IDH Mutations in Chondrosarcoma: Case Closed or Not?
Abstract
Simple Summary
Abstract
1. Introduction
2. Frequency and Prognostic Value of IDH1 and IDH2 Mutations
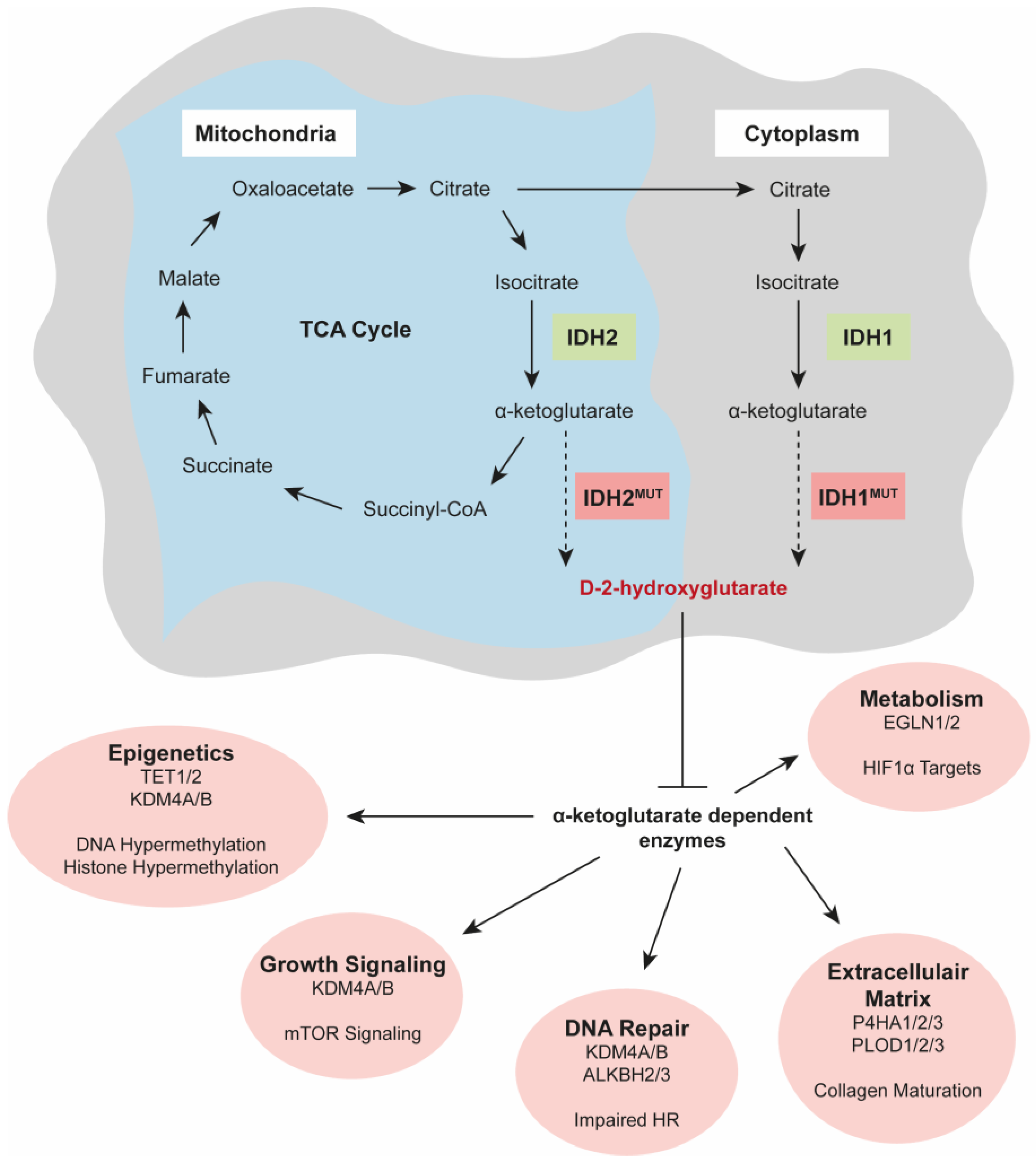
3. Oncogenic Activities of IDH Mutations
4. Inhibition of the IDHMUT Protein
5. Synthetic Lethal Interactions with the IDH Mutation
6. Putting the IDH Mutation into Context to Define Underlying Vulnerabilities
7. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Bovée, J.V.M.G.; Bloem, J.L.; Flanagan, A.M.; Nielsen, G.P.; Yoshida, A. Central Atypical Cartilaginous Tumour/Chondrosarcoma, Grade 1. In WHO Classification of Tumours—Soft Tissue and Bone Tumours; WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 370–372. [Google Scholar]
- Bovée, J.V.M.G.; Bloem, J.L.; Flanagan, A.M.; Nielsen, G.P.; Yoshida, A. Central Chondrosarcoma, Grades 2 and 3. In WHO Classification of Tumours—Soft Tissue and Bone Tumours; WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 375–378. [Google Scholar]
- Bovée, J.V.M.G.; Bloem, J.L.; Flanagan, A.M.; Nielsen, G.P.; Yoshida, A. Enchondroma. In WHO Classification of Tumours—Soft Tissue and Bone Tumours; WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 353–355. [Google Scholar]
- Bovée, J.V.M.G.; Alman, B.A. Enchondromatosis. In WHO Classification of Tumours—Soft Tissue and Bone Tumours; WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 506–509. [Google Scholar]
- Inwards, C.Y.; Bloem, J.L.; Hogendoorn, P.C.W. Dedifferentiated Chondrosarcoma. In WHO Classification of Tumours—Soft Tissue and Bone Tumours; WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 388–390. [Google Scholar]
- Gelderblom, H.; Hogendoorn, P.C.W.; Dijkstra, S.D.; van Rijswijk, C.S.; Krol, A.D.; Taminiau, A.H.M.; Bovee, J.V.M.G. The Clinical Approach Towards Chondrosarcoma. Oncologist 2008, 13, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Pansuriya, T.C.; Van Eijk, R.; D’Adamo, P.; Van Ruler, M.A.J.H.; Kuijjer, M.L.; Oosting, J.; Cleton-Jansen, A.M.; Van Oosterwijk, J.G.; Verbeke, S.L.J.; Meijer, D.; et al. Somatic Mosaic IDH1 and IDH2 Mutations Are Associated with Enchondroma and Spindle Cell Hemangioma in Ollier Disease and Maffucci Syndrome. Nat. Genet. 2011, 43, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Amary, M.F.; Damato, S.; Halai, D.; Eskandarpour, M.; Berisha, F.; Bonar, F.; McCarthy, S.; Fantin, V.R.; Straley, K.S.; Lobo, S.; et al. Ollier Disease and Maffucci Syndrome Are Caused by Somatic Mosaic Mutations of IDH1 and IDH2. Nat. Genet. 2011, 43, 1262–1265. [Google Scholar] [CrossRef] [PubMed]
- Amary, M.F.; Bacsi, K.; Maggiani, F.; Damato, S.; Halai, D.; Berisha, F.; Pollock, R.; O’Donnell, P.; Grigoriadis, A.; Diss, T.; et al. IDH1 and IDH2 Mutations Are Frequent Events in Central Chondrosarcoma and Central and Periosteal Chondromas but Not in Other Mesenchymal Tumours. J. Pathol. 2011, 224, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Fritchie, K.; Wei, S.; Ali, N.; Curless, K.; Shen, T.; Brini, A.T.; Latif, F.; Sumathi, V.; Siegal, G.P.; et al. Diagnostic Utility of IDH1/2 Mutations to Distinguish Dedifferentiated Chondrosarcoma from Undifferentiated Pleomorphic Sarcoma of Bone. Hum. Pathol. 2017, 65, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Hirata, M.; Sasaki, M.; Cairns, R.A.; Inoue, S.; Puviindran, V.; Li, W.Y.; Snow, B.E.; Jones, L.D.; Wei, Q.; Sato, S.; et al. Mutant IDH Is Sufficient to Initiate Enchondromatosis in Mice. Proc. Natl. Acad. Sci. USA 2015, 112, 2829–2834. [Google Scholar] [CrossRef]
- Suijker, J.; Baelde, H.J.; Roelofs, H.; Cleton-Jansen, A.M.; Bovée, J.V.M.G. The Oncometabolite D-2-Hydroxyglutarate Induced by Mutant IDH1 or -2 Blocks Osteoblast Differentiation in Vitro and in Vivo. Oncotarget 2015, 6, 14832–14842. [Google Scholar] [CrossRef]
- Jin, Y.; Elalaf, H.; Watanabe, M.; Tamaki, S.; Hineno, S.; Matsunaga, K.; Woltjen, K.; Kobayashi, Y.; Nagata, S.; Ikeya, M.; et al. Mutant Idh1 Dysregulates the Differentiation of Mesenchymal Stem Cells in Association with Gene-Specific Histone Modifications to Cartilage- and Bone-Related Genes. PLoS ONE 2015, 10, e0131998. [Google Scholar] [CrossRef]
- Clark, O.; Yen, K.; Mellinghoff, I.K. Molecular Pathways: Isocitrate Dehydrogenase Mutations in Cancer. Clin. Cancer Res. 2016, 22, 1837–1842. [Google Scholar] [CrossRef]
- Cleven, A.H.G.; Suijker, J.; Agrogiannis, G.; Briaire-de Bruijn, I.H.; Frizzell, N.; Hoekstra, A.S.; Wijers-Koster, P.M.; Cleton-Jansen, A.-M.; Bovée, J.V.M.G. IDH1 or -2 Mutations Do Not Predict Outcome and Do Not Cause Loss of 5-Hydroxymethylcytosine or Altered Histone Modifications in Central Chondrosarcomas. Clin. Sarcoma Res. 2017, 7, 8. [Google Scholar] [CrossRef]
- Tesileanu, C.M.S.; Vallentgoed, W.R.; Sanson, M.; Taal, W.; Clement, P.M.; Wick, W.; Brandes, A.A.; Baurain, J.F.; Chinot, O.L.; Wheeler, H.; et al. Non-IDH1-R132H IDH1/2 Mutations Are Associated with Increased DNA Methylation and Improved Survival in Astrocytomas, Compared to IDH1-R132H Mutations. Acta Neuropathol. 2021, 141, 945–957. [Google Scholar] [CrossRef]
- Ward, P.S.; Lu, C.; Cross, J.R.; Abdel-Wahab, O.; Levine, R.L.; Schwartz, G.K.; Thompson, C.B. The Potential for Isocitrate Dehydrogenase Mutations to Produce 2-Hydroxyglutarate Depends on Allele Specificity and Subcellular Compartmentalization. J. Biol. Chem. 2013, 288, 3804–3815. [Google Scholar] [CrossRef]
- Pusch, S.; Schweizer, L.; Beck, A.C.; Lehmler, J.M.; Weissert, S.; Balss, J.; Miller, A.K.; von Deimling, A. D-2-Hydroxyglutarate Producing Neo-Enzymatic Activity Inversely Correlates with Frequency of the Type of Isocitrate Dehydrogenase 1 Mutations Found in Glioma. Acta Neuropathol. Commun. 2014, 2, 19. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Radivoyevitch, T.; Maciejewski, J.P.; van Noorden, C.J.F.; Bleeker, F.E. The Driver and Passenger Effects of Isocitrate Dehydrogenase 1 and 2 Mutations in Oncogenesis and Survival Prolongation. Biochim. Biophys. Acta 2014, 1846, 326–341. [Google Scholar] [CrossRef]
- Jin, J.; Hu, C.; Yu, M.; Chen, F.; Ye, L.; Yin, X.; Zhuang, Z.; Tong, H. Prognostic Value of Isocitrate Dehydrogenase Mutations in Myelodysplastic Syndromes: A Retrospective Cohort Study and Meta-Analysis. PLoS ONE 2014, 9, e100206. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Ravandi, F.; Agresta, S.; Konopleva, M.; Takahashi, K.; Kadia, T.; Routbort, M.; Patel, K.P.; Brandt, M.; Pierce, S.; et al. Characteristics, Clinical Outcome, and Prognostic Significance of IDH Mutations in AML. Am. J. Hematol. 2015, 90, 732–736. [Google Scholar] [CrossRef]
- Xia, L.; Wu, B.; Fu, Z.; Feng, F.; Qiao, E.; Li, Q.; Sun, C.; Ge, M. Prognostic Role of IDH Mutations in Gliomas: A Meta-Analysis of 55 Observational Studies. Oncotarget 2015, 6, 17354–17365. [Google Scholar] [CrossRef]
- Goyal, L.; Govindan, A.; Sheth, R.A.; Nardi, V.; Blaszkowsky, L.S.; Faris, J.E.; Clark, J.W.; Ryan, D.P.; Kwak, E.L.; Allen, J.N.; et al. Prognosis and Clinicopathologic Features of Patients with Advanced Stage Isocitrate Dehydrogenase (IDH) Mutant and IDH Wild-Type Intrahepatic Cholangiocarcinoma. Oncologist 2015, 20, 1019–1027. [Google Scholar] [CrossRef]
- Lugowska, I.; Teterycz, P.; Mikula, M.; Kulecka, M.; Kluska, A.; Balabas, A.; Piatkowska, M.; Wagrodzki, M.; Pienkowski, A.; Rutkowski, P.; et al. IDH1/2 Mutations Predict Shorter Survival in Chondrosarcoma. J. Cancer 2018, 9, 998–1005. [Google Scholar] [CrossRef]
- Zhu, G.G.; Nafa, K.; Agaram, N.; Zehir, A.; Benayed, R.; Sadowska, J.; Borsu, L.; Kelly, C.; Tap, W.D.; Fabbri, N.; et al. Genomic Profiling Identifies Association of IDH1/IDH2 Mutation with Longer Relapse-Free and Metastasis-Free Survival in High-Grade Chondrosarcoma. Clin. Cancer Res. 2020, 26, 419–427. [Google Scholar] [CrossRef]
- Wang, F.; Travins, J.; DeLaBarre, B.; Penard-Lacronique, V.; Schalm, S.; Hansen, E.; Straley, K.; Kernytsky, A.; Liu, W.; Gliser, C.; et al. Targeted Inhibition of Mutant IDH2 in Leukemia Cells Induces Cellular Differentiation. Science 2013, 340, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Wakimoto, H.; Iafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, H.; Tateishi, K.; Kato, H.; Nakatsuka, T.; Yamamoto, K.; Tanaka, Y.; Ijichi, H.; Takahara, N.; Mizuno, S.; Kogure, H.; et al. Isocitrate Dehydrogenase 1 Mutation Sensitizes Intrahepatic Cholangiocarcinoma to the BET Inhibitor JQ1. Cancer Sci. 2018, 109, 3602–3610. [Google Scholar] [CrossRef] [PubMed]
- Suijker, J.; Oosting, J.; Koornneef, A.; Struys, E.A.; Salomons, G.S.; Schaap, F.G.; Waaijer, C.J.F.; Wijers-Koster, P.M.; Briaire-de Bruijn, I.H.; Haazen, L.; et al. Inhibition of Mutant IDH1 Decreases D-2-HG Levels without Affecting Tumorigenic Properties of Chondrosarcoma Cell Lines. Oncotarget 2015, 6, 12505–12519. [Google Scholar] [CrossRef]
- Li, L.; Paz, A.C.; Wilky, B.A.; Johnson, B.; Galoian, K.; Rosenberg, A.; Hu, G.; Tinoco, G.; Bodamer, O.; Trent, J.C. Treatment with a Small Molecule Mutant IDH1 Inhibitor Suppresses Tumorigenic Activity and Decreases Production of the Oncometabolite 2-Hydroxyglutarate in Human Chondrosarcoma Cells. PLoS ONE 2015, 10, e0133813. [Google Scholar] [CrossRef]
- Nakagawa, M.; Nakatani, F.; Matsunaga, H.; Seki, T.; Endo, M.; Ogawara, Y.; Machida, Y.; Katsumoto, T.; Yamagata, K.; Hattori, A.; et al. Selective Inhibition of Mutant IDH1 by DS-1001b Ameliorates Aberrant Histone Modifications and Impairs Tumor Activity in Chondrosarcoma. Oncogene 2019, 38, 6835–6849. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Stein, E.M.; de Botton, S.; Roboz, G.J.; Altman, J.K.; Mims, A.S.; Swords, R.; Collins, R.H.; Mannis, G.N.; Pollyea, D.A.; et al. Durable Remissions with Ivosidenib in IDH1 -Mutated Relapsed or Refractory AML. N. Engl. J. Med. 2018, 378, 2386–2398. [Google Scholar] [CrossRef]
- Intlekofer, A.M.; Shih, A.H.; Wang, B.; Nazir, A.; Rustenburg, A.S.; Albanese, S.K.; Patel, M.; Famulare, C.; Correa, F.M.; Takemoto, N.; et al. Acquired Resistance to IDH Inhibition through Trans or Cis Dimer-Interface Mutations. Nature 2018, 559, 125–129. [Google Scholar] [CrossRef]
- Harding, J.J.; Lowery, M.A.; Shih, A.H.; Schvartzman, J.M.; Hou, S.; Famulare, C.; Patel, M.; Roshal, M.; Do, R.K.; Zehir, A.; et al. Isoform Switching as a Mechanism of Acquired Resistance to Mutant Isocitrate Dehydrogenase Inhibition. Cancer Discov. 2018, 8, 1540–1547. [Google Scholar] [CrossRef]
- Choe, S.; Wang, H.; DiNardo, C.D.; Stein, E.M.; de Botton, S.; Roboz, G.J.; Altman, J.K.; Mims, A.S.; Watts, J.M.; Pollyea, D.A.; et al. Molecular Mechanisms Mediating Relapse Following Ivosidenib Monotherapy in IDH1-Mutant Relapsed or Refractory AML. Blood Adv. 2020, 4, 1894–1905. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; Ellingson, B.M.; Touat, M.; Maher, E.; De La Fuente, M.I.; Holdhoff, M.; Cote, G.M.; Burris, H.; Janku, F.; Young, R.J.; et al. Ivosidenib in Isocitrate Dehydrogenase 1-Mutated Advanced Glioma. J. Clin. Oncol. 2020, 38, 3398–3406. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Macarulla, T.; Javle, M.M.; Kelley, R.K.; Lubner, S.J.; Adeva, J.; Cleary, J.M.; Catenacci, D.V.; Borad, M.J.; Bridgewater, J.; et al. Ivosidenib in IDH1-Mutant, Chemotherapy-Refractory Cholangiocarcinoma (ClarIDHy): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Study. Lancet Oncol. 2020, 21, 796–807. [Google Scholar] [CrossRef]
- Tap, W.D.; Villalobos, V.M.; Cote, G.M.; Burris, H.; Janku, F.; Mir, O.; Beeram, M.; Wagner, A.J.; Jiang, L.; Wu, B.; et al. Phase I Study of the Mutant IDH1 Inhibitor Ivosidenib: Safety and Clinical Activity in Patients with Advanced Chondrosarcoma. J. Clin. Oncol. 2020, 38, 1693–1701. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.T.; et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef]
- Chowdhury, R.; Yeoh, K.K.; Tian, Y.M.; Hillringhaus, L.; Bagg, E.A.; Rose, N.R.; Leung, I.K.H.; Li, X.S.; Woon, E.C.Y.; Yang, M.; et al. The Oncometabolite 2-Hydroxyglutarate Inhibits Histone Lysine Demethylases. EMBO Rep. 2011, 12, 463–469. [Google Scholar] [CrossRef]
- Gagné, L.M.; Boulay, K.; Topisirovic, I.; Huot, M.É.; Mallette, F.A. Oncogenic Activities of IDH1/2 Mutations: From Epigenetics to Cellular Signaling. Trends Cell Biol. 2017, 27, 738–752. [Google Scholar] [CrossRef]
- Hvinden, I.C.; Cadoux-Hudson, T.; Schofield, C.J.; McCullagh, J.S.O. Metabolic Adaptations in Cancers Expressing Isocitrate Dehydrogenase Mutations. Cell Rep. Med. 2021, 2, 100469. [Google Scholar] [CrossRef]
- Venneker, S.; Kruisselbrink, A.B.; Baranski, Z.; Palubeckaite, I.; Briaire-de Bruijn, I.H.; Oosting, J.; French, P.J.; Danen, E.H.J.; Bovée, J.V.M.G. Beyond the Influence of IDH Mutations: Exploring Epigenetic Vulnerabilities in Chondrosarcoma. Cancers 2020, 12, 3589. [Google Scholar] [CrossRef]
- Guilhamon, P.; Eskandarpour, M.; Halai, D.; Wilson, G.A.; Feber, A.; Teschendorff, A.E.; Gomez, V.; Hergovich, A.; Tirabosco, R.; Fernanda Amary, M.; et al. Meta-Analysis of IDH-Mutant Cancers Identifies EBF1 as an Interaction Partner for TET2. Nat. Commun. 2013, 4, 2166. [Google Scholar] [CrossRef]
- Liu, S.; Cadoux-Hudson, T.; Schofield, C.J. Isocitrate Dehydrogenase Variants in Cancer—Cellular Consequences and Therapeutic Opportunities. Curr. Opin. Chem. Biol. 2020, 57, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, R.J.; Radivoyevitch, T.; Nagata, Y.; Khurshed, M.; Przychodzen, B.; Makishima, H.; Xu, M.; Bleeker, F.E.; Wilmink, J.W.; Carraway, H.E.; et al. Idh1/2 Mutations Sensitize Acute Myeloid Leukemia to Parp Inhibition and This Is Reversed by Idh1/2-Mutant Inhibitors. Clin. Cancer Res. 2018, 24, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chou, A.P.; Chen, W.; Chen, R.; Deng, Y.; Phillips, H.S.; Selfridge, J.; Zurayk, M.; Lou, J.J.; Everson, R.G.; et al. Overexpression of Isocitrate Dehydrogenase Mutant Proteins Renders Glioma Cells More Sensitive to Radiation. Neuro. Oncol. 2013, 15, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Kessler, J.; Güttler, A.; Wichmann, H.; Rot, S.; Kappler, M.; Bache, M.; Vordermark, D. IDH1R132H Mutation Causes a Less Aggressive Phenotype and Radiosensitizes Human Malignant Glioma Cells Independent of the Oxygenation Status. Radiother. Oncol. 2015, 116, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Kwintkiewicz, J.; Liu, Y.; Tech, K.; Frady, L.N.; Su, Y.T.; Bautista, W.; Moon, S.I.; MacDonald, J.; Ewend, M.G.; et al. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res. 2017, 77, 1709–1718. [Google Scholar] [CrossRef]
- Tateishi, K.; Higuchi, F.; Miller, J.J.; Koerner, M.V.A.; Lelic, N.; Shankar, G.M.; Tanaka, S.; Fisher, D.E.; Batchelor, T.T.; Iafrate, A.J.; et al. The Alkylating Chemotherapeutic Temozolomide Induces Metabolic Stress in IDH1-Mutant Cancers and Potentiates NAD+depletion-Mediated Cytotoxicity. Cancer Res. 2017, 77, 4102–4115. [Google Scholar] [CrossRef]
- Sulkowski, P.L.; Corso, C.D.; Robinson, N.D.; Scanlon, S.E.; Purshouse, K.R.; Bai, H.; Liu, Y.; Sundaram, R.K.; Hegan, D.C.; Fons, N.R.; et al. 2-Hydroxyglutarate Produced by Neomorphic IDH Mutations Suppresses Homologous Recombination and Induces PARP Inhibitor Sensitivity. Sci. Transl. Med. 2017, 9, eaal2463. [Google Scholar] [CrossRef]
- Wang, Y.; Wild, A.T.; Turcan, S.; Wu, W.H.; Sigel, C.; Klimstra, D.S.; Ma, X.; Gong, Y.; Holland, E.C.; Huse, J.T.; et al. Targeting Therapeutic Vulnerabilities with PARP Inhibition and Radiation in IDH-Mutant Gliomas and Cholangiocarcinomas. Sci. Adv. 2020, 6, eaaz3221. [Google Scholar] [CrossRef]
- Chan, S.M.; Thomas, D.; Corces-Zimmerman, M.R.; Xavy, S.; Rastogi, S.; Hong, W.J.; Zhao, F.; Medeiros, B.C.; Tyvoll, D.A.; Majeti, R. Isocitrate Dehydrogenase 1 and 2 Mutations Induce BCL-2 Dependence in Acute Myeloid Leukemia. Nat. Med. 2015, 21, 178–184. [Google Scholar] [CrossRef]
- Karpel-Massler, G.; Ishida, C.T.; Bianchetti, E.; Zhang, Y.; Shu, C.; Tsujiuchi, T.; Banu, M.A.; Garcia, F.; Roth, K.A.; Bruce, J.N.; et al. Induction of Synthetic Lethality in IDH1-Mutated Gliomas through Inhibition of Bcl-XL. Nat. Commun. 2017, 8, 1067. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Lu, C.; Cross, J.R.; Morris IV, J.P.; Shroff, A.S.; Ward, P.S.; Bradner, J.E.; Thompson, C.; Lowe, S.W. Cancer-Associated IDH2 Mutants Drive an Acute Myeloid Leukemia That Is Susceptible to Brd4 Inhibition. Genes Dev. 2013, 27, 1974–1985. [Google Scholar] [CrossRef]
- Turcan, S.; Fabius, A.W.; Borodovsky, A.; Pedraza, A.; Brennan, C.; Huse, J.; Viale, A.; Riggins, G.J.; Chan, T.A. Efficient Induction of Differentiation and Growth Inhibition in IDH1 Mutant Glioma Cells by the DNMT Inhibitor Decitabine. Oncotarget 2013, 4, 1729–1736. [Google Scholar] [CrossRef]
- Batsios, G.; Viswanath, P.; Subramani, E.; Najac, C.; Gillespie, A.M.; Santos, R.D.; Molloy, A.R.; Pieper, R.O.; Ronen, S.M. PI3K/MTOR Inhibition of IDH1 Mutant Glioma Leads to Reduced 2HG Production That Is Associated with Increased Survival. Sci. Rep. 2019, 9, 10521. [Google Scholar] [CrossRef]
- Emadi, A.; Jun, S.A.; Tsukamoto, T.; Fathi, A.T.; Minden, M.D.; Dang, C. V Inhibition of Glutaminase Selectively Suppresses the Growth of Primary Acute Myeloid Leukemia Cells with IDH Mutations. Exp. Hematol. 2014, 42, 247–251. [Google Scholar] [CrossRef]
- Seltzer, M.J.; Bennett, B.D.; Joshi, A.D.; Gao, P.; Thomas, A.G.; Ferraris, D.V.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Rabinowitz, J.D.; et al. Inhibition of Glutaminase Preferentially Slows Growth of Glioma Cells with Mutant IDH1. Cancer Res. 2010, 70, 8981–8987. [Google Scholar] [CrossRef]
- de Jong, Y.; Ingola, M.; Briaire-de Bruijn, I.H.; Kruisselbrink, A.B.; Venneker, S.; Palubeckaite, I.; Heijs, B.P.A.M.; Cleton-Jansen, A.-M.; Haas, R.L.M.; Bovée, J.V.M.G. Radiotherapy Resistance in Chondrosarcoma Cells; a Possible Correlation with Alterations in Cell Cycle Related Genes. Clin. Sarcoma Res. 2019, 9, 9. [Google Scholar] [CrossRef]
- de Jong, Y.; Monderer, D.; Brandinelli, E.; Monchanin, M.; van den Akker, B.E.; van Oosterwijk, J.G.; Blay, J.Y.; Dutour, A.; Bovée, J.V.M.G. Bcl-Xl as the Most Promising Bcl-2 Family Member in Targeted Treatment of Chondrosarcoma. Oncogenesis 2018, 7, 74. [Google Scholar] [CrossRef]
- Addie, R.D.; de Jong, Y.; Alberti, G.; Kruisselbrink, A.B.; Que, I.; Baelde, H.; Bovée, J.V.M.G. Exploration of the Chondrosarcoma Metabolome; the MTOR Pathway as an Important pro-Survival Pathway. J. Bone Oncol. 2019, 15, 100222. [Google Scholar] [CrossRef]
- Peterse, E.F.P.; van den Akker, B.E.W.M.; Niessen, B.; Oosting, J.; Suijker, J.; de Jong, Y.; Danen, E.H.J.; Cleton-Jansen, A.-M.; Bovée, J.V.M.G. NAD Synthesis Pathway Interference Is a Viable Therapeutic Strategy for Chondrosarcoma. Mol. Cancer Res. 2017, 15, 1714–1721. [Google Scholar] [CrossRef]
- Peterse, E.F.P.; Niessen, B.; Addie, R.D.; De Jong, Y.; Cleven, A.H.G.; Kruisselbrink, A.B.; Van Den Akker, B.E.W.M.; Molenaar, R.J.; Cleton-Jansen, A.M.; Bovée, J.V.M.G. Targeting Glutaminolysis in Chondrosarcoma in Context of the IDH1/2 Mutation. Br. J. Cancer 2018, 118, 1074–1083. [Google Scholar] [CrossRef]
- Venneker, S.; Kruisselbrink, A.B.; Briaire-de Bruijn, I.H.; de Jong, Y.; van Wijnen, A.J.; Danen, E.H.J.; Bovée, J.V.M.G. Inhibition of PARP Sensitizes Chondrosarcoma Cell Lines to Chemo- and Radiotherapy Irrespective of the IDH1 or IDH2 Mutation Status. Cancers 2019, 11, 1918. [Google Scholar] [CrossRef] [PubMed]
- Palubeckaitė, I.; Venneker, S.; Van Den Akker, B.E.W.M.; Briaire-de Bruijn, I.H.; Bovée, J.V.M.G. Does PARP Inhibition Sensitize Chondrosarcoma Cell Lines to Chemotherapy or Radiotherapy? Results From a Three-Dimensional Spheroid Cell Model. Clin. Orthop. Relat. Res. 2022, 481, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Garrett, M.; Sperry, J.; Braas, D.; Yan, W.; Le, T.M.; Mottahedeh, J.; Ludwig, K.; Eskin, A.; Qin, Y.; Levy, R.; et al. Metabolic Characterization of Isocitrate Dehydrogenase (IDH) Mutant and IDH Wildtype Gliomaspheres Uncovers Cell Type-Specific Vulnerabilities. Cancer Metab. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Reitman, Z.J.; Duncan, C.G.; Spasojevic, I.; Gooden, D.M.; Rasheed, B.A.; Yang, R.; Lopez, G.Y.; He, Y.; McLendon, R.E.; et al. Disruption of Wild-Type IDH1 Suppresses D-2-Hydroxyglutarate Production in IDH1-Mutated Gliomas. Cancer Res. 2013, 73, 496–501. [Google Scholar] [CrossRef]
- Núñez, F.J.; Mendez, F.M.; Kadiyala, P.; Alghamri, M.S.; Savelieff, M.G.; Garcia-Fabiani, M.B.; Haase, S.; Koschmann, C.; Calinescu, A.-A.; Kamran, N.; et al. IDH1-R132H Acts as a Tumor Suppressor in Glioma via Epigenetic up-Regulation of the DNA Damage Response. Sci. Transl. Med. 2019, 11, eaaq1427. [Google Scholar] [CrossRef]
- Raffel, S.; Falcone, M.; Kneisel, N.; Hansson, J.; Wang, W.; Lutz, C.; Bullinger, L.; Poschet, G.; Nonnenmacher, Y.; Barnert, A.; et al. BCAT1 Restricts AkG Levels in AML Stem Cells Leading to IDHmut-like DNA Hypermethylation. Nature 2017, 551, 384–388. [Google Scholar] [CrossRef]
- Yang, Z.; Hu, N.; Wang, W.; Hu, W.; Zhou, S.; Shi, J.; Li, M.; Jing, Z.; Chen, C.; Zhang, X.; et al. Loss of FBXW7 Correlates with Increased IDH1 Expression in Glioma and Enhances IDH1-Mutant Cancer Cell Sensitivity to Radiation. Cancer Res. 2022, 82, 497–509. [Google Scholar] [CrossRef]
- Hu, C.; Wang, K.; Damon, C.; Fu, Y.; Ma, T.; Kratz, L.; Lal, B.; Ying, M.; Xia, S.; Cahill, D.P.; et al. ATRX Loss Promotes Immunosuppressive Mechanisms in IDH1 Mutant Glioma. Neuro. Oncol. 2021, noab292. [Google Scholar] [CrossRef]
- Wilson, E.R.; Helton, N.M.; Heath, S.E.; Fulton, R.S.; Payton, J.E.; Welch, J.S.; Walter, M.J.; Westervelt, P.; DiPersio, J.F.; Link, D.C.; et al. Focal Disruption of DNA Methylation Dynamics at Enhancers in IDH-Mutant AML Cells. Leukemia 2022, 36, 935–945. [Google Scholar] [CrossRef]
- Lee, S.D.; Song, J.; LeBlanc, V.G.; Marra, M.A. Integrative Multi-Omic Analysis Reveals Neurodevelopmental Gene Dysregulation in CIC-Knockout and IDH1-Mutant Cells. J. Pathol. 2022, 256, 297–309. [Google Scholar] [CrossRef]
- Middeke, J.M.; Metzeler, K.H.; Röllig, C.; Krämer, M.; Eckardt, J.-N.; Stasik, S.; Greif, P.A.; Spiekermann, K.; Rothenberg-Thurley, M.; Krug, U.; et al. Differential Impact of IDH1/2 Mutational Subclasses on Outcome in Adult AML: Results from a Large Multicenter Study. Blood Adv. 2022, 6, 1394–1405. [Google Scholar] [CrossRef]
- Duchmann, M.; Micol, J.B.; Duployez, N.; Raffoux, E.; Thomas, X.; Marolleau, J.P.; Braun, T.; Adès, L.; Chantepie, S.; Lemasle, E.; et al. Prognostic Significance of Concurrent Gene Mutations in Intensively Treated Patients with IDH-Mutated AML: An ALFA Study. Blood 2021, 137, 2827–2837. [Google Scholar] [CrossRef]
- Li, K.K.-W.; Shi, Z.-F.; Malta, T.M.; Chan, A.K.-Y.; Cheng, S.; Kwan, J.S.H.; Yang, R.R.; Poon, W.S.; Mao, Y.; Noushmehr, H.; et al. Identification of Subsets of IDH-Mutant Glioblastomas with Distinct Epigenetic and Copy Number Alterations and Stratified Clinical Risks. Neuro-Oncol. Adv. 2019, 1, vdz015. [Google Scholar] [CrossRef]
- Yang, R.R.; Shi, Z.F.; Zhang, Z.; Chan, A.K.Y.; Aibaidula, A.; Wang, W.-W.; Kwan, J.S.H.; Poon, W.S.; Chen, H.; Li, W.-C.; et al. IDH Mutant Lower Grade (WHO Grades II/III) Astrocytomas Can Be Stratified for Risk by CDKN2A, CDK4 and PDGFRA Copy Number Alterations. Brain Pathol. 2020, 30, 541–553. [Google Scholar] [CrossRef]
- Chan, A.K.Y.; Shi, Z.F.; Li, K.K.W.; Wang, W.W.; Chen, H.; Chung, N.Y.F.; Chan, D.T.M.; Poon, W.S.; Loong, H.H.F.; Liu, X.Z.; et al. Combinations of Single-Gene Biomarkers Can Precisely Stratify 1,028 Adult Gliomas for Prognostication. Front. Oncol. 2022, 12, 839302. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; Reville, P.K.; Kantarjian, H.; Jabbour, E.; Borthakur, G.; Daver, N.; Issa, G.; Furudate, K.; Tanaka, T.; Pierce, S.; et al. Contemporary Outcomes in IDH-mutated AML: The Impact of Co-occurring NPM1 Mutations and Venetoclax-based Treatment. Am. J. Hematol. 2022, 97, 1443–1452. [Google Scholar] [CrossRef]
- Schrage, Y.M.; Lam, S.; Jochemsen, A.G.; Cleton-Jansen, A.M.; Taminiau, A.H.M.; Hogendoorn, P.C.W.; Bovée, J.V.M.G. Central Chondrosarcoma Progression Is Associated with PRb Pathway Alterations: CDK4 down-Regulation and P16 Overexpression Inhibit Cell Growth in Vitro. J. Cell Mol. Med. 2009, 13, 2843–2852. [Google Scholar] [CrossRef]
- Tarpey, P.S.; Behjati, S.; Cooke, S.L.; Van Loo, P.; Wedge, D.C.; Pillay, N.; Marshall, J.; Meara, S.O.; Davies, H.; Zainal, S.N.; et al. Frequent Mutation of the Major Cartilage Collagen Gene COL2A1 in Chondrosarcoma. Nat. Genet. 2013, 45, 923–926. [Google Scholar] [CrossRef]
- Totoki, Y.; Yoshida, A.; Hosoda, F.; Nakamura, H.; Hama, N.; Ogura, K.; Yoshida, A.; Fujiwara, T.; Arai, Y.; Toguchida, J.; et al. Unique Mutation Portraits and Frequent COL2A1 Gene Alteration in Chondrosarcoma. Genome Res. 2014, 24, 1411–1420. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Van Oosterwijk, J.G.; Sicinska, E.; Moss, S.; Remillard, S.P.; Van Wezel, T.; Bühnemann, C.; Hassan, A.B.; Demetri, G.D.; Bovée, J.V.M.G.; et al. Functional Profiling of Receptor Tyrosine Kinases and Downstream Signaling in Human Chondrosarcomas Identifies Pathways for Rational Targeted Therapy. Clin. Cancer Res. 2013, 19, 3796–3807. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Yang, C.; Seger, N.; Hesla, A.C.; Tsagkozis, P.; Larsson, O.; Lin, Y.; Haglund, F. TERT Promoter Mutation Is an Objective Clinical Marker for Disease Progression in Chondrosarcoma. Mod. Pathol. 2021, 34, 2020–2027. [Google Scholar] [CrossRef]
- Cross, W.; Lyskjær, I.; Lesluyes, T.; Hargreaves, S.; Strobl, A.-C.; Davies, C.; Waise, S.; Hames-Fathi, S.; Oukrif, D.; Ye, H.; et al. A Genetic Model for Central Chondrosarcoma Evolution Correlates with Patient Outcome. Genome Med. 2022, 14, 99. [Google Scholar] [CrossRef] [PubMed]
- Koelsche, C.; Schrimpf, D.; Stichel, D.; Sill, M.; Sahm, F.; Reuss, D.E.; Blattner, M.; Worst, B.; Heilig, C.E.; Beck, K.; et al. Sarcoma Classification by DNA Methylation Profiling. Nat. Commun. 2021, 12, 498. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, R.; Ayadi, M.; Gomez-Brouchet, A.; Armenoult, L.; Banneau, G.; Elarouci, N.; Tallegas, M.; Decouvelaere, A.V.; Aubert, S.; Rédini, F.; et al. Integrated Molecular Characterization of Chondrosarcoma Reveals Critical Determinants of Disease Progression. Nat. Commun. 2019, 10, 4622. [Google Scholar] [CrossRef] [PubMed]
| AML | Glioma | Cholangiocarcinoma | Chondrosarcoma | |
|---|---|---|---|---|
| Frequency of IDHMUT | ~10–15% [14] | >70% [14] | ~15–20% [14] | ~50% [7,8,9] |
| Most common IDHMUT variant | IDH2 p.R140Q (~40%) Weak D-2-HG Producer [16,17] | IDH1 p.R132H (~90%) Weak D-2-HG Producer [16,18] | IDH1 p.R132C (~60%) Strong D-2-HG Producer [16,18] | IDH1 p.R132C (~60%) Strong D-2-HG Producer [15,16,18] |
| IDHMUT inhibition in vitro | Differentiation [26] | No effect [27] | No effect [28] | Controversial [27,29,30,31] |
| IDHMUT inhibition clinical trials | ~40% response, secondary resistance [32,33,34,35] | Less promising, prolonged disease control in subset [36] | Less promising, prolonged disease control in subset [37] | Durable disease control in subset [38] |
| IDHMUT effect on outcome | No difference (in MDS: worse prognosis) [20,21] | Better prognosis, due to favourable response? [22] | Beneficial? [23] | Controversial [15,24,25] |
| AML | Glioma | Cholangiocarcinoma | Chondrosarcoma | |
|---|---|---|---|---|
| Radiotherapy | Molenaar 2018 [47] * | Li 2013 [48] Kessler 2015 [49] | De Jong 2019 [61] * | |
| Temozolomide | Lu 2017 [50] Tateishi 2017 [51] * | Venneker 2019 [66] * | ||
| PARP inh. | Sulkowski 2017 [52] Molenaar 2018 [47] * | Sulkowski 2017 [52] | Wang 2020 [53] | Venneker 2019 [66] * Palubeckaitė [67] * |
| Bcl-2/Bcl-xL inh. | Chan 2015 [54] | Karpel-Massler 2017 [55] | De Jong 2018 [62] * | |
| BET inh. | Chen 2013 [56] | Fujiwara 2018 [28] | Fujiwara 2018 [28] * | |
| DNMT inh. | Turcan 2013 [57] * | |||
| mTOR inh. | Batsios 2019 [58] | Addie 2019 [63] * | ||
| NAMPT inh. | Tateishi 2015 [27] * | Peterse 2017 [64] * | ||
| Glutaminolysis inh. | Emadi 2014 [59] * | Seltzer 2010 [60] | Peterse 2018 [65] * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venneker, S.; Bovée, J.V.M.G. IDH Mutations in Chondrosarcoma: Case Closed or Not? Cancers 2023, 15, 3603. https://doi.org/10.3390/cancers15143603
Venneker S, Bovée JVMG. IDH Mutations in Chondrosarcoma: Case Closed or Not? Cancers. 2023; 15(14):3603. https://doi.org/10.3390/cancers15143603
Chicago/Turabian StyleVenneker, Sanne, and Judith V. M. G. Bovée. 2023. "IDH Mutations in Chondrosarcoma: Case Closed or Not?" Cancers 15, no. 14: 3603. https://doi.org/10.3390/cancers15143603
APA StyleVenneker, S., & Bovée, J. V. M. G. (2023). IDH Mutations in Chondrosarcoma: Case Closed or Not? Cancers, 15(14), 3603. https://doi.org/10.3390/cancers15143603







