Life Challenges Using Personalized Prognostic Scoring Systems in Acute Myeloid Leukemia. Cancer Med. 2023, 12, 5656–5660. [Google Scholar] [CrossRef]
- Herrmann, L.; Bischof, L.; Backhaus, D.; Brauer, D.; Franke, G.-N.; Vucinic, V.; Platzbecker, U.; Schwind, S.; Jentzsch, M. Outcome Prediction by the Knowledge Bank Approach in Acute Myeloid Leukemia Patients Undergoing Allogeneic Stem Cell Transplantation. Am. J. Hematol. 2022, 97, E382–E384. [Google Scholar] [CrossRef]
- Letai, A.; Bhola, P.; Welm, A.L. Functional Precision Oncology: Testing Tumors with Drugs to Identify Vulnerabilities and Novel Combinations. Cancer Cell 2022, 40, 26–35. [Google Scholar] [CrossRef]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional Genomic Landscape of Acute Myeloid Leukaemia. Nature 2018, 562, 526–531. [Google Scholar] [CrossRef]
- Dal Bello, R.; Pasanisi, J.; Joudinaud, R.; Duchmann, M.; Pardieu, B.; Ayaka, P.; Di Feo, G.; Sodaro, G.; Chauvel, C.; Kim, R.; et al. A Multiparametric Niche-like Drug Screening Platform in Acute Myeloid Leukemia. Blood Cancer J. 2022, 12, 95. [Google Scholar] [CrossRef]
- Walter, R.B.; Estey, E.H. Selection of Initial Therapy for Newly-Diagnosed Adult Acute Myeloid Leukemia: Limitations of Predictive Models. Blood Rev. 2020, 44, 100679. [Google Scholar] [CrossRef] [PubMed]
| Detection Methods | |
| qPCR | NPM1-mutated AML, CBF AML (RUNX1::RUNX1T1 or CBFB::MYH11) |
| MFC * | AML lacking a molecular marker |
| NGS | At present, there are insufficient data to recommend it as a stand-alone technique |
| Timing Assessment | |
| qPCR-MRD | In PB, after two cycles of chemotherapy; in BM, at the end of consolidation; and in BM, every 3 months; or in PB, every 4–6 weeks for 24 months after the end of consolidation |
| MFC-MRD | In BM, after two cycles of chemotherapy, at the end of consolidation and prior to HCT |
| MRD-Driven Treatment Decisions | |
| Additional consolidation strategies |
|
| No change in treatment | Patients with NPM1-mutated or CBF AML who have stable molecular MRD detection at low level (MRD-LL) |
| ELN 2017 | ELN 2022 | Comments |
|---|---|---|
| Favorable Risk | ||
| t(8;21)(q22;q22.1)/RUNX1::RUNX1T1 | t(8;21)(q22;q22.1)/RUNX1::RUNX1T1 | |
| inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB::MYH11 | inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB::MYH11 | |
| Mutated NPM1 without FLT3-ITD or with FLT3-ITD low | Mutated NPM1 without FLT3-ITD (and without adverse-risk cytogenetics) | FLT3-ITD allelic ratio is no longer considered due to the impact of midostaurin-based regimens and the absence of a standardized assay to assess it |
| Biallelic mutated CEBPA | bZIP in-frame mutated CEBPA | Mono- or biallelic mutational state lost its prognostic weight in the latter classification, with inframe bZIP mutations gaining a predominant role |
| Intermediate Risk | ||
| Mutated NPM1 with FLT3-ITD high | Mutated NPM1 with FLT3-ITD (and without adverse-risk cytogenetics) | |
| Wild-type NPM1 without FLT3-ITD or with FLT3-ITD low (without adverse-risk genetic lesions) | Wild-type NPM1 with FLT3-ITD (without adverse-risk genetic lesions or favorable cytogenetics) | FLT3-ITD showed an independent prognostic impact, globally placing patients at intermediate risk |
| t(9;11)(p21.3;q23.3)/MLLT3::KMT2A | t(9;11)(p21.3;q23.3)/MLLT3::KMT2A | |
| Cytogenetic abnormalities not classified as favorable or adverse | Cytogenetic abnormalities not classified as favorable or adverse | |
| Adverse Risk | ||
| t(6;9)(p23;q34.1); DEK::NUP214 | t(6;9)(p23;q34.1); DEK::NUP214 | |
| t(v;11q23.3); KMT2A rearranged | t(v;11q23.3); KMT2A-rearranged | |
| t(9;22)(q34.1;q11.2); BCR::ABL1 | t(9;22)(q34.1;q11.2); BCR::ABL1 | |
| t(8;16)(p11.2;p13.3)/KAT6A::CREBBP | New cytogenetic abnormality included in the ELN 2022 classification | |
| inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2); GATA2, MECOM(EVI1) | inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2)/GATA2, MECOM(EVI1) | |
| t(3q26.2;v)/MECOM(EVI1)-rearranged | New cytogenetic abnormality included in the ELN 2022 classification | |
| −5 or del(5q); −7; −17/abn(17p) | −5 or del(5q); −7; −17/abn(17p) | |
| Complex karyotype, monosomal karyotype | Complex karyotype, monosomal karyotype | Multiple trisomies or polysomies no longer define CK |
| Mutated RUNX1, ASXL1 | Mutated ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and/or ZRSR2 | Additional gene mutations are added, irrespective of prior MDS history |
| Mutated TP53 | Mutated TP53 | At least a 10% VAF is required to classify patients as TP53-mutated |
| Wild-type NPM1 and FLT3-ITD high | FLT3-ITD define an intermediate risk, irrespective of its allelic ratio or concurrent NPM1 mutations | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boscaro, E.; Urbino, I.; Catania, F.M.; Arrigo, G.; Secreto, C.; Olivi, M.; D’Ardia, S.; Frairia, C.; Giai, V.; Freilone, R.; et al. Modern Risk Stratification of Acute Myeloid Leukemia in 2023: Integrating Established and Emerging Prognostic Factors. Cancers 2023, 15, 3512. https://doi.org/10.3390/cancers15133512
Boscaro E, Urbino I, Catania FM, Arrigo G, Secreto C, Olivi M, D’Ardia S, Frairia C, Giai V, Freilone R, et al. Modern Risk Stratification of Acute Myeloid Leukemia in 2023: Integrating Established and Emerging Prognostic Factors. Cancers. 2023; 15(13):3512. https://doi.org/10.3390/cancers15133512
Chicago/Turabian StyleBoscaro, Eleonora, Irene Urbino, Federica Maria Catania, Giulia Arrigo, Carolina Secreto, Matteo Olivi, Stefano D’Ardia, Chiara Frairia, Valentina Giai, Roberto Freilone, and et al. 2023. "Modern Risk Stratification of Acute Myeloid Leukemia in 2023: Integrating Established and Emerging Prognostic Factors" Cancers 15, no. 13: 3512. https://doi.org/10.3390/cancers15133512
APA StyleBoscaro, E., Urbino, I., Catania, F. M., Arrigo, G., Secreto, C., Olivi, M., D’Ardia, S., Frairia, C., Giai, V., Freilone, R., Ferrero, D., Audisio, E., & Cerrano, M. (2023). Modern Risk Stratification of Acute Myeloid Leukemia in 2023: Integrating Established and Emerging Prognostic Factors. Cancers, 15(13), 3512. https://doi.org/10.3390/cancers15133512







