RET-Altered Cancers—A Tumor-Agnostic Review of Biology, Diagnosis and Targeted Therapy Activity
Abstract
Simple Summary
Abstract
1. Introduction
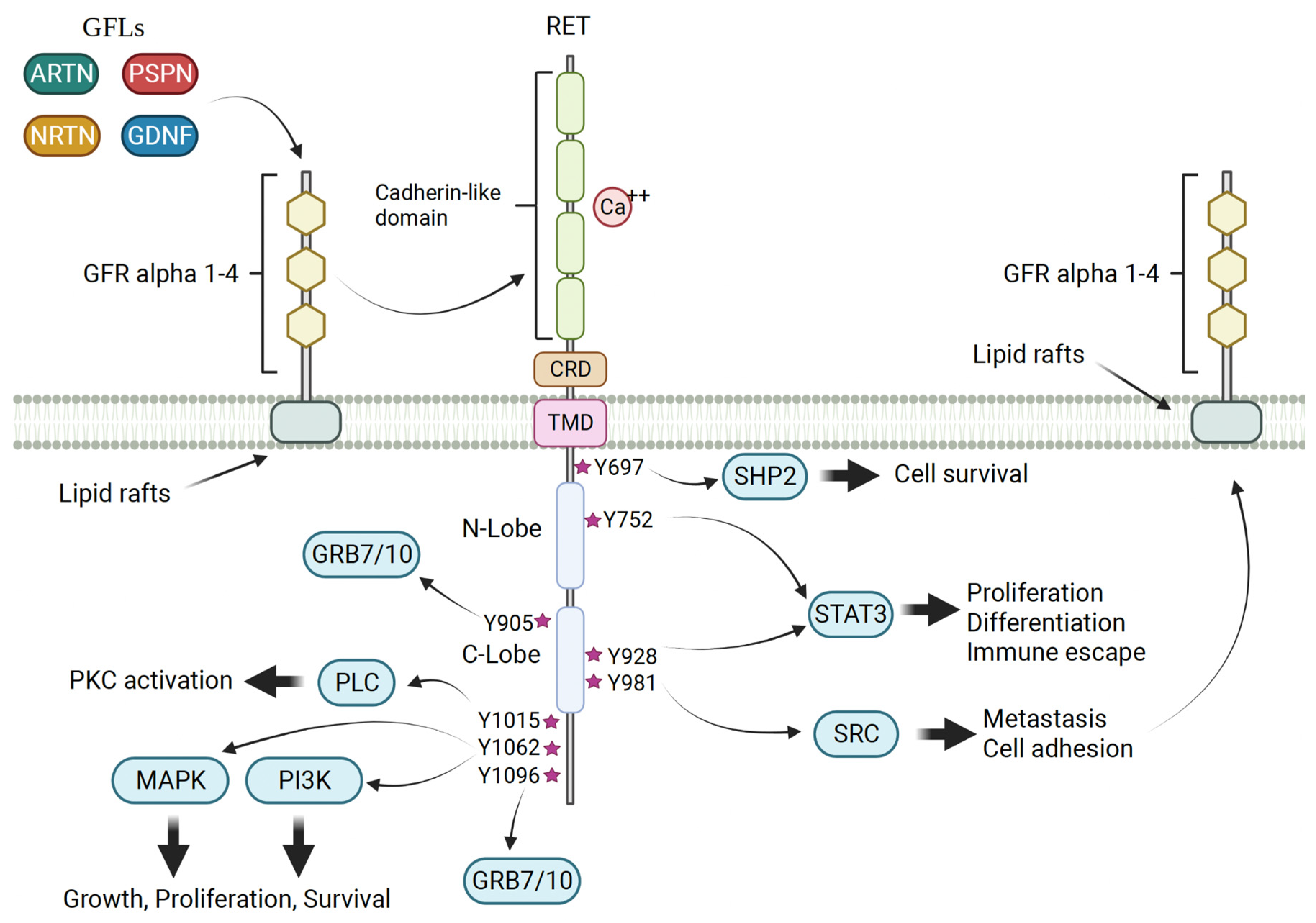
2. RET Biology
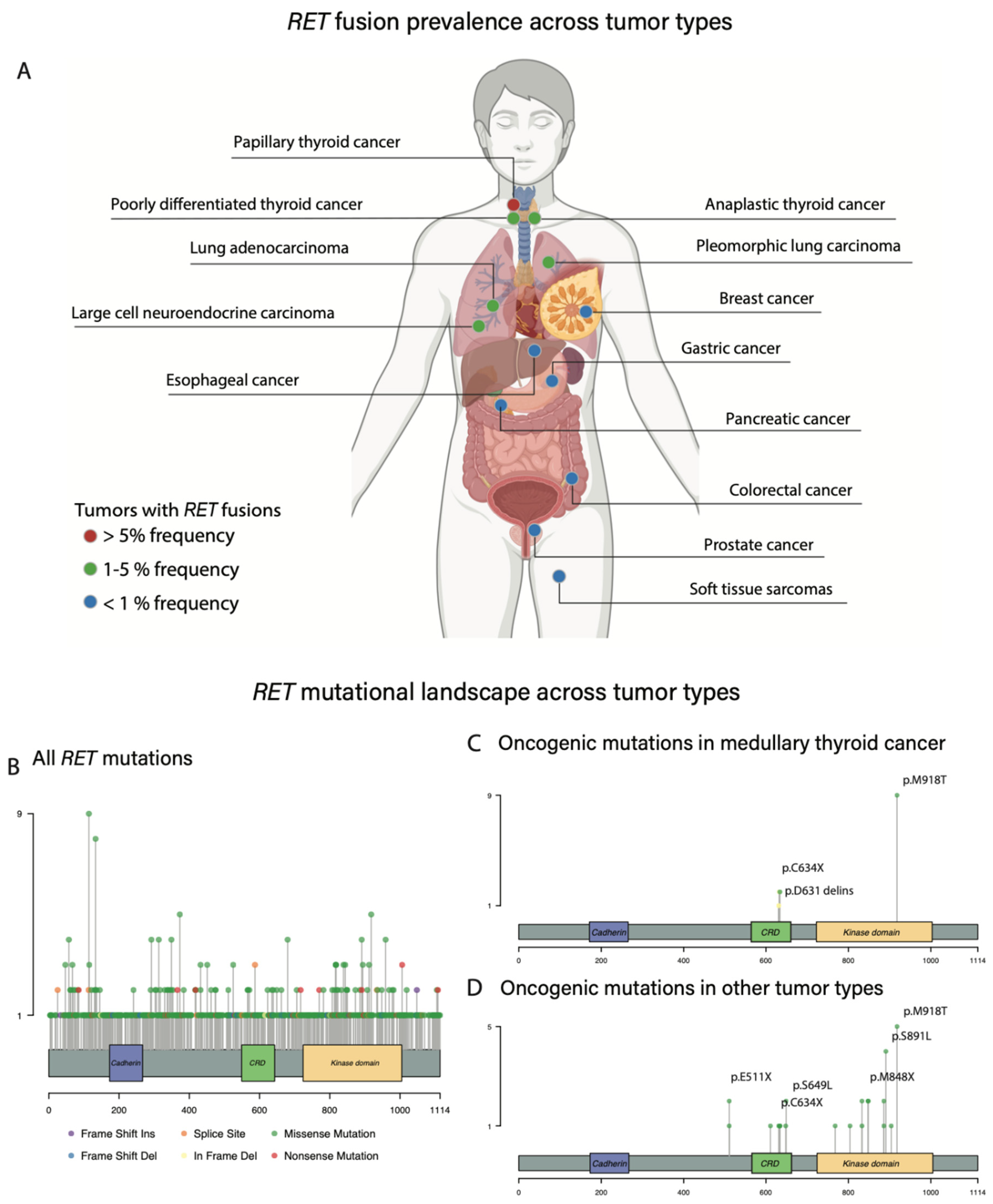
3. Oncogenic RET Alterations
3.1. Activating RET Mutations: MTC
3.2. RET Rearrangements: Thyroid Cancer
3.3. RET Rearrangements: NSCLC
3.4. RET Activating Mutations across Other Histologies
3.5. RET Fusions across Other Histologies
3.6. RET Amplifications
4. Standard Methods for Detection
4.1. Immunohistochemistry (IHC)
4.2. FISH
4.3. DNA PCR-Based Assays
4.4. NGS: DNA
4.5. NGS: RNA
5. RET-Altered Cancers: Pre-Targeted Therapy Era
6. Multikinase RET Inhibitors
6.1. Cabozantinib
6.2. Vandetanib
6.3. Limitations of Multitargeted RET Inhibitors
7. Selective RET Inhibitors
7.1. Selpercatinib (LOXO-292)
7.2. Pralsetinib (BLU-667)
| Selpercatinib | ||||||
|---|---|---|---|---|---|---|
| Study | Study design | Tumor site | Patients, n | ORR, n (%) | Responses (n) | DOR range (mo) |
| LIBRETTO-001 trial (2022) [84] | Prospective | Pancreas | 12 | 6 (55%) | PR (6), SD (6) | 3–38 * |
| Colorectal | 10 | 2 (20%) | PR (2), SD (8) | 6–13 | ||
| Salivary | 4 | 2 (50%) | PR (2), SD (2) | 6–29 * | ||
| Unknown primary | 3 | 1 (33%) | PR (1), SD (2) | 9 * | ||
| Breast | 2 | 2 (100%) | CR (1), PR (1) | 2–17 | ||
| Sarcoma | 2 | 2 (100%) | PR (2) | 15 * | ||
| Xanthogranuloma | 2 | NE | N/A | N/A | ||
| Carcinoid (lung) | 1 | 1 (100%) | PR (1) | 24 * | ||
| Skin carcinoma | 1 | NE | N/A | N/A | ||
| Cholangiocarcinoma | 1 | 1 (100%) | PR (1) | 6 * | ||
| Ovarian | 1 | 1 (100%) | PR (1) | 15 * | ||
| Pulmonary carcinosarcoma | 1 | NE | N/A | N/A | ||
| Neuroendocrine (rectum) | 1 | NE | N/A | N/A | ||
| Small intestine | 1 | 1 (100%) | CR (1) | 25 | ||
| Durham et al. (2019) [80] | Case report | Xanthogranuloma | 1 | N/A | Clinical response | N/A |
| Kander et al. (2021) [81] | Case report | Carcinoid (lung) | 1 | N/A | PR | 6 * |
| Watanabe et al. (2021) [148] | Case report | Breast, ER+/HER2- | 1 | N/A | CR | 10 * |
| Mweempwa et al. (2021) [77] | Case report | Pheochromocytoma | 1 | N/A | PR | 5 * |
| Arora et al. (2023) [147] | Case report | Neuroendocrine (lung) | 1 | N/A | PR | 12 * |
| Pralsetinib | ||||||
| ARROW trial (2022) [36,58] | Prospective | Pancreas | 4 | 4 (100%) | CR (1), PR (3) | 3–27 * |
| Cholangiocarcinoma | 3 | 2 (67%) | PR (2), SD (1) | 8–19 | ||
| Neuroendocrine | 3 | 2 (67%) | PR (2), SD (1) | 11–14 * | ||
| Sarcoma | 3 | 2 (67%) | CR (1), PR (1), SD (1) | 11 | ||
| Head and neck | 2 | 1 (50% | PR (1), SD (1) | 9 | ||
| Colorectal | 2 | 0 (0%) | SD (2) | N/A | ||
| Small cell lung cancer | 2 | 1 (50%) | PR (1), SD (1) | 9 * | ||
| Unknown primary | 1 | 1 (100%) | CR (1) | 5 | ||
| Stomach | 1 | 0 (0%) | PD (1) | N/A | ||
| Ovarian | 1 | 0 (0% | PD (1) | N/A | ||
| Thymic | 1 | 0 (0%) | SD (1) | N/A | ||
| Wu et al. (2022) [156] | Case report | Sarcomatoid carcinoma (lung) | 1 | N/A | PR | NE |
| Zhang et al. (2023) [155] | Case report | Pancreas | 1 | N/A | PR | 12 * |
| Zhao et al. (2023) [157] | Case report | Breast, triple negative | 1 | N/A | PR | 8 * |
8. Acquired Resistance
9. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mullard, A. FDA releases tissue-agnostic cancer drug draft guidance. Nat. Rev. Drug Discov. 2022, 21, 868. [Google Scholar] [CrossRef] [PubMed]
- Ishizaka, Y.; Itoh, F.; Tahira, T.; Ikeda, I.; Sugimura, T.; Tucker, J.; Fertitta, A.; Carrano, A.V.; Nagao, M. Human ret proto-oncogene mapped to chromosome 10q11.2. Oncogene 1989, 4, 1519–1521. [Google Scholar] [PubMed]
- Chi, X.; Michos, O.; Shakya, R.; Riccio, P.; Enomoto, H.; Licht, J.D.; Asai, N.; Takahashi, M.; Ohgami, N.; Kato, M.; et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell 2009, 17, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, T.; Takahashi, M.; Asai, N.; Iwashita, T.; Matsuyama, M.; Asai, J. Spatial and temporal expression of the ret proto-oncogene product in embryonic, infant and adult rat tissues. Oncogene 1995, 10, 191–198. [Google Scholar] [PubMed]
- Tomuschat, C.; Puri, P. RET gene is a major risk factor for Hirschsprung’s disease: A meta-analysis. Pediatr. Surg. Int. 2015, 31, 701–710. [Google Scholar] [CrossRef]
- Fitze, G.; Cramer, J.; Ziegler, A.; Schierz, M.; Schreiber, M.; Kuhlisch, E.; Roesner, D.; Schackert, H.K. Association between c135G/A genotype and RET proto-oncogene germline mutations and phenotype of Hirschsprung’s disease. Lancet 2002, 359, 1200–1205. [Google Scholar] [CrossRef]
- Hwang, D.Y.; Dworschak, G.C.; Kohl, S.; Saisawat, P.; Vivante, A.; Hilger, A.C.; Reutter, H.M.; Soliman, N.A.; Bogdanovic, R.; Kehinde, E.O.; et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014, 85, 1429–1433. [Google Scholar] [CrossRef]
- Fitze, G.; Paditz, E.; Schläfke, M.; Kuhlisch, E.; Roesner, D.; Schackert, H.K. Association of germline mutations and polymorphisms of the RET proto-oncogene with idiopathic congenital central hypoventilation syndrome in 33 patients. J. Med. Genet. 2003, 40, e10. [Google Scholar] [CrossRef]
- Vega, Q.C.; Worby, C.A.; Lechner, M.S.; Dixon, J.E.; Dressler, G.R. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc. Natl. Acad. Sci. USA 1996, 93, 10657–10661. [Google Scholar] [CrossRef]
- Kotzbauer, P.T.; Lampe, P.A.; Heuckeroth, R.O.; Golden, J.P.; Creedon, D.J.; Johnson, E.M., Jr.; Milbrandt, J. Neurturin, a relative of glial-cell-line-derived neurotrophic factor. Nature 1996, 384, 467–470. [Google Scholar] [CrossRef]
- Milbrandt, J.; de Sauvage, F.J.; Fahrner, T.J.; Baloh, R.H.; Leitner, M.L.; Tansey, M.G.; Lampe, P.A.; Heuckeroth, R.O.; Kotzbauer, P.T.; Simburger, K.S.; et al. Persephin, a novel neurotrophic factor related to GDNF and neurturin. Neuron 1998, 20, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Structural studies of GDNF family ligands with their receptors-Insights into ligand recognition and activation of receptor tyrosine kinase RET. Biochim. Biophys. Acta 2013, 1834, 2205–2212. [Google Scholar] [CrossRef]
- Mulligan, L.M. GDNF and the RET Receptor in Cancer: New Insights and Therapeutic Potential. Front. Physiol. 2018, 9, 1873. [Google Scholar] [CrossRef] [PubMed]
- Schuringa, J.J.; Wojtachnio, K.; Hagens, W.; Vellenga, E.; Buys, C.H.; Hofstra, R.; Kruijer, W. MEN2A-RET-induced cellular transformation by activation of STAT3. Oncogene 2001, 20, 5350–5358. [Google Scholar] [CrossRef] [PubMed]
- Andreozzi, F.; Melillo, R.M.; Carlomagno, F.; Oriente, F.; Miele, C.; Fiory, F.; Santopietro, S.; Castellone, M.D.; Beguinot, F.; Santoro, M.; et al. Protein kinase Cα activation by RET: Evidence for a negative feedback mechanism controlling RET tyrosine kinase. Oncogene 2003, 22, 2942–2949. [Google Scholar] [CrossRef][Green Version]
- Worby, C.A.; Vega, Q.C.; Zhao, Y.; Chao, H.H.; Seasholtz, A.F.; Dixon, J.E. Glial cell line-derived neurotrophic factor signals through the RET receptor and activates mitogen-activated protein kinase. J. Biol. Chem. 1996, 271, 23619–23622. [Google Scholar] [CrossRef]
- Maeda, K.; Murakami, H.; Yoshida, R.; Ichihara, M.; Abe, A.; Hirai, M.; Murohara, T.; Takahashi, M. Biochemical and biological responses induced by coupling of Gab1 to phosphatidylinositol 3-kinase in RET-expressing cells. Biochem. Biophys. Res. Commun. 2004, 323, 345–354. [Google Scholar] [CrossRef]
- Romei, C.; Ciampi, R.; Elisei, R. A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat. Rev. Endocrinol. 2016, 12, 192–202. [Google Scholar] [CrossRef]
- Mulligan, L.M. RET revisited: Expanding the oncogenic portfolio. Nat. Rev. Cancer 2014, 14, 173–186. [Google Scholar] [CrossRef]
- Fusco, A.; Grieco, M.; Santoro, M.; Berlingieri, M.T.; Pilotti, S.; Pierotti, M.A.; Della Porta, G.; Vecchio, G. A new oncogene in human thyroid papillary carcinomas and their lymph-nodal metastases. Nature 1987, 328, 170–172. [Google Scholar] [CrossRef]
- Takahashi, M.; Ritz, J.; Cooper, G.M. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 1985, 42, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Yang, D.; Velcheti, V.; Drilon, A.; Meric-Bernstam, F. State-of-the-Art Strategies for Targeting RET-Dependent Cancers. J. Clin. Oncol. 2020, 38, 1209–1221. [Google Scholar] [CrossRef]
- Moline, J.; Eng, C. Multiple endocrine neoplasia type 2: An overview. Genet. Med. 2011, 13, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Jhiang, S.M. The RET proto-oncogene in human cancers. Oncogene 2000, 19, 5590–5597. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, J.E.; Gild, M.L.; Clifton-Bligh, R.J.; Robinson, B.G. Multiple endocrine neoplasia: An update. Intern. Med. J. 2019, 49, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Moura, M.M.; Cavaco, B.M.; Pinto, A.E.; Domingues, R.; Santos, J.R.; Cid, M.O.; Bugalho, M.J.; Leite, V. Correlation of RET somatic mutations with clinicopathological features in sporadic medullary thyroid carcinomas. Br. J. Cancer 2009, 100, 1777–1783. [Google Scholar] [CrossRef]
- Dvorakova, S.; Vaclavikova, E.; Sykorova, V.; Vcelak, J.; Novak, Z.; Duskova, J.; Ryska, A.; Laco, J.; Cap, J.; Kodetova, D.; et al. Somatic mutations in the RET proto-oncogene in sporadic medullary thyroid carcinomas. Mol. Cell Endocrinol. 2008, 284, 21–27. [Google Scholar] [CrossRef]
- Kurzrock, R.; Sherman, S.I.; Ball, D.W.; Forastiere, A.A.; Cohen, R.B.; Mehra, R.; Pfister, D.G.; Cohen, E.E.; Janisch, L.; Nauling, F.; et al. Activity of XL184 (Cabozantinib), an oral tyrosine kinase inhibitor, in patients with medullary thyroid cancer. J. Clin. Oncol. 2011, 29, 2660–2666. [Google Scholar] [CrossRef]
- Kohno, T.; Ichikawa, H.; Totoki, Y.; Yasuda, K.; Hiramoto, M.; Nammo, T.; Sakamoto, H.; Tsuta, K.; Furuta, K.; Shimada, Y.; et al. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 2012, 18, 375–377. [Google Scholar] [CrossRef]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Suzuki, R.; Sakata, S.; Hatano, S.; Asaka, R.; Hamanaka, W.; Ninomiya, H.; Uehara, H.; et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012, 18, 378–381. [Google Scholar] [CrossRef]
- Nikiforov, Y.E. RET/PTC rearrangement in thyroid tumors. Endocr. Pathol. 2002, 13, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Wang, L.; Hasanovic, A.; Suehara, Y.; Lipson, D.; Stephens, P.; Ross, J.; Miller, V.; Ginsberg, M.; Zakowski, M.F.; et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013, 3, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Hamatani, K.; Eguchi, H.; Koyama, K.; Mukai, M.; Nakachi, K.; Kusunoki, Y. A novel RET rearrangement (ACBD5/RET) by pericentric inversion, inv(10)(p12.1;q11.2), in papillary thyroid cancer from an atomic bomb survivor exposed to high-dose radiation. Oncol. Rep. 2014, 32, 1809–1814. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velcheti, V.; Thawani, R.; Khunger, M.; Mukhopadhyay, S.; Chute, D.J.; Schrock, A.B.; Ali, S.M. FRMD4A/RET: A Novel RET Oncogenic Fusion Variant in Non-Small Cell Lung Carcinoma. J. Thorac. Oncol. 2017, 12, e15–e16. [Google Scholar] [CrossRef]
- Lee, M.S.; Kim, R.N.; Hoseok, I.; Oh, D.Y.; Song, J.Y.; Noh, K.W.; Kim, Y.J.; Yang, J.W.; Lira, M.E.; Lee, C.H.; et al. Identification of a novel partner gene, KIAA1217, fused to RET: Functional characterization and inhibitor sensitivity of two isoforms in lung adenocarcinoma. Oncotarget 2016, 7, 36101–36114. [Google Scholar] [CrossRef]
- Subbiah, V.; Cassier, P.A.; Siena, S.; Garralda, E.; Paz-Ares, L.; Garrido, P.; Nadal, E.; Vuky, J.; Lopes, G.; Kalemkerian, G.P.; et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion-positive solid tumors from the phase 1/2 ARROW trial. Nat. Med. 2022, 28, 1640–1645. [Google Scholar] [CrossRef]
- Wirth, L.J.; Sherman, E.; Robinson, B.; Solomon, B.; Kang, H.; Lorch, J.; Worden, F.; Brose, M.; Patel, J.; Leboulleux, S.; et al. Efficacy of Selpercatinib in RET-Altered Thyroid Cancers. N. Engl. J. Med. 2020, 383, 825–835. [Google Scholar] [CrossRef]
- Drilon, A.; Oxnard, G.R.; Tan, D.S.W.; Loong, H.H.F.; Johnson, M.; Gainor, J.; McCoach, C.E.; Gautschi, O.; Besse, B.; Cho, B.C.; et al. Efficacy of Selpercatinib in RET Fusion–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 813–824. [Google Scholar] [CrossRef]
- Ameziane-El-Hassani, R.; Boufraqech, M.; Lagente-Chevallier, O.; Weyemi, U.; Talbot, M.; Métivier, D.; Courtin, F.; Bidart, J.-M.; El Mzibri, M.; Schlumberger, M. Role of H2O2 in RET/PTC1 Chromosomal Rearrangement Produced by Ionizing Radiation in Human Thyroid CellsRole of H2O2 in RET/PTC1 Rearrangement Formation. Cancer Res. 2010, 70, 4123–4132. [Google Scholar] [CrossRef]
- Dillon, L.W.; Pierce, L.C.; Lehman, C.E.; Nikiforov, Y.E.; Wang, Y.-H. DNA topoisomerases participate in fragility of the oncogene RET. PLoS ONE 2013, 8, e75741. [Google Scholar] [CrossRef]
- Kramara, J.; Osia, B.; Malkova, A. Break-induced replication: The where, the why, and the how. Trends Genet. 2018, 34, 518–531. [Google Scholar] [CrossRef]
- Shaw, A.T.; Hsu, P.P.; Awad, M.M.; Engelman, J.A. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat. Rev. Cancer 2013, 13, 772–787. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Stringer, J.R.; Blough, R.; Medvedovic, M.; Fagin, J.A.; Nikiforov, Y.E. Proximity of Chromosomal Loci That Participate in Radiation-Induced Rearrangements in Human Cells. Science 2000, 290, 138–141. [Google Scholar] [CrossRef]
- Gandhi, M.; Medvedovic, M.; Stringer, J.R.; Nikiforov, Y.E. Interphase chromosome folding determines spatial proximity of genes participating in carcinogenic RET/PTC rearrangements. Oncogene 2006, 25, 2360–2366. [Google Scholar] [CrossRef]
- Santoro, M.; Carlomagno, F. Central role of RET in thyroid cancer. Cold Spring Harb. Perspect. Biol. 2013, 5, a009233. [Google Scholar] [CrossRef] [PubMed]
- Parimi, V.; Tolba, K.; Danziger, N.; Kuang, Z.; Sun, D.; Lin, D.I.; Hiemenz, M.C.; Schrock, A.B.; Ross, J.S.; Oxnard, G.R.; et al. Genomic landscape of 891 RET fusions detected across diverse solid tumor types. Npj Precis. Oncol. 2023, 7, 10. [Google Scholar] [CrossRef]
- Kato, S.; Subbiah, V.; Marchlik, E.; Elkin, S.K.; Carter, J.L.; Kurzrock, R. RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4871 Patients. Clin. Cancer Res. 2017, 23, 1988–1997. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.R.; Aypar, U.; Rosen, E.Y.; Mata, D.A.; Benayed, R.; Mullaney, K.; Jayakumaran, G.; Zhang, Y.; Frosina, D.; Drilon, A.; et al. A Performance Comparison of Commonly Used Assays to Detect RET Fusions. Clin. Cancer Res. 2021, 27, 1316–1328. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The landscape of kinase fusions in cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef]
- Santoro, M.; Thomas, G.A.; Vecchio, G.; Williams, G.H.; Fusco, A.; Chiappetta, G.; Pozcharskaya, V.; Bogdanova, T.I.; Demidchik, E.P.; Cherstvoy, E.D.; et al. Gene rearrangement and Chernobyl related thyroid cancers. Br. J. Cancer 2000, 82, 315–322. [Google Scholar] [CrossRef]
- Unger, K.; Zurnadzhy, L.; Walch, A.; Mall, M.; Bogdanova, T.; Braselmann, H.; Hieber, L.; Tronko, N.; Hutzler, P.; Jeremiah, S.; et al. RET rearrangements in post-Chernobyl papillary thyroid carcinomas with a short latency analysed by interphase FISH. Br. J. Cancer 2006, 94, 1472–1477. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Voskoboĭnik, L.G.; Kostiuchenko, N.M.; Pushkar’ov, V.M.; Bogdanova, T.I.; Tron’ko, M.D. Analysis of the expression of RET/PTC oncogenes in post-chernobyl papillary thyroid carcinomas of patients from different age groups. Ukr. Biokhim. Zh. (1999) 2010, 82, 79–84. [Google Scholar] [PubMed]
- Ju, Y.S.; Lee, W.C.; Shin, J.Y.; Lee, S.; Bleazard, T.; Won, J.K.; Kim, Y.T.; Kim, J.I.; Kang, J.H.; Seo, J.S. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012, 22, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hu, H.; Pan, Y.; Li, Y.; Ye, T.; Li, C.; Luo, X.; Wang, L.; Li, H.; Zhang, Y.; et al. RET Fusions Define a Unique Molecular and Clinicopathologic Subtype of Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Su, C.; Li, X.; Fan, L.; Zheng, L.; Fei, K.; Zhou, C. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer 2013, 119, 1486–1494. [Google Scholar] [CrossRef]
- Lipson, D.; Capelletti, M.; Yelensky, R.; Otto, G.; Parker, A.; Jarosz, M.; Curran, J.A.; Balasubramanian, S.; Bloom, T.; Brennan, K.W.; et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 2012, 18, 382–384. [Google Scholar] [CrossRef]
- Offin, M.; Guo, R.; Wu, S.L.; Sabari, J.; Land, J.D.; Ni, A.; Montecalvo, J.; Halpenny, D.F.; Buie, L.W.; Pak, T.; et al. Immunophenotype and Response to Immunotherapy of RET-Rearranged Lung Cancers. JCO Precis. Oncol. 2019, 3, 1–8. [Google Scholar] [CrossRef]
- Griesinger, F.; Curigliano, G.; Thomas, M.; Subbiah, V.; Baik, C.S.; Tan, D.S.W.; Lee, D.H.; Misch, D.; Garralda, E.; Kim, D.W.; et al. Safety and efficacy of pralsetinib in RET fusion-positive non-small-cell lung cancer including as first-line therapy: Update from the ARROW trial. Ann. Oncol. 2022, 33, 1168–1178. [Google Scholar] [CrossRef]
- Rich, T.A.; Reckamp, K.L.; Chae, Y.K.; Doebele, R.C.; Iams, W.T.; Oh, M.; Raymond, V.M.; Lanman, R.B.; Riess, J.W.; Stinchcombe, T.E.; et al. Analysis of Cell-Free DNA from 32,989 Advanced Cancers Reveals Novel Co-occurring Activating RET Alterations and Oncogenic Signaling Pathway Aberrations. Clin. Cancer Res. 2019, 25, 5832–5842. [Google Scholar] [CrossRef]
- Hess, L.M.; Han, Y.; Zhu, Y.E.; Bhandari, N.R.; Sireci, A. Characteristics and outcomes of patients with RET-fusion positive non-small lung cancer in real-world practice in the United States. BMC Cancer 2021, 21, 28. [Google Scholar] [CrossRef]
- Drilon, A.; Hu, Z.I.; Lai, G.G.Y.; Tan, D.S.W. Targeting RET-driven cancers: Lessons from evolving preclinical and clinical landscapes. Nat. Rev. Clin. Oncol. 2018, 15, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Plodkowski, A.J.; Drilon, A.; Halpenny, D.F.; O’Driscoll, D.; Blair, D.; Litvak, A.M.; Zheng, J.; Moskowitz, C.S.; Ginsberg, M.S. From genotype to phenotype: Are there imaging characteristics associated with lung adenocarcinomas harboring RET and ROS1 rearrangements? Lung Cancer 2015, 90, 321–325. [Google Scholar] [CrossRef][Green Version]
- Lee, S.E.; Lee, B.; Hong, M.; Song, J.Y.; Jung, K.; Lira, M.E.; Mao, M.; Han, J.; Kim, J.; Choi, Y.L. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod. Pathol. 2015, 28, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Yakes, F.M.; Chen, J.; Tan, J.; Yamaguchi, K.; Shi, Y.; Yu, P.; Qian, F.; Chu, F.; Bentzien, F.; Cancilla, B.; et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 2011, 10, 2298–2308. [Google Scholar] [CrossRef]
- Drilon, A.; Lin, J.J.; Filleron, T.; Ni, A.; Milia, J.; Bergagnini, I.; Hatzoglou, V.; Velcheti, V.; Offin, M.; Li, B.; et al. Frequency of Brain Metastases and Multikinase Inhibitor Outcomes in Patients with RET-Rearranged Lung Cancers. J. Thorac. Oncol. 2018, 13, 1595–1601. [Google Scholar] [CrossRef]
- Rotow, J.; Patel, J.D.; Hanley, M.P.; Yu, H.; Awad, M.; Goldman, J.W.; Nechushtan, H.; Scheffler, M.; Kuo, C.S.; Rajappa, S.; et al. Osimertinib and selpercatinib efficacy, safety, and resistance in a multicenter, prospectively treated cohort of EGFR-mutant and RET fusion-positive lung cancers. Clin. Cancer Res. 2023, 29, OF1–OF9. [Google Scholar] [CrossRef] [PubMed]
- Dabir, S.; Babakoohi, S.; Kluge, A.; Morrow, J.J.; Kresak, A.; Yang, M.; MacPherson, D.; Wildey, G.; Dowlati, A. RET Mutation and Expression in Small-Cell Lung Cancer. J. Thorac. Oncol. 2014, 9, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, N.G. Pancreatic Acinar Cell Carcinoma. Adv. Anat. Pathol. 2001, 8, 144–159. [Google Scholar]
- Chmielecki, J.; Hutchinson, K.E.; Frampton, G.M.; Chalmers, Z.R.; Johnson, A.; Shi, C.; Elvin, J.; Ali, S.M.; Ross, J.S.; Basturk, O.; et al. Comprehensive Genomic Profiling of Pancreatic Acinar Cell Carcinomas Identifies Recurrent RAF Fusions and Frequent Inactivation of DNA Repair Genes. Cancer Discov. 2014, 4, 1398–1405. [Google Scholar] [CrossRef]
- Chou, A.; Brown, I.S.; Kumarasinghe, M.P.; Perren, A.; Riley, D.; Kim, Y.; Pajic, M.; Steinmann, A.; Rathi, V.; Jamieson, N.B.; et al. RET gene rearrangements occur in a subset of pancreatic acinar cell carcinomas. Mod. Pathol. 2020, 33, 657–664. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Di Nicolantonio, F.; Schrock, A.B.; Lee, J.; Morano, F.; Fucà, G.; Nikolinakos, P.; Drilon, A.; Hechtman, J.F.; Christiansen, J.; et al. RET fusions in a small subset of advanced colorectal cancers at risk of being neglected. Ann. Oncol. 2018, 29, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Paratala, B.S.; Chung, J.H.; Williams, C.B.; Yilmazel, B.; Petrosky, W.; Williams, K.; Schrock, A.B.; Gay, L.M.; Lee, E.; Dolfi, S.C.; et al. RET rearrangements are actionable alterations in breast cancer. Nat. Commun. 2018, 9, 4821. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, I.; Bishop, J.A.; Chiosea, S.I.; Seethala, R.R.; Perez-Ordonez, B.; Zhang, L.; Sung, Y.S.; Chen, C.L.; Assaad, A.; Oliai, B.R.; et al. Recurrent RET Gene Rearrangements in Intraductal Carcinomas of Salivary Gland. Am. J. Surg. Pathol. 2018, 42, 442–452. [Google Scholar] [CrossRef]
- Su, Y.J.; Lee, Y.H.; Jin, Y.T.; Hsieh, M.S. Using pan-TRK and RET Immunohistochemistry for the Detection of Fusion Types of Salivary Gland Secretory Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2022, 30, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Skálová, A.; Banečkova, M.; Thompson, L.D.R.; Ptáková, N.; Stevens, T.M.; Brcic, L.; Hyrcza, M.; Michal, M., Jr.; Simpson, R.H.W.; Santana, T.; et al. Expanding the Molecular Spectrum of Secretory Carcinoma of Salivary Glands with a Novel VIM-RET Fusion. Am. J. Surg. Pathol. 2020, 44, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; He, J.; Yelensky, R.; Esteve-Puig, R.; Botton, T.; Yeh, I.; Lipson, D.; Otto, G.; Brennan, K.; Murali, R.; et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat. Commun. 2014, 5, 3116. [Google Scholar] [CrossRef]
- Mweempwa, A.; Xu, H.; Vissers, J.H.A.; Tothill, R.W.; Pattison, A.D.; Fellowes, A.P.; Thomas, D.M.; Richardson, G.; Hicks, R.J.; Grimmond, S.M.; et al. Novel RET Fusion RET-SEPTIN9 Predicts Response to Selective RET Inhibition with Selpercatinib in Malignant Pheochromocytoma. JCO Precis. Oncol. 2021, 5, 1160–1165. [Google Scholar] [CrossRef]
- Estrada-Zuniga, C.M.; Cheng, Z.M.; Ethiraj, P.; Guo, Q.; Gonzalez-Cantú, H.; Adderley, E.; Lopez, H.; Landry, B.N.; Zainal, A.; Aronin, N.; et al. A RET::GRB2 fusion in pheochromocytoma defies the classic paradigm of RET oncogenic fusions. Cell Rep. Med. 2022, 3, 100686. [Google Scholar] [CrossRef]
- Davis, J.L.; Vargas, S.O.; Rudzinski, E.R.; López Marti, J.M.; Janeway, K.; Forrest, S.; Winsnes, K.; Pinto, N.; Yang, S.E.; VanSandt, M.; et al. Recurrent RET gene fusions in paediatric spindle mesenchymal neoplasms. Histopathology 2020, 76, 1032–1041. [Google Scholar] [CrossRef]
- Durham, B.H.; Lopez Rodrigo, E.; Picarsic, J.; Abramson, D.; Rotemberg, V.; De Munck, S.; Pannecoucke, E.; Lu, S.X.; Pastore, A.; Yoshimi, A.; et al. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat. Med. 2019, 25, 1839–1842. [Google Scholar] [CrossRef]
- Kander, E.M.; Shah, M.H.; Zhou, Y.; Goyal, A.; Palmer, J.D.; Owen, D.H.; Shilo, K.; Patel, G.; Raval, R.R.; Gonzalez, J.; et al. Response to the Selective RET Inhibitor Selpercatinib (LOXO-292) in a Patient with RET Fusion-positive Atypical Lung Carcinoid. Clin. Lung Cancer 2021, 22, e442–e445. [Google Scholar] [CrossRef] [PubMed]
- Tsuta, K.; Kohno, T.; Yoshida, A.; Shimada, Y.; Asamura, H.; Furuta, K.; Kushima, R. RET-rearranged non-small-cell lung carcinoma: A clinicopathological and molecular analysis. Br. J. Cancer 2014, 110, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Besse, B.; Subbiah, V.; Drilon, A.; Shah, M.; Wirth, L.J.; Bauer, T.M.; Velcheti, V.; Lakhani, N.; Boni, V.; Solomon, B.J.; et al. Detection and clearance of RET variants in plasma cell free DNA (cfDNA) from patients (pts) treated with LOXO-292. Ann. Oncol. 2018, 29, viii33. [Google Scholar] [CrossRef]
- Subbiah, V.; Wolf, J.; Konda, B.; Kang, H.; Spira, A.; Weiss, J.; Takeda, M.; Ohe, Y.; Khan, S.; Ohashi, K.; et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): A phase 1/2, open-label, basket trial. Lancet Oncol. 2022, 23, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.; Torres-Cruz, J.; Pack, S.D.; Koch, C.A.; Vortmeyer, A.O.; Mannan, P.; Lubensky, I.A.; Gagel, R.F.; Zhuang, Z. Amplification and overexpression of mutant RET in multiple endocrine neoplasia type 2-associated medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2003, 88, 459–463. [Google Scholar] [CrossRef]
- Nakashima, M.; Takamura, N.; Namba, H.; Saenko, V.; Meirmanov, S.; Matsumoto, N.; Hayashi, T.; Maeda, S.; Sekine, I. RET oncogene amplification in thyroid cancer: Correlations with radiation-associated and high-grade malignancy. Hum. Pathol. 2007, 38, 621–628. [Google Scholar] [CrossRef]
- Platt, A.; Morten, J.; Ji, Q.; Elvin, P.; Womack, C.; Su, X.; Donald, E.; Gray, N.; Read, J.; Bigley, G.; et al. A retrospective analysis of RET translocation, gene copy number gain and expression in NSCLC patients treated with vandetanib in four randomized Phase III studies. BMC Cancer 2015, 15, 171. [Google Scholar] [CrossRef]
- Gandhi, M.M.; Ricciuti, B.; Gildenberg, M.; Singh, A.; Li, Y.Y.; Gagné, A.; Wang, X.; Fitzgerald, K.; Aizer, A.; Nishino, M.; et al. Amplification of wild-type RET and clinical response to selpercatinib for non–small-cell lung cancer (NSCLC). J. Clin. Oncol. 2023, 41, 9123. [Google Scholar] [CrossRef]
- Yang, X.; Shi, J.; Chen, X.; Jiang, Y.; Zhao, H. Efficacy of Cabozantinib and Nivolumab in Treating Hepatocellular Carcinoma with RET Amplification, High Tumor Mutational Burden, and PD-L1 Expression. Oncologist 2020, 25, 470–474. [Google Scholar] [CrossRef]
- Czech, C.; Chen, A.; Morgan, K.P.; Zamora, C.; El-Refai, S.; Poynter, N.; Khagi, S. Response to Selpercatinib in a Patient with Recurrent Glioblastoma and RET Amplification. J. Natl. Compr. Cancer Netw. 2022, 20, 966–971. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Zhang, X.; Xu, Z.; Yan, Y.; Hu, K. An integrative pan cancer analysis of RET aberrations and their potential clinical implications. Sci. Rep. 2022, 12, 13913. [Google Scholar] [CrossRef]
- Singh, N.; Temin, S.; Baker, S.; Blanchard, E.; Brahmer, J.R.; Celano, P.; Duma, N.; Ellis, P.M.; Elkins, I.B.; Haddad, R.Y.; et al. Therapy for Stage IV Non–Small-Cell Lung Cancer with Driver Alterations: ASCO Living Guideline. J. Clin. Oncol. 2022, 40, 3310–3322. [Google Scholar] [CrossRef]
- Drilon, A.; Wang, L.; Arcila, M.E.; Balasubramanian, S.; Greenbowe, J.R.; Ross, J.S.; Stephens, P.; Lipson, D.; Miller, V.A.; Kris, M.G.; et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. 2015, 21, 3631–3639. [Google Scholar] [CrossRef] [PubMed]
- Belli, C.; Penault-Llorca, F.; Ladanyi, M.; Normanno, N.; Scoazec, J.Y.; Lacroix, L.; Reis-Filho, J.S.; Subbiah, V.; Gainor, J.F.; Endris, V.; et al. ESMO recommendations on the standard methods to detect RET fusions and mutations in daily practice and clinical research. Ann. Oncol. 2021, 32, 337–350. [Google Scholar] [CrossRef]
- Feng, J.; Li, Y.; Wei, B.; Guo, L.; Li, W.; Xia, Q.; Zhao, C.; Zheng, J.; Zhao, J.; Sun, R.; et al. Clinicopathologic characteristics and diagnostic methods of RET rearrangement in Chinese non-small cell lung cancer patients. Transl. Lung Cancer Res. 2022, 11, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.F.; Brachold, J.M.; Tischler, A.S. Ret protein expression in adrenal medullary hyperplasia and pheochromocytoma. Endocr. Pathol. 2003, 14, 351–361. [Google Scholar] [CrossRef]
- Luo, Y.; Tsuchiya, K.D.; Il Park, D.; Fausel, R.; Kanngurn, S.; Welcsh, P.; Dzieciatkowski, S.; Wang, J.; Grady, W.M. RET is a potential tumor suppressor gene in colorectal cancer. Oncogene 2013, 32, 2037–2047. [Google Scholar] [CrossRef]
- Furugaki, K.; Mochizuki, M.; Kohno, M.; Shu, S.; Harada, N.; Yoshimura, Y. Expression of C-terminal ALK, RET, or ROS1 in lung cancer cells with or without fusion. BMC Cancer 2019, 19, 301. [Google Scholar] [CrossRef]
- Hechtman, J.F.; Benayed, R.; Hyman, D.M.; Drilon, A.; Zehir, A.; Frosina, D.; Arcila, M.E.; Dogan, S.; Klimstra, D.S.; Ladanyi, M.; et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am. J. Surg. Pathol. 2017, 41, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lu, Y.; Ye, Q.; Zhang, M.; Zheng, L.; Yin, X.; Gavine, P.; Sun, Z.; Ji, Q.; Zhu, G.; et al. An evaluation and recommendation of the optimal methodologies to detect RET gene rearrangements in papillary thyroid carcinoma. Genes Chromosomes Cancer 2015, 54, 168–176. [Google Scholar] [CrossRef]
- Zhu, Z.; Ciampi, R.; Nikiforova, M.N.; Gandhi, M.; Nikiforov, Y.E. Prevalence of RET/PTC rearrangements in thyroid papillary carcinomas: Effects of the detection methods and genetic heterogeneity. J. Clin. Endocrinol. Metab. 2006, 91, 3603–3610. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef]
- Qiu, Z.; Ye, B.; Wang, K.; Zhou, P.; Zhao, S.; Li, W.; Tian, P. Unique Genetic Characteristics and Clinical Prognosis of Female Patients with Lung Cancer Harboring RET Fusion Gene. Sci. Rep. 2020, 10, 10387. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Bergagnini, I.; Delasos, L.; Sabari, J.; Woo, K.M.; Plodkowski, A.; Wang, L.; Hellmann, M.D.; Joubert, P.; Sima, C.S.; et al. Clinical outcomes with pemetrexed-based systemic therapies in RET-rearranged lung cancers. Ann. Oncol. 2016, 27, 1286–1291. [Google Scholar] [CrossRef]
- Saito, M.; Ishigame, T.; Tsuta, K.; Kumamoto, K.; Imai, T.; Kohno, T. A mouse model of KIF5B-RET fusion-dependent lung tumorigenesis. Carcinogenesis 2014, 35, 2452–2456. [Google Scholar] [CrossRef]
- Lin, C.; Wang, S.; Xie, W.; Zheng, R.; Gan, Y.; Chang, J. Apatinib inhibits cellular invasion and migration by fusion kinase KIF5B-RET via suppressing RET/Src signaling pathway. Oncotarget 2016, 7, 59236–59244. [Google Scholar] [CrossRef]
- Huang, Q.; Schneeberger, V.E.; Luetteke, N.; Jin, C.; Afzal, R.; Budzevich, M.M.; Makanji, R.J.; Martinez, G.V.; Shen, T.; Zhao, L.; et al. Preclinical Modeling of KIF5B-RET Fusion Lung Adenocarcinoma. Mol. Cancer Ther. 2016, 15, 2521–2529. [Google Scholar] [CrossRef]
- Kodama, T.; Tsukaguchi, T.; Satoh, Y.; Yoshida, M.; Watanabe, Y.; Kondoh, O.; Sakamoto, H. Alectinib shows potent antitumor activity against RET-rearranged non-small cell lung cancer. Mol. Cancer Ther. 2014, 13, 2910–2918. [Google Scholar] [CrossRef]
- Li, G.G.; Somwar, R.; Joseph, J.; Smith, R.S.; Hayashi, T.; Martin, L.; Franovic, A.; Schairer, A.; Martin, E.; Riely, G.J.; et al. Antitumor Activity of RXDX-105 in Multiple Cancer Types with RET Rearrangements or Mutations. Clin. Cancer Res. 2017, 23, 2981–2990. [Google Scholar] [CrossRef]
- Verbeek, H.H.; Alves, M.M.; de Groot, J.W.; Osinga, J.; Plukker, J.T.; Links, T.P.; Hofstra, R.M. The effects of four different tyrosine kinase inhibitors on medullary and papillary thyroid cancer cells. J. Clin. Endocrinol. Metab. 2011, 96, E991–E995. [Google Scholar] [CrossRef]
- Bentzien, F.; Zuzow, M.; Heald, N.; Gibson, A.; Shi, Y.; Goon, L.; Yu, P.; Engst, S.; Zhang, W.; Huang, D.; et al. In vitro and in vivo activity of cabozantinib (XL184), an inhibitor of RET, MET, and VEGFR2, in a model of medullary thyroid cancer. Thyroid. 2013, 23, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, F.; Anaganti, S.; Guida, T.; Salvatore, G.; Troncone, G.; Wilhelm, S.M.; Santoro, M. BAY 43-9006 inhibition of oncogenic RET mutants. J. Natl. Cancer Inst. 2006, 98, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Elisei, R.; Schlumberger, M.J.; Müller, S.P.; Schöffski, P.; Brose, M.S.; Shah, M.H.; Licitra, L.; Jarzab, B.; Medvedev, V.; Kreissl, M.C.; et al. Cabozantinib in progressive medullary thyroid cancer. J. Clin. Oncol. 2013, 31, 3639–3646. [Google Scholar] [CrossRef] [PubMed]
- Sherman, S.I.; Clary, D.O.; Elisei, R.; Schlumberger, M.J.; Cohen, E.E.; Schöffski, P.; Wirth, L.J.; Mangeshkar, M.; Aftab, D.T.; Brose, M.S. Correlative analyses of RET and RAS mutations in a phase 3 trial of cabozantinib in patients with progressive, metastatic medullary thyroid cancer. Cancer 2016, 122, 3856–3864. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Rekhtman, N.; Arcila, M.; Wang, L.; Ni, A.; Albano, M.; Van Voorthuysen, M.; Somwar, R.; Smith, R.S.; Montecalvo, J.; et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: An open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 2016, 17, 1653–1660. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.K.; Ahn, M.J.; Kim, D.W.; Sun, J.M.; Keam, B.; Kim, T.M.; Heo, D.S.; Ahn, J.S.; Choi, Y.L.; et al. Vandetanib in pretreated patients with advanced non-small cell lung cancer-harboring RET rearrangement: A phase II clinical trial. Ann. Oncol. 2017, 28, 292–297. [Google Scholar] [CrossRef]
- Wells, S.A., Jr.; Robinson, B.G.; Gagel, R.F.; Dralle, H.; Fagin, J.A.; Santoro, M.; Baudin, E.; Elisei, R.; Jarzab, B.; Vasselli, J.R.; et al. Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: A randomized, double-blind phase III trial. J. Clin. Oncol. 2012, 30, 134–141. [Google Scholar] [CrossRef]
- Kreissl, M.C.; Bastholt, L.; Elisei, R.; Haddad, R.; Hauch, O.; Jarząb, B.; Robinson, B.; Colzani, R.; Foster, M.; Weiss, R.; et al. Efficacy and Safety of Vandetanib in Progressive and Symptomatic Medullary Thyroid Cancer: Post Hoc Analysis From the ZETA Trial. J. Clin. Oncol. 2020, 38, 2773–2781. [Google Scholar] [CrossRef]
- Langmuir, P.B.; Yver, A. Vandetanib for the treatment of thyroid cancer. Clin. Pharmacol. Ther. 2012, 91, 71–80. [Google Scholar] [CrossRef]
- Subbiah, V.; Velcheti, V.; Tuch, B.B.; Ebata, K.; Busaidy, N.L.; Cabanillas, M.E.; Wirth, L.J.; Stock, S.; Smith, S.; Lauriault, V.; et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann. Oncol. 2018, 29, 1869–1876. [Google Scholar] [CrossRef]
- Wirth, L.J.; Kohno, T.; Udagawa, H.; Matsumoto, S.; Ishii, G.; Ebata, K.; Tuch, B.B.; Zhu, E.Y.; Nguyen, M.; Smith, S.; et al. Emergence and Targeting of Acquired and Hereditary Resistance to Multikinase RET Inhibition in Patients with RET-Altered Cancer. JCO Precis. Oncol. 2019, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Stevens, S.E.; Lin, J.J.; Nagy, R.; Ferris, L.; Shaw, A.T.; Gainor, J.F. Emergence of a RET V804M Gatekeeper Mutation During Treatment with Vandetanib in RET-Rearranged NSCLC. J. Thorac. Oncol. 2018, 13, e226–e227. [Google Scholar] [CrossRef] [PubMed]
- Horiike, A.; Takeuchi, K.; Uenami, T.; Kawano, Y.; Tanimoto, A.; Kaburaki, K.; Tambo, Y.; Kudo, K.; Yanagitani, N.; Ohyanagi, F.; et al. Sorafenib treatment for patients with RET fusion-positive non-small cell lung cancer. Lung Cancer 2016, 93, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, S.; Yanagitani, N.; Seto, T.; Hattori, Y.; Ohashi, K.; Morise, M.; Matsumoto, S.; Yoh, K.; Goto, K.; Nishio, M.; et al. Phase 1/2 study of alectinib in RET-rearranged previously-treated non-small cell lung cancer (ALL-RET). Transl. Lung Cancer Res. 2020, 10, 314–325. [Google Scholar] [CrossRef]
- Gautschi, O.; Milia, J.; Filleron, T.; Wolf, J.; Carbone, D.P.; Owen, D.; Camidge, R.; Narayanan, V.; Doebele, R.C.; Besse, B.; et al. Targeting RET in Patients with RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J. Clin. Oncol. 2017, 35, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Wedge, S.R.; Ogilvie, D.J.; Dukes, M.; Kendrew, J.; Chester, R.; Jackson, J.A.; Boffey, S.J.; Valentine, P.J.; Curwen, J.O.; Musgrove, H.L.; et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002, 62, 4645–4655. [Google Scholar]
- Okamoto, K.; Kodama, K.; Takase, K.; Sugi, N.H.; Yamamoto, Y.; Iwata, M.; Tsuruoka, A. Antitumor activities of the targeted multi-tyrosine kinase inhibitor lenvatinib (E7080) against RET gene fusion-driven tumor models. Cancer Lett. 2013, 340, 97–103. [Google Scholar] [CrossRef]
- Drilon, A.; Fu, S.; Patel, M.R.; Fakih, M.; Wang, D.; Olszanski, A.J.; Morgensztern, D.; Liu, S.V.; Cho, B.C.; Bazhenova, L.; et al. A Phase I/Ib Trial of the VEGFR-Sparing Multikinase RET Inhibitor RXDX-105. Cancer Discov. 2019, 9, 384–395. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Schindler, T.; Bornmann, W.; Pellicena, P.; Miller, W.T.; Clarkson, B.; Kuriyan, J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 2000, 289, 1938–1942. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Saleh, T.; Rossi, P.; Kalodimos, C.G. Conformational states dynamically populated by a kinase determine its function. Science 2020, 370, eabc2754. [Google Scholar] [CrossRef] [PubMed]
- Meharena, H.S.; Chang, P.; Keshwani, M.M.; Oruganty, K.; Nene, A.K.; Kannan, N.; Taylor, S.S.; Kornev, A.P. Deciphering the Structural Basis of Eukaryotic Protein Kinase Regulation. PLoS Biol. 2013, 11, e1001680. [Google Scholar] [CrossRef] [PubMed]
- van Linden, O.P.; Kooistra, A.J.; Leurs, R.; de Esch, I.J.; de Graaf, C. KLIFS: A knowledge-based structural database to navigate kinase-ligand interaction space. J. Med. Chem. 2014, 57, 249–277. [Google Scholar] [CrossRef]
- Solomon, B.J.; Tan, L.; Lin, J.J.; Wong, S.Q.; Hollizeck, S.; Ebata, K.; Tuch, B.B.; Yoda, S.; Gainor, J.F.; Sequist, L.V.; et al. RET Solvent Front Mutations Mediate Acquired Resistance to Selective RET Inhibition in RET-Driven Malignancies. J. Thorac. Oncol. 2020, 15, 541–549. [Google Scholar] [CrossRef]
- Knowles, P.P.; Murray-Rust, J.; Kjaer, S.; Scott, R.P.; Hanrahan, S.; Santoro, M.; Ibáñez, C.F.; McDonald, N.Q. Structure and chemical inhibition of the RET tyrosine kinase domain. J. Biol. Chem. 2006, 281, 33577–33587. [Google Scholar] [CrossRef]
- Subbiah, V.; Shen, T.; Terzyan, S.S.; Liu, X.; Hu, X.; Patel, K.P.; Hu, M.; Cabanillas, M.; Behrang, A.; Meric-Bernstam, F.; et al. Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations. Ann. Oncol. 2021, 32, 261–268. [Google Scholar] [CrossRef]
- Plaza-Menacho, I.; Mologni, L.; McDonald, N.Q. Mechanisms of RET signaling in cancer: Current and future implications for targeted therapy. Cell Signal 2014, 26, 1743–1752. [Google Scholar] [CrossRef]
- Plenker, D.; Riedel, M.; Brägelmann, J.; Dammert, M.A.; Chauhan, R.; Knowles, P.P.; Lorenz, C.; Keul, M.; Bührmann, M.; Pagel, O.; et al. Drugging the catalytically inactive state of RET kinase in RET-rearranged tumors. Sci. Transl. Med. 2017, 9, eaah6144. [Google Scholar] [CrossRef]
- Vodopivec, D.M.; Hu, M.I. RET kinase inhibitors for RET-altered thyroid cancers. Ther. Adv. Med. Oncol. 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Subbiah, V.; Gautschi, O.; Tomasini, P.; de Braud, F.; Solomon, B.J.; Shao-Weng Tan, D.; Alonso, G.; Wolf, J.; Park, K.; et al. Selpercatinib in Patients with RET Fusion-Positive Non-Small-Cell Lung Cancer: Updated Safety and Efficacy From the Registrational LIBRETTO-001 Phase I/II Trial. J. Clin. Oncol. 2023, 41, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Gainor, J.F.; Oxnard, G.R.; Tan, D.S.W.; Owen, D.H.; Cho, B.C.; Loong, H.H.; McCoach, C.E.; Weiss, J.; Kim, Y.J.; et al. Intracranial Efficacy of Selpercatinib in RET Fusion-Positive Non–Small Cell Lung Cancers on the LIBRETTO-001 Trial. Clin. Cancer Res. 2021, 27, 4160–4167. [Google Scholar] [CrossRef]
- Murciano-Goroff, Y.R.; Falcon, C.J.; Lin, S.T.; Chacko, C.; Grimaldi, G.; Liu, D.; Wilhelm, C.; Iasonos, A.; Drilon, A. Central Nervous System Disease in Patients with RET Fusion-Positive NSCLC Treated with Selpercatinib. J. Thorac. Oncol. 2023, 18, 620–627. [Google Scholar] [CrossRef]
- Baek, H.S.; Ha, J.; Ha, S.; Bae, J.S.; Jung, C.K.; Lim, D.J. Initial Experiences of Selective RET Inhibitor Selpercatinib in Adults with Metastatic Differentiated Thyroid Carcinoma and Medullary Thyroid Carcinoma: Real-World Case Series in Korea. Curr. Oncol. 2023, 30, 3020–3031. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Selpercatinib: First Approval. Drugs 2020, 80, 1119–1124. [Google Scholar] [CrossRef]
- Arora, A.; Zaemes, J.; Ozdemirli, M.; Kim, C. Response to selpercatinib in a patient with RET fusion-positive pulmonary large-cell neuroendocrine carcinoma: A case report. Front. Oncol. 2023, 13, 1134151. [Google Scholar] [CrossRef]
- Watanabe, S.; Takeda, M.; Otani, T.; Yoshida, T.; Sakai, K.; Olek, E.; Rothenberg, S.M.; Kherani, J.; French, P.P.; Nishio, K.; et al. Complete Response to Selective RET Inhibition with Selpercatinib (LOXO-292) in a Patient with RET Fusion-Positive Breast Cancer. JCO Precis. Oncol. 2021, 5, 103–106. [Google Scholar] [CrossRef]
- McCoach, C.E.; Rolfo, C.; Drilon, A.; Lacouture, M.; Besse, B.; Goto, K.; Zhu, V.W.; Tan, D.S.W.; Farajian, S.; Potter, L.A.; et al. Hypersensitivity Reactions to Selpercatinib Treatment with or without Prior Immune Checkpoint Inhibitor Therapy in Patients with NSCLC in LIBRETTO-001. J. Thorac. Oncol. 2022, 17, 768–778. [Google Scholar] [CrossRef]
- Kalchiem-Dekel, O.; Falcon, C.J.; Bestvina, C.M.; Liu, D.; Kaplanis, L.A.; Wilhelm, C.; Eichholz, J.; Harada, G.; Wirth, L.J.; Digumarthy, S.R.; et al. Brief Report: Chylothorax and Chylous Ascites During RET Tyrosine Kinase Inhibitor Therapy. J. Thorac. Oncol. 2022, 17, 1130–1136. [Google Scholar] [CrossRef]
- Boucai, L.; Salas-Lucia, F.; Krishnamoorthy, G.P.; Sherman, E.; Rudin, C.M.; Drilon, A.; Bianco, A.C.; Fagin, J.A. Selpercatinib-Induced Hypothyroidism Through Off-Target Inhibition of Type 2 Iodothyronine Deiodinase. JCO Precis. Oncol. 2022, 6, e2100496. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Curigliano, G.; Kim, D.-W.; Lee, D.H.; Besse, B.; Baik, C.S.; Doebele, R.C.; Cassier, P.A.; Lopes, G.; Tan, D.S.W.; et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): A multi-cohort, open-label, phase 1/2 study. Lancet Oncol. 2021, 22, 959–969. [Google Scholar] [CrossRef]
- Subbiah, V.; Hu, M.I.; Wirth, L.J.; Schuler, M.; Mansfield, A.S.; Curigliano, G.; Brose, M.S.; Zhu, V.W.; Leboulleux, S.; Bowles, D.W.; et al. Pralsetinib for patients with advanced or metastatic RET-altered thyroid cancer (ARROW): A multi-cohort, open-label, registrational, phase 1/2 study. Lancet Diabetes Endocrinol. 2021, 9, 491–501. [Google Scholar] [CrossRef]
- Popat, S.; Liu, S.V.; Scheuer, N.; Hsu, G.G.; Lockhart, A.; Ramagopalan, S.V.; Griesinger, F.; Subbiah, V. Addressing challenges with real-world synthetic control arms to demonstrate the comparative effectiveness of Pralsetinib in non-small cell lung cancer. Nat. Commun. 2022, 13, 3500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, H.; Cai, Z.; Zhang, S.; Jiang, C. RET rearrangement-positive pancreatic cancer has remarkable response to pralsetinib: A case report. Front. Oncol. 2023, 13, 1078076. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yan, Z.; Pan, J.; Chang, X.; Huang, B.; Luo, D.; Meng, R.; Shi, H.; Fan, J.; Nie, X. Partial response to pralsetinib in an advanced pulmonary sarcomatoid carcinoma patient harboring a KIF5B-RET rearrangement: A case report. World J. Surg. Oncol. 2022, 20, 386. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, W.; Zhuo, X.; Liu, L.; Zhang, J.; Jiang, F.; Shen, Y.; Lei, Y.; Hou, D.; Lin, X.; et al. Response to Pralsetinib in Multi-Drug-Resistant Breast Cancer with CCDC6-RET Mutation. Oncologist 2023, 28, e416–e424. [Google Scholar] [CrossRef]
- Nakaoku, T.; Kohno, T.; Araki, M.; Niho, S.; Chauhan, R.; Knowles, P.P.; Tsuchihara, K.; Matsumoto, S.; Shimada, Y.; Mimaki, S.; et al. A secondary RET mutation in the activation loop conferring resistance to vandetanib. Nat. Commun. 2018, 9, 625. [Google Scholar] [CrossRef]
- Lin, J.J.; Liu, S.V.; McCoach, C.E.; Zhu, V.W.; Tan, A.C.; Yoda, S.; Peterson, J.; Do, A.; Prutisto-Chang, K.; Dagogo-Jack, I.; et al. Mechanisms of resistance to selective RET tyrosine kinase inhibitors in RET fusion-positive non-small-cell lung cancer. Ann. Oncol. 2020, 31, 1725–1733. [Google Scholar] [CrossRef]
- Cao, S.; Tan, C.; Fei, A.; Hu, G.; Fu, M.; Lv, J. Insights into pralsetinib resistance to the non-gatekeeper RET kinase G810C mutation through molecular dynamics simulations. J. Mol. Model. 2022, 29, 24. [Google Scholar] [CrossRef]
- Shen, T.; Hu, X.; Liu, X.; Subbiah, V.; Mooers, B.H.M.; Wu, J. The L730V/I RET roof mutations display different activities toward pralsetinib and selpercatinib. Npj Precis. Oncol. 2021, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.Y.; Won, H.H.; Zheng, Y.; Cocco, E.; Selcuklu, D.; Gong, Y.; Friedman, N.D.; de Bruijn, I.; Sumer, O.; Bielski, C.M.; et al. The evolution of RET inhibitor resistance in RET-driven lung and thyroid cancers. Nat. Commun. 2022, 13, 1450. [Google Scholar] [CrossRef]
- Gazeu, A.; Aubert, M.; Pissaloux, D.; Lantuejoul, S.; Pérol, M.; Ikhlef, N.; Bouhamama, A.; Franceschi, T.; Swalduz, A. Small-Cell Lung Cancer Transformation as a Mechanism of Resistance to Pralsetinib in RET-Rearranged Lung Adenocarcinoma: A Case Report. Clin. Lung Cancer 2023, 24, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Dimou, A.; Lo, Y.C.; Merrell, K.W.; Halling, K.C.; Mansfield, A.S. Small Cell Transformation in a Patient with RET Fusion-Positive Lung Adenocarcinoma on Pralsetinib. JCO Precis. Oncol. 2022, 6, e2200478. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desilets, A.; Repetto, M.; Yang, S.-R.; Sherman, E.J.; Drilon, A. RET-Altered Cancers—A Tumor-Agnostic Review of Biology, Diagnosis and Targeted Therapy Activity. Cancers 2023, 15, 4146. https://doi.org/10.3390/cancers15164146
Desilets A, Repetto M, Yang S-R, Sherman EJ, Drilon A. RET-Altered Cancers—A Tumor-Agnostic Review of Biology, Diagnosis and Targeted Therapy Activity. Cancers. 2023; 15(16):4146. https://doi.org/10.3390/cancers15164146
Chicago/Turabian StyleDesilets, Antoine, Matteo Repetto, Soo-Ryum Yang, Eric J. Sherman, and Alexander Drilon. 2023. "RET-Altered Cancers—A Tumor-Agnostic Review of Biology, Diagnosis and Targeted Therapy Activity" Cancers 15, no. 16: 4146. https://doi.org/10.3390/cancers15164146
APA StyleDesilets, A., Repetto, M., Yang, S.-R., Sherman, E. J., & Drilon, A. (2023). RET-Altered Cancers—A Tumor-Agnostic Review of Biology, Diagnosis and Targeted Therapy Activity. Cancers, 15(16), 4146. https://doi.org/10.3390/cancers15164146






