The Three-Dimensional In Vitro Cell Culture Models in the Study of Oral Cancer Immune Microenvironment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Tumor Immune Microenvironment of Oral Cancer
2.1. Interaction between the Immune System and Tumor Cells
2.2. Immune and Non-Immune Markers
2.3. Stromal Cell and Extracellular Matrix (ECM) on Cancer Immunity
3. In Vitro Models in Oral Cancer
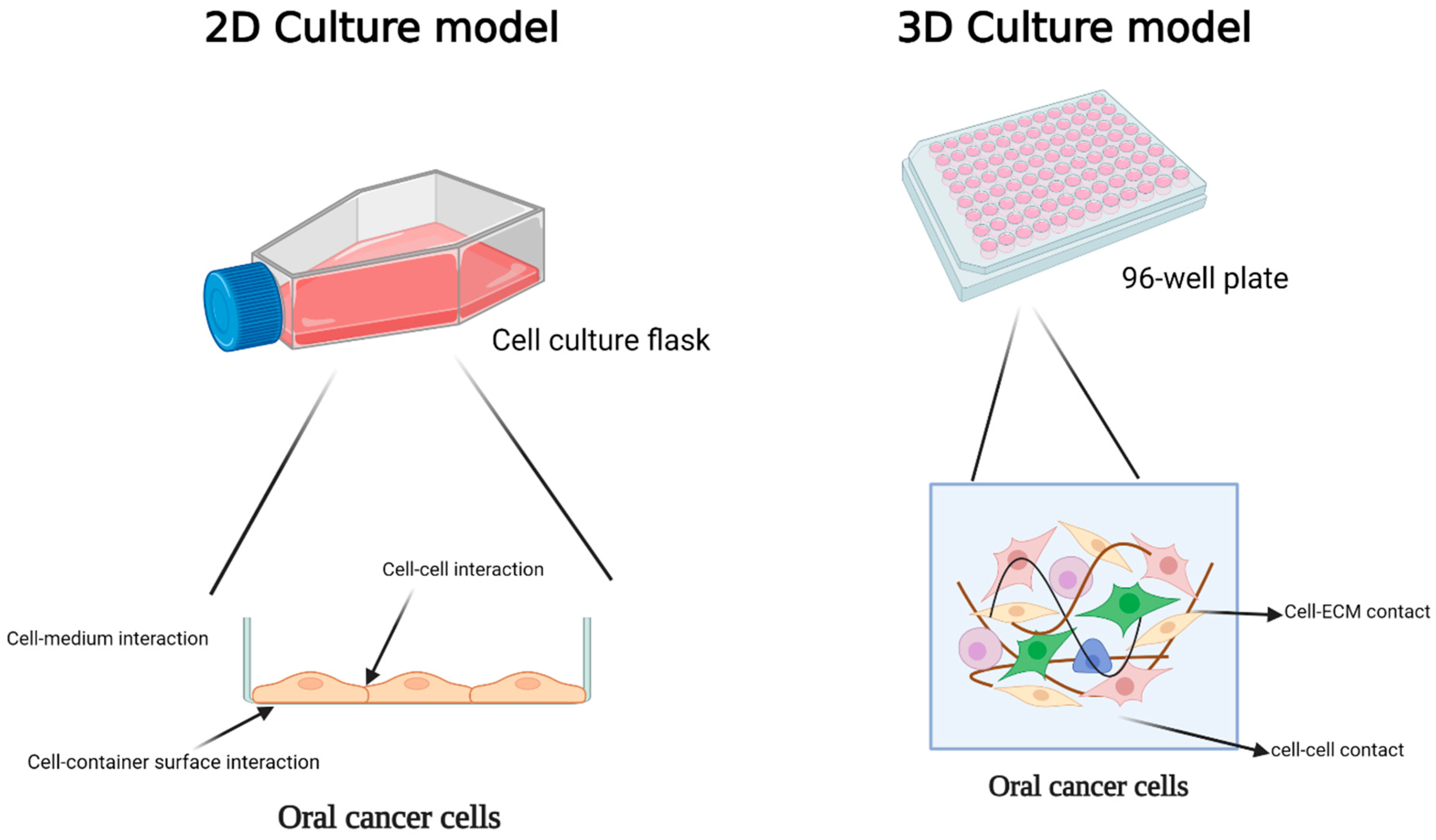
3.1. Two-Dimensional Models
3.2. Three-Dimensional Models
3.2.1. Scaffold-Free Strategy
Spheroid
Organoid
3.2.2. Scaffold-Based Strategy
Limitation and Improvement of 3D Model
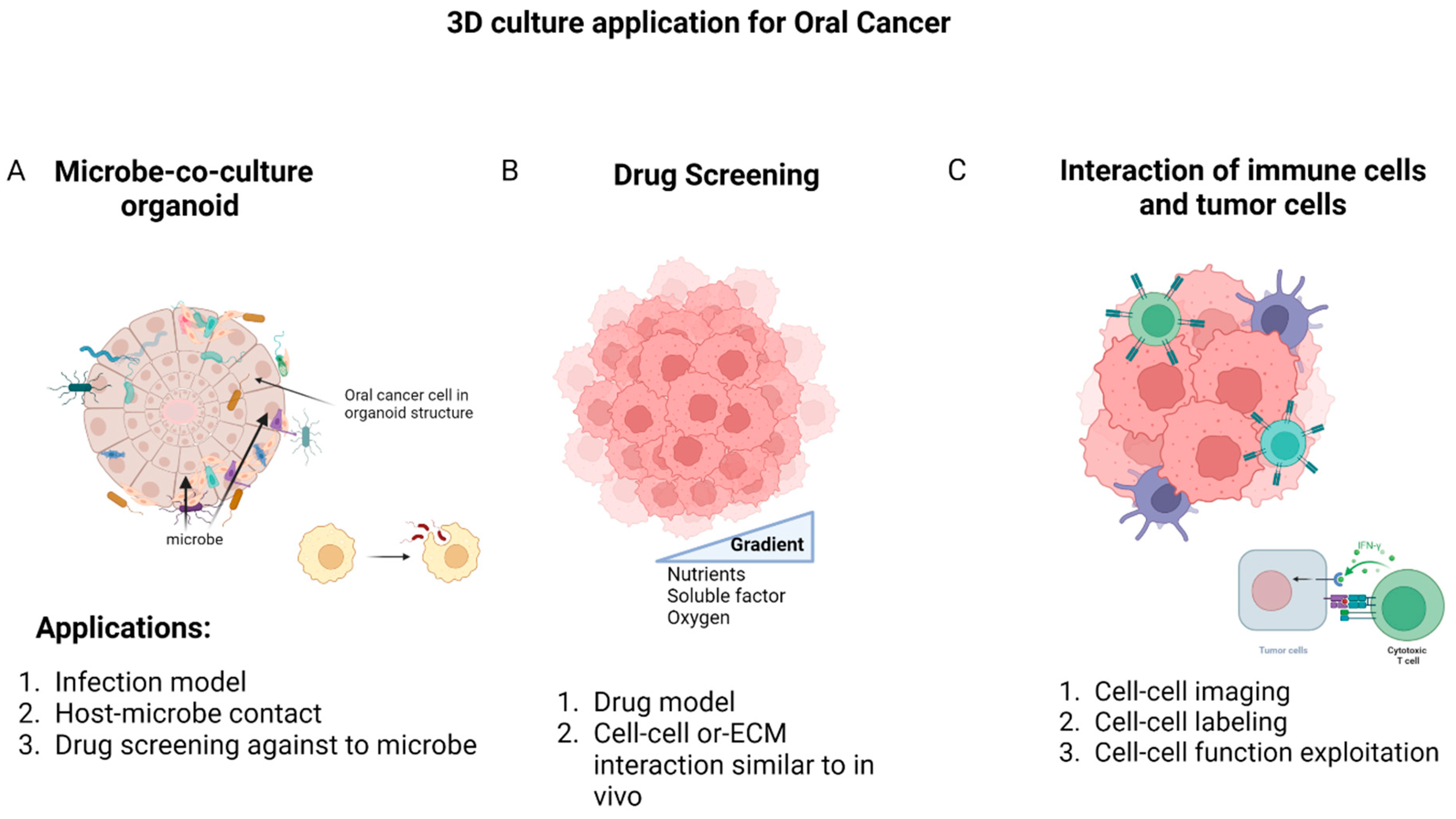
4. Application of 3D Model in Oral Cancer
4.1. Oral Microbiota Study
4.2. Drug Discovery
4.3. Cell–Cell Interactions
| 3D Model | Aim | Result | References |
|---|---|---|---|
| Co-culture of monocytes with spheroids originating Malignant/benign HNC | The connection between the response of this cytokine co-culture and the prediction of outcomes. | The secretion of IL-6 during in vitro co-culture with monocytes and BF 1-spheroids serves as a prognostic indicator for recurrence and overall prognosis, whereas co-culture with monocytes and MF 2-spheroids predicts the likelihood of recurrence. | [227] |
| Co-culture of HNC cell line with fibroblasts in spheroid form | Generation of a spheroid model of EGFR-expressing HNC. | The upregulation of chemokine expression by anti-EGFR mAb 3 promotes the infiltration of leukocytes into tumor spheroids. This unique mechanism of action of anti-EGFR mAb could potentially enhance the anti-tumor effects of the antibody in living organisms. | [228] |
| Spheroid form of HNC cell line culture with leukocytes from PBMC 4 | The evaluation of utilizing a 3D tumor cell culture model, specifically spheroids, as a suitable representation of micro-metastases. | The utilization of the spheroid model demonstrates the manifestation of pathophysiological traits, intricacy, and heterogeneity of tumor tissue observed in vivo, which significantly impacts the effectiveness of therapeutic interventions. | [229] |
| Co culture of HNC spheroids with TAMs | The signaling of CD44, influenced by TAMs, has the potential to facilitate stemness through the PI3K-4EBP1-SOX2 pathway. This effect may occur by regulating the availability of HA 5, which is the primary ligand for CD44. | The results establish a mechanistic connection between CD44 in tumor cells, TAMs, and the properties of CSCs 6 at the interface between tumor and stroma. This connection highlights a crucial area for targeting and discovering drugs. | [231] |
| Co-culture of HNC cell line with HDFs | The understanding of how cancer cells, fibroblasts, and the surrounding collagen matrices interact and promote cancer cell invasion in different environments with varying concentrations of collagen. | The presence of HDFs played a crucial role in facilitating the invasion of HNC cells into the surrounding extracellular matrix characterized by high collagen concentration, elevated storage modulus, and narrow pore sizes. | [230] |
| Co-culture of HNC cell line with CAFS | Assessing the impact of CAFs on the treatment response and migratory behavior of HNC. | The presence of CAFs resulted in enhanced cell proliferation within the tumor spheroids, which was accompanied by elevated EGFR expression. Notably, spheroids exhibiting heightened EGFR expression displayed an augmented response to cetuximab treatment. | [210] |
| HNC spheroids | Role of ERK1/2-Nanog pathway in tumorigenesis in HNC. | HNSCCs sustain a population of CSCs by utilizing the ERK1/2 signaling pathway and Nanog. | [232] |
| Oral mucosal Organoids and HNC patient-derived tumoroids | In vitro 3D model for HNC. | Drug screening for both existing and experimental therapeutic treatments for HNC. | [163] |
| HNC spheroids | The correlation of CD44 and HIF-1α expression. | By focusing on HIF-1α, the impact of NOTCH1-induced stemness, which controls the reaction to chemotherapy or radiotherapy as well as the malignancy in CD44+ HNSCCs, was reduced. Targeting the signaling of HIF-1α/NOTCH1 could potentially serve as a therapeutic approach for the treatment of HNSCC. | [237] |
| Co-culture of OSCC cell line with CAFS | Role of stromal NNMT 7 in TME. | The harmful cancer-promoting effects caused by stromal NNMT were reduced when fibroblasts were treated with inhibitors targeting collagen production, such as losartan, tranilast, and halofuginone. | [238] |
5. Potential Application of 3D Model in Studying TAM Functions in Oral Cancer
6. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Patterson, R.H.; Fischman, V.G.; Wasserman, I.; Siu, J.; Shrime, M.G.; Fagan, J.J.; Koch, W.; Alkire, B.C. Global Burden of Head and Neck Cancer: Economic Consequences, Health, and the Role of Surgery. Otolaryngol. Head Neck Surg. 2020, 162, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018; Volume 3, p. 2019.
- Chaturvedi, A.K.; Anderson, W.F.; Lortet-Tieulent, J.; Curado, M.P.; Ferlay, J.; Franceschi, S.; Rosenberg, P.S.; Bray, F.; Gillison, M.L. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J. Clin. Oncol. 2013, 31, 4550–4559. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Ghani, W.M.N.; Ramanathan, A.; Prime, S.S.; Yang, Y.H.; Razak, I.A.; Rahman, Z.A.A.; Abraham, M.T.; Mustafa, W.M.W.; Tay, K.K.; Kallarakkal, T.G.; et al. Survival of Oral Cancer Patients in Different Ethnicities. Cancer Investig. 2019, 37, 275–287. [Google Scholar] [CrossRef]
- Jou, A.; Hess, J. Epidemiology and Molecular Biology of Head and Neck Cancer. Oncol. Res. Treat. 2017, 40, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Tomita, H.; Hirata, A.; Ishida, K.; Hisamatsu, K.; Hatano, Y.; Kanayama, T.; Niwa, A.; Noguchi, K.; Kato, K.; et al. Promotion of cell proliferation by the proto-oncogene DEK enhances oral squamous cell carcinogenesis through field cancerization. Cancer Med. 2017, 6, 2424–2439. [Google Scholar] [CrossRef] [PubMed]
- Vigneswaran, N.; Williams, M.D. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac. Surg. Clin. N. Am. 2014, 26, 123–141. [Google Scholar] [CrossRef]
- Daraei, P.; Moore, C.E. Racial Disparity Among the Head and Neck Cancer Population. J. Cancer Educ. 2015, 30, 546–551. [Google Scholar] [CrossRef]
- Sathiasekar, A.C.; Mathew, D.G.; Lal, M.S.J.; Prakash, A.A.A.; Kumar, K.U.G. Oral Field Cancerization and Its Clinical Implications in the Management in Potentially Malignant Disorders. J. Pharm. Bioallied Sci. 2017, 9 (Suppl. S1), S23–S25. [Google Scholar] [PubMed]
- Hadler-Olsen, E.; Wirsing, A.M. Tissue-infiltrating immune cells as prognostic markers in oral squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2019, 120, 714–727. [Google Scholar] [CrossRef]
- Freeman, P.; Mielgo, A. Cancer-Associated Fibroblast Mediated Inhibition of CD8+ Cytotoxic T Cell Accumulation in Tumours: Mechanisms and Therapeutic Opportunities. Cancers 2020, 12, 2687. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Nazemi, M.; Rainero, E. Cross-talk between the tumor microenvironment, extracellular matrix, and cell metabolism in cancer. Front. Oncol. 2020, 10, 239. [Google Scholar] [CrossRef]
- Diskin, B.; Adam, S.; Cassini, M.F.; Sanchez, G.; Liria, M.; Aykut, B.; Buttar, C.; Li, E.; Sundberg, B.; Salas, R.D.; et al. PD-L1 engagement on T cells promotes self-tolerance and suppression of neighboring macrophages and effector T cells in cancer. Nat. Immunol. 2020, 21, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.V.; Chisamore, M.J.; Chaney, M.F.; Maradeo, M.E.; Anderson, J.; Baltus, G.A.; Pinheiro, E.M.; Uebele, V.N. Assessment of Clinical Activity of PD-1 Checkpoint Inhibitor Combination Therapies Reported in Clinical Trials. JAMA Netw. Open 2020, 3, e1920833. [Google Scholar] [CrossRef]
- Arner, E.N.; Rathmell, J.C. Metabolic programming and immune suppression in the tumor microenvironment. Cancer Cell 2023, 41, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Bule, P.; Aguiar, S.I.; Aires-Da-Silva, F.; Dias, J.N.R. Chemokine-Directed Tumor Microenvironment Modulation in Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 9804. [Google Scholar] [CrossRef] [PubMed]
- Curry, J.M.; Sprandio, J.; Cognetti, D.; Luginbuhl, A.; Barad, V.; Pribitkin, E.; Tuluc, M. Tumor microenvironment in head and neck squamous cell carcinoma. Semin. Oncol. 2014, 41, 217–234. [Google Scholar] [CrossRef]
- Liu, C.; Wang, M.; Zhang, H.; Li, C.; Zhang, T.; Liu, H.; Zhu, S.; Chen, J. Tumor microenvironment and immunotherapy of oral cancer. Eur. J. Med. Res. 2022, 27, 198. [Google Scholar] [CrossRef]
- Kondoh, N.; Mizuno-Kamiya, M. The Role of Immune Modulatory Cytokines in the Tumor Microenvironments of Head and Neck Squamous Cell Carcinomas. Cancers 2022, 14, 2884. [Google Scholar] [CrossRef]
- Niklander, S.E. Inflammatory Mediators in Oral Cancer: Pathogenic Mechanisms and Diagnostic Potential. Front. Oral Health 2021, 2, 642238. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, N.; Guan, X.; Wu, H.; Sun, Z.; Zeng, H. Immunosuppression Induced by Chronic Inflammation and the Progression to Oral Squamous Cell Carcinoma. Mediat. Inflamm. 2016, 2016, 5715719. [Google Scholar] [CrossRef]
- Kim, R.; Emi, M.; Tanabe, K. Cancer immunoediting from immune surveillance to immune escape. Immunology 2007, 121, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Tavakoli, F.; Sartakhti, J.S.; Manshaei, M.H.; Basanta, D. Cancer immunoediting: A game theoretical approach. In Silico Biol. 2021, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aragon-Sanabria, V.; Kim, G.B.; Dong, C. From Cancer Immunoediting to New Strategies in Cancer Immunotherapy: The Roles of Immune Cells and Mechanics in Oncology. Adv. Exp. Med. Biol. 2018, 1092, 113–138. [Google Scholar] [CrossRef]
- Kubick, B.J.; Fan, X.; Crouch, A.; McCarthy, R.; Roop, D.R. Tracing the Equilibrium Phase of Cancer Immunoediting in Epidermal Neoplasms via Longitudinal Intravital Imaging. J. Investig. Dermatol. 2020, 140, 891–900.e810. [Google Scholar] [CrossRef] [PubMed]
- Okubo, M.; Kioi, M.; Nakashima, H.; Sugiura, K.; Mitsudo, K.; Aoki, I.; Taniguchi, H.; Tohnai, I. M2-polarized macrophages contribute to neovasculogenesis, leading to relapse of oral cancer following radiation. Sci. Rep. 2016, 6, 27548. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, Q.; Ren, X. Novel strategies for cancer immunotherapy: Counter-immunoediting therapy. J. Hematol. Oncol. 2023, 16, 38. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Ionna, F.; Longo, F.; Della Vittoria Scarpati, G.; De Angelis, C.; Ottaiano, A.; Botti, G.; Caponigro, F. Immune Response Against Head and Neck Cancer: Biological Mechanisms and Implication on Therapy. Transl. Oncol. 2020, 13, 262–274. [Google Scholar] [CrossRef]
- Suárez-Sánchez, F.J.; Lequerica-Fernández, P.; Suárez-Canto, J.; Rodrigo, J.P.; Rodriguez-Santamarta, T.; Domínguez-Iglesias, F.; García-Pedrero, J.M.; de Vicente, J.C. Macrophages in Oral Carcinomas: Relationship with Cancer Stem Cell Markers and PD-L1 Expression. Cancers 2020, 12, 1764. [Google Scholar] [CrossRef] [PubMed]
- Almangush, A.; Heikkinen, I.; Mäkitie, A.A.; Coletta, R.D.; Läärä, E.; Leivo, I.; Salo, T. Prognostic biomarkers for oral tongue squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2017, 117, 856–866. [Google Scholar] [CrossRef]
- Rivera, C.; Oliveira, A.K.; Costa, R.A.P.; De Rossi, T.; Leme, A.F.P. Prognostic biomarkers in oral squamous cell carcinoma: A systematic review. Oral Oncol. 2017, 72, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Karmakar, T.; Arora, N.; Mukherjee, G. Immune and genomic signatures in oral (head and neck) cancer. Heliyon 2018, 4, e00880. [Google Scholar] [CrossRef]
- Dikova, V.; Jantus-Lewintre, E.; Bagan, J. Potential Non-Invasive Biomarkers for Early Diagnosis of Oral Squamous Cell Carcinoma. J. Clin. Med. 2021, 10, 1658. [Google Scholar] [CrossRef]
- Grimm, M.; Feyen, O.; Hofmann, H.; Teriete, P.; Biegner, T.; Munz, A.; Reinert, S. Immunophenotyping of patients with oral squamous cell carcinoma in peripheral blood and associated tumor tissue. Tumour Biol. 2016, 37, 3807–3816. [Google Scholar] [CrossRef]
- Németh, Z.; Velich, N.; Bogdan, S.; Ujpál, M.; Szabó, G.; Suba, Z.S. The prognostic role of clinical, morphological and molecular markers in oral squamous cell tumors. Neoplasma 2005, 52, 95–102. [Google Scholar] [PubMed]
- Gustafson, M.P.; Lin, Y.; Bleeker, J.S.; Warad, D.; Tollefson, M.K.; Crispen, P.L.; Bulur, P.A.; Harrington, S.M.; Laborde, R.R.; Gastineau, D.A.; et al. Intratumoral CD14+ Cells and Circulating CD14+HLA-DRlo/neg Monocytes Correlate with Decreased Survival in Patients with Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2015, 21, 4224–4233. [Google Scholar] [CrossRef]
- Motomura, Y.; Kanno, S.; Asano, K.; Tanaka, M.; Hasegawa, Y.; Katagiri, H.; Saito, T.; Hara, H.; Nishio, H.; Hara, T.; et al. Identification of Pathogenic Cardiac CD11c+ Macrophages in Nod1-Mediated Acute Coronary Arteritis. Arter. Thromb. Vasc. Biol. 2015, 35, 1423–1433. [Google Scholar] [CrossRef]
- Weber, M.; Büttner-Herold, M.; Hyckel, P.; Moebius, P.; Distel, L.; Ries, J.; Amann, K.; Neukam, F.W.; Wehrhan, F. Small oral squamous cell carcinomas with nodal lymphogenic metastasis show increased infiltration of M2 polarized macrophages—An immunohistochemical analysis. J. Craniomaxillofac. Surg. 2014, 42, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Moriyama, M.; Kubota, K.; Ishiguro, N.; Sakamoto, M.; Chinju, A.; Mochizuki, K.; Sakamoto, T.; Kaneko, N.; Munemura, R.; et al. CD206+ tumor-associated macrophages promote proliferation and invasion in oral squamous cell carcinoma via EGF production. Sci. Rep. 2019, 9, 14611. [Google Scholar] [CrossRef]
- Tan-Garcia, A.; Wai, L.E.; Zheng, D.; Ceccarello, E.; Jo, J.; Banu, N.; Khakpoor, A.; Chia, A.; Tham, C.Y.L.; Tan, A.T.; et al. Intrahepatic CD206+ macrophages contribute to inflammation in advanced viral-related liver disease. J. Hepatol. 2017, 67, 490–500. [Google Scholar] [CrossRef]
- Yamagata, Y.; Tomioka, H.; Sakamoto, K.; Sato, K.; Harada, H.; Ikeda, T.; Kayamori, K. CD163-Positive Macrophages Within the Tumor Stroma Are Associated With Lymphangiogenesis and Lymph Node Metastasis in Oral Squamous Cell Carcinoma. J. Oral Maxillofac. Surg. 2017, 75, 2144–2153. [Google Scholar] [CrossRef]
- Jardim, J.F.; Gondak, R.; Galvis, M.M.; Pinto, C.A.L.; Kowalski, L.P. A decreased peritumoral CD1a+ cell number predicts a worse prognosis in oral squamous cell carcinoma. Histopathology 2018, 72, 905–913. [Google Scholar] [CrossRef]
- Peña-Cardelles, J.F.; Pozo-Kreilinger, J.J.; Roncador, G.; Esteban-Hernández, J.; Cebrián-Carretero, J.L.; Moro-Rodríguez, J.E. Expression of clec9a in the oral cancer microenvironment. A preliminary immunohistochemical pilot study. Med. Oral Patol. Oral Cir. Bucal 2021, 26, e651–e660. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.; Shurin, M.R.; Collins, B.; Kogan, D.; Tourkova, I.L.; Shurin, G.V. Comparative analysis of CD1a, S-100, CD83, and CD11c human dendritic cells in normal, premalignant, and malignant tissues. Histol. Histopathol. 2005, 20, 1165–1172. [Google Scholar] [PubMed]
- Agarwal, R.; Chaudhary, M.; Bohra, S.; Bajaj, S. Evaluation of natural killer cell (CD57) as a prognostic marker in oral squamous cell carcinoma: An immunohistochemistry study. J. Oral Maxillofac. Pathol. 2016, 20, 173–177. [Google Scholar] [CrossRef]
- Mele, D.; Pessino, G.; Trisolini, G.; Luchena, A.; Benazzo, M.; Morbini, P.; Mantovani, S.; Oliviero, B.; Mondelli, M.U.; Varchetta, S. Impaired intratumoral natural killer cell function in head and neck carcinoma. Front. Immunol. 2022, 13, 997806. [Google Scholar] [CrossRef]
- Taghavi, N.; Bagheri, S.; Akbarzadeh, A. Prognostic implication of CD57, CD16, and TGF-β expression in oral squamous cell carcinoma. J. Oral Pathol. Med. 2016, 45, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Bag, S.; Chakraborty, P.; Dey, D.; Roy, S.; Jain, P.; Roy, P.; Soong, R.; Majumder, P.P.; Dutt, S. Density of CD3+ and CD8+ cells in gingivo-buccal oral squamous cell carcinoma is associated with lymph node metastases and survival. PLoS ONE 2020, 15, e0242058. [Google Scholar] [CrossRef]
- Spanier, G.; Ugele, I.; Nieberle, F.; Symeou, L.; Schmidhofer, S.; Brand, A.; Meier, J.; Spoerl, S.; Krupar, R.; Rümmele, P.; et al. The predictive power of CD3+ T cell infiltration of oral squamous cell tumors is limited to non-diabetic patients. Cancer Lett. 2021, 499, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.L.; Korzinkin, M.; Zhavoronkov, A.; Ozerov, I.V.; Walker, M.T.; Higgins, K.; Lingen, M.W.; Izumchenko, E.P. A Savage Effector T cell responses unleashed by regulatory T cell ablation exacerbate oral squamous cell carcinoma. Cell Rep. Med. 2021, 2, 100399. [Google Scholar] [CrossRef]
- Kondo, Y.; Suzuki, S.; Takahara, T.; Ono, S.; Goto, M.; Miyabe, S.; Sugita, Y.; Ogawa, T.; Ito, H.; Satou, A.; et al. Improving function of cytotoxic T-lymphocytes by transforming growth factor-β inhibitor in oral squamous cell carcinoma. Cancer Sci. 2021, 112, 4037–4049. [Google Scholar] [CrossRef] [PubMed]
- Maggioni, D.; Pignataro, L.; Garavello, W. T-helper and T-regulatory cells modulation in head and neck squamous cell carcinoma. Oncoimmunology 2017, 6, e1325066. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.; Xie, F. T-regulatory cell/T helper 17 cell imbalance functions as prognostic biomarker of oral squamous cell carcinoma—CONSORT. Medicine 2020, 99, e23145. [Google Scholar] [CrossRef]
- Lechner, A.; Schlößer, H.A.; Thelen, M.; Wennhold, K.; Rothschild, S.I.; Gilles, R.; Quaas, A.; Siefer, O.G.; Huebbers, C.U.; Cukuroglu, E.; et al. Tumor-associated B cells and humoral immune response in head and neck squamous cell carcinoma. Oncoimmunology 2019, 8, 1535293. [Google Scholar] [CrossRef]
- Suárez-Sánchez, F.J.; Lequerica-Fernández, P.; Rodrigo, J.P.; Hermida-Prado, F.; Suárez-Canto, J.; Rodríguez-Santamarta, T.; Domínguez-Iglesias, F.; García-Pedrero, J.M.; de Vicente, J.C. Tumor-Infiltrating CD20+ B Lymphocytes: Significance and Prognostic Implications in Oral Cancer Microenvironment. Cancers 2021, 13, 395. [Google Scholar] [CrossRef]
- Hao, Z.; Li, R.; Wang, Y.; Li, S.; Hong, Z.; Han, Z. Landscape of Myeloid-derived Suppressor Cell in Tumor Immunotherapy. Biomark. Res. 2021, 9, 77. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, C.; Wang, Y.; Zhang, W.; Liu, L.; Cheng, J. Prognostic role of CD11b+ myeloid-derived suppressor cells in oral squamous cell carcinoma. Arch. Med. Sci. 2023, 19, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Fan, H.Y.; Tang, Y.L.; Wang, S.S.; Cao, M.X.; Wang, H.F.; Dai, L.L.; Wang, K.; Yu, X.H.; Wu, J.B.; et al. Myeloid derived suppressor cells contribute to the malignant progression of oral squamous cell carcinoma. PLoS ONE 2020, 15, e0229089. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yang, P.; Xiao, Y.; Zhang, Y.; Liu, J.; Xie, D.; Cai, M.; Zhang, X. Overexpression of α-sma-positive fibroblasts (CAFs) in Nasopharyngeal Carcinoma Predicts Poor Prognosis. J. Cancer 2017, 8, 3897–3902. [Google Scholar] [CrossRef]
- Lao, X.M.; Liang, Y.J.; Su, Y.X.; Zhang, S.E.; Zhou, X.I.; Liao, G.Q. Distribution and significance of interstitial fibrosis and stroma-infiltrating B cells in tongue squamous cell carcinoma. Oncol. Lett. 2016, 11, 2027–2034. [Google Scholar] [CrossRef]
- Patel, A.K.; Vipparthi, K.; Thatikonda, V.; Arun, I.; Bhattacharjee, S.; Sharan, R.; Arun, P.; Singh, S. A subtype of cancer-associated fibroblasts with lower expression of alpha-smooth muscle actin suppresses stemness through BMP4 in oral carcinoma. Oncogenesis 2018, 7, 78. [Google Scholar] [CrossRef]
- Gaur, P.; Qadir, G.A.; Upadhyay, S.; Singh, A.K.; Shukla, N.K.; Das, S.N. Skewed immunological balance between Th17 (CD4+IL17A+) and Treg (CD4+CD25+FOXP3+) cells in human oral squamous cell carcinoma. Cell. Oncol. 2012, 35, 335–343. [Google Scholar] [CrossRef]
- Lim, K.P.; Chun, N.A.; Ismail, S.M.; Abraham, M.T.; Yusoff, M.N.; Zain, R.B.; Ngeow, W.C.; Ponniah, S.; Cheong, S.C. CD4+CD25hiCD127low regulatory T cells are increased in oral squamous cell carcinoma patients. PLoS ONE 2014, 9, e103975. [Google Scholar] [CrossRef] [PubMed]
- Kukreja, I.; Kapoor, P.; Deshmukh, R.; Kulkarni, V. VEGF and CD34: A correlation between tumor angiogenesis and microvessel density—An immunohistochemical study. J. Oral Maxillofac. Pathol. 2013, 17, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, A.; Ali, A.; Zaffar, Z.; Mokeem, S.; Mokeem, S.S.; Ahmed, N.; Al-Hamoudi, N.; Vohra, F.; Javed, F.; Abduljabbar, T. Expression of CD34 and α-SMA Markers in Oral Squamous Cell Carcinoma Differentiation. A Histological and Histo-Chemical Study. Int. J. Environ. Res. Public. Health 2020, 18, 192. [Google Scholar] [CrossRef]
- Teófilo, C.R.; Junior, A.E.C.F.; Batista, A.C.; Jamacaru, F.V.F.; Sousa, F.B.; Mota, M.R.L.; Silva, M.F.E.; Silva, P.G.B.; Alves, A. Mast Cells and Blood Vessels Profile in Oral Carcinogenesis: An Immunohistochemistry Study. Asian Pac. J. Cancer Prev. 2020, 21, 1097–1102. [Google Scholar] [CrossRef]
- Panneerselvam, K.; Ishikawa, S.; Krishnan, R.; Sugimoto, M. Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites 2022, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Santosh, A.B.; Jones, T.; Harvey, J. A review on oral cancer biomarkers: Understanding the past and learning from the present. J. Cancer Res. Ther. 2016, 12, 486–492. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014, 4, 6802. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Yen, C.Y.; Chen, C.H.; Tsai, J.H.; Tang, J.Y.; Chang, Y.T.; Kao, Y.H.; Wang, Y.Y.; Yuan, S.F.; Lee, S.Y. Evaluation of the mRNA expression levels of integrins α3, α5, β1 and β6 as tumor biomarkers of oral squamous cell carcinoma. Oncol. Lett. 2018, 16, 4773–4781. [Google Scholar] [CrossRef]
- Kurokawa, A.; Nagata, M.; Kitamura, N.; Noman, A.A.; Ohnishi, M.; Ohyama, T.; Kobayashi, T.; Shingaki, S.; Takagi, R. Diagnostic value of integrin α3, β4, and β5 gene expression levels for the clinical outcome of tongue squamous cell carcinoma. Cancer 2008, 112, 1272–1281. [Google Scholar] [CrossRef]
- Nagata, M.; Noman, A.A.; Suzuki, K.; Kurita, H.; Ohnishi, M.; Ohyama, T.; Kitamura, N.; Kobayashi, T.; Uematsu, K.; Takahashi, K.; et al. ITGA3 and ITGB4 expression biomarkers estimate the risks of locoregional and hematogenous dissemination of oral squamous cell carcinoma. BMC Cancer 2013, 13, 410. [Google Scholar] [CrossRef] [PubMed]
- Basheer, H.A.; Pakanavicius, E.; Cooper, P.A.; Shnyder, S.D.; Martin, L.; Hunter, K.D.; Vinader, V.; Afarinkia, K. Hypoxia modulates CCR7 expression in head and neck cancers. Oral Oncol. 2018, 80, 64–73. [Google Scholar] [CrossRef]
- Guo, N.; Liu, F.; Yang, L.; Huang, J.; Ding, X.; Sun, C. Chemokine receptor 7 enhances cell chemotaxis and migration of metastatic squamous cell carcinoma of head and neck through activation of matrix metalloproteinase-9. Oncol. Rep. 2014, 32, 794–800. [Google Scholar] [CrossRef]
- Shang, Z.J.; Liu, K.; Shao, Z. Expression of chemokine receptor CCR7 is associated with cervical lymph node metastasis of oral squamous cell carcinoma. Oral Oncol. 2009, 45, 480–485. [Google Scholar] [CrossRef]
- Ohmura, G.; Tsujikawa, T.; Yaguchi, T.; Kawamura, N.; Mikami, S.; Sugiyama, J.; Nakamura, K.; Kobayashi, A.; Iwata, T.; Nakano, H.; et al. Aberrant Myosin 1b Expression Promotes Cell Migration and Lymph Node Metastasis of HNSCC. Mol. Cancer Res. 2015, 13, 721–731. [Google Scholar] [CrossRef]
- Sun, Z.; Guo, X.; Chen, H.; Ling, J.; Zhao, H.; Chang, A.; Zhuo, X. MYO1B as a prognostic biomarker and a therapeutic target in Arecoline-associated oral carcinoma. Mol. Carcinogen. 2023, 62, 920–939. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Okato, A.; Idichi, T.; Arai, T.; Sugawara, S.; Katada, K.; Okamoto, Y.; et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int. J. Oncol. 2018, 52, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Maman, S.; Witz, I.P. A history of exploring cancer in context. Nat. Rev. Cancer 2018, 18, 359–376. [Google Scholar] [CrossRef] [PubMed]
- McMillin, D.W.; Negri, J.M.; Mitsiades, C.S. The role of tumour-stromal interactions in modifying drug response: Challenges and opportunities. Nat. Rev. Drug Discov. 2013, 12, 217–228. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; de Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Jain, R.K.; Langer, R. Engineering and physical sciences in oncology: Challenges and opportunities. Nat. Rev. Cancer 2017, 17, 659–675. [Google Scholar] [CrossRef]
- Li, M.; He, L.; Zhu, J.; Zhang, P.; Liang, S. Targeting tumor-associated macrophages for cancer treatment. Cell Biosci. 2022, 12, 85. [Google Scholar] [CrossRef]
- Yan, M.; Jurasz, P. The role of platelets in the tumor microenvironment: From solid tumors to leukemia. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 392–400. [Google Scholar] [CrossRef]
- Ramamonjisoa, N.; Ackerstaff, E. Characterization of the Tumor Microenvironment and Tumor-Stroma Interaction by Non-Invasive Preclinical Imaging. Front. Oncol. 2017, 7, 3. [Google Scholar] [CrossRef]
- Bellomo, C.; Caja, L.; Moustakas, A. Transforming growth factor β as regulator of cancer stemness and metastasis. Br. J. Cancer 2016, 115, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Sapudom, J.; Müller, C.D.; Nguyen, K.-T.; Martin, S.; Anderegg, U.; Pompe, T. Matrix Remodeling and Hyaluronan Production by Myofibroblasts and Cancer-Associated Fibroblasts in 3D Collagen Matrices. Gels 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tao, D.; Zhang, P.; Liu, X.; Zhang, Y.; Cheng, J.; Yuan, H.; Liu, L.; Jiang, H. Hyaluronan synthase 2 expressed by cancer-associated fibroblasts promotes oral cancer invasion. J. Exp. Clin. Cancer Res. 2016, 35, 181. [Google Scholar] [CrossRef]
- Elmusrati, A.A.; Pilborough, A.E.; Khurram, S.A.; Lambert, D.W. Cancer-associated fibroblasts promote bone invasion in oral squamous cell carcinoma. Br. J. Cancer 2017, 117, 867–875. [Google Scholar] [CrossRef]
- Lin, N.N.; Wang, P.; Zhao, D.; Zhang, F.J.; Yang, K.; Chen, R. Significance of oral cancer-associated fibroblasts in angiogenesis, lymphangiogenesis, and tumor invasion in oral squamous cell carcinoma. J. Oral Pathol. Med. 2017, 46, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Russmueller, G.; Moser, D.; Würger, T.; Wrba, F.; Christopoulos, P.; Kostakis, G.; Seemann, R.; Stadler, V.; Wimmer, G.; Kornek, G.; et al. Upregulation of osteoprotegerin expression correlates with bone invasion and predicts poor clinical outcome in oral cancer. Oral Oncol. 2015, 51, 247–253. [Google Scholar] [CrossRef]
- Sambandam, Y.; Ethiraj, P.; Hathaway-Schrader, J.D.; Novince, C.M.; Panneerselvam, E.; Sundaram, K.; Reddy, S.V. Autoregulation of RANK ligand in oral squamous cell carcinoma tumor cells. J. Cell. Physiol. 2018, 233, 6125–6134. [Google Scholar] [CrossRef]
- Bae, J.Y.; Kim, E.K.; Yang, D.H.; Zhang, X.; Park, Y.J.; Lee, D.Y.; Che, C.M.; Kim, J. Reciprocal interaction between carcinoma-associated fibroblasts and squamous carcinoma cells through interleukin-1α induces cancer progression. Neoplasia 2014, 16, 928–938. [Google Scholar] [CrossRef]
- Korbecki, J.; Bosiacki, M.; Barczak, K.; Łagocka, R.; Chlubek, D.; Baranowska-Bosiacka, I. The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers. Cells 2023, 12, 1406. [Google Scholar] [CrossRef]
- Shishido, K.; Kuroishi, T.; Sugawara, S. P2 purinergic receptor signaling and interleukin-1 synergistically induce interleukin-6 production in a human oral squamous carcinoma cell line. J. Oral Biosci. 2021, 63, 80–90. [Google Scholar] [CrossRef]
- Brady, G.; Crean, S.J.; Naik, P.; Kapas, S. Upregulation of IGF-2 and IGF-1 receptor expression in oral cancer cell lines. Int. J. Oncol. 2007, 31, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Cacheux, W.; Lièvre, A.; Richon, S.; Vacher, S.; El Alam, E.; Briaux, A.; El Botty, R.; Mariani, P.; Buecher, B.; Schnitzler, A.; et al. Dangles-Marie Interaction between IGF2-PI3K axis and cancer-associated-fibroblasts promotes anal squamous carcinogenesis. Int. J. Cancer 2019, 145, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Ferreira Mendes, J.M.; de Faro Valverde, L.; Vidal, M.T.A.; Paredes, B.D.; Coelho, P.; Allahdadi, K.J.; Coletta, R.D.; Souza, B.S.F.; Rocha, C.A.G. Effects of IGF-1 on Proliferation, Angiogenesis, Tumor Stem Cell Populations and Activation of AKT and Hedgehog Pathways in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 6487. [Google Scholar] [CrossRef] [PubMed]
- Kayamori, K.; Katsube, K.; Sakamoto, K.; Ohyama, Y.; Hirai, H.; Yukimori, A.; Ohata, Y.; Akashi, T.; Saitoh, M.; Harada, K.; et al. NOTCH3 Is Induced in Cancer-Associated Fibroblasts and Promotes Angiogenesis in Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0154112. [Google Scholar] [CrossRef]
- Porcheri, C.; Meisel, C.T.; Mitsiadis, T. Multifactorial Contribution of Notch Signaling in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 1520. [Google Scholar] [CrossRef]
- Sun, W.; Gaykalova, D.A.; Ochs, M.F.; Mambo, E.; Arnaoutakis, D.; Liu, Y.; Loyo, M.; Agrawal, N.; Howard, J.; Li, R.; et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014, 74, 1091–1104. [Google Scholar] [CrossRef]
- Karakasheva, T.A.; Lin, E.W.; Tang, Q.; Qiao, E.; Waldron, T.J.; Soni, M.; Klein-Szanto, A.J.; Sahu, V.; Basu, D.; Ohashi, S.; et al. IL-6 Mediates Cross-Talk between Tumor Cells and Activated Fibroblasts in the Tumor Microenvironment. Cancer Res. 2018, 78, 4957–4970. [Google Scholar] [CrossRef]
- Mirkeshavarz, M.; Ganjibakhsh, M.; Aminishakib, P.; Farzaneh, P.; Mahdavi, N.; Vakhshiteh, F.; Karimi, A.; Gohari, N.S.; Kamali, F.; Kharazifard, M.J.; et al. Interleukin-6 secreted by oral cancer- associated fibroblast accelerated VEGF expression in tumor and stroma cells. Cell. Mol. Biol. 2017, 63, 131–136. [Google Scholar] [CrossRef]
- Qin, X.; Yan, M.; Wang, X.; Xu, Q.; Wang, X.; Zhu, X.; Shi, J.; Li, Z.; Zhang, J.; Chen, W. Cancer-associated Fibroblast-derived IL-6 Promotes Head and Neck Cancer Progression via the Osteopontin-NF-κB Signaling Pathway. Theranostics 2018, 8, 921–940. [Google Scholar] [CrossRef]
- Li, X.; Bu, W.; Meng, L.; Liu, X.; Wang, S.; Jiang, L.; Ren, M.; Fan, Y.; Sun, H. CXCL12/CXCR4 pathway orchestrates CSC-like properties by CAF recruited tumor associated macrophage in OSCC. Exp. Cell Res. 2019, 378, 131–138. [Google Scholar] [CrossRef]
- Wu, M.H.; Hong, H.C.; Hong, T.M.; Chiang, W.F.; Jin, Y.T.; Chen, Y.L. Targeting galectin-1 in carcinoma-associated fibroblasts inhibits oral squamous cell carcinoma metastasis by downregulating MCP-1/CCL2 expression. Clin. Cancer Res. 2011, 17, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.W.; Hanafi, Z.B.; Chew, L.C.Y.; Mei, Y.; Liu, H. IL-1α Processing, Signaling and Its Role in Cancer Progression. Cells 2021, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.-W.; Che, Z.M.; Kim, J.; Kim, K.; Kim, K.-Y.; Williams, D.; Kim, J. Tumor-stromal crosstalk in invasion of oral squamous cell carcinoma: A pivotal role of CCL7. Int. J. Cancer 2010, 127, 332–344. [Google Scholar] [CrossRef]
- Sergi, B. Remodeling the tumor microenvironment to overcome treatment resistance in HPV-negative head and neck cancer. Cancer Drug Resist. 2023, 6, 291–313. [Google Scholar] [CrossRef]
- Takahashi, H.; Sakakura, K.; Kudo, T.; Toyoda, M.; Kaira, K.; Oyama, T.; Chikamatsu, K. Cancer-associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget 2017, 8, 8633–8647. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Moriyama, M.; Furukawa, S.; Rafiul, H.; Maruse, Y.; Jinno, T.; Tanaka, A.; Ohta, M.; Ishiguro, N.; Yamauchi, M.; et al. CD163+CD204+ tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci. Rep. 2017, 7, 1755. [Google Scholar] [CrossRef]
- Wu, T.; Tang, C.; Tao, R.; Yong, X.; Jiang, Q.; Feng, C. PD-L1-Mediated Immunosuppression in Oral Squamous Cell Carcinoma: Relationship With Macrophage Infiltration and Epithelial to Mesenchymal Transition Markers. Front. Immunol. 2021, 12, 693881. [Google Scholar] [CrossRef]
- O’Sullivan, C.; Lewis, C.E.; Harris, A.L.; McGee, J.O. Secretion of epidermal growth factor by macrophages associated with breast carcinoma. Lancet 1993, 342, 148–149. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.D.; Haridas, N.; Patel, J.B.; Shah, F.D.; Shukla, S.N.; Shah, P.M.; Patel, P.S. Matrix metalloproteinases and their inhibitors: Correlation with invasion and metastasis in oral cancer. Indian J. Clin. Biochem. 2010, 25, 250–259. [Google Scholar] [CrossRef]
- Alves, A.; Diel, L.; Ramos, G.; Pinto, A.; Bernardi, L.; Yates, J., 3rd; Lamers, M. Tumor microenvironment and Oral Squamous Cell Carcinoma: A crosstalk between the inflammatory state and tumor cell migration. Oral Oncol. 2021, 112, 105038. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.X.; Wang, S.; Zhao, H.; Liu, N.; Chen, D.; Sun, M.; Zheng, J.H. Sonic hedgehog signaling may promote invasion and metastasis of oral squamous cell carcinoma by activating MMP-9 and E-cadherin expression. Med. Oncol. 2014, 31, 41. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; He, M.Y.; Zhu, L.F.; Yang, C.C.; Zhou, M.L.; Wang, Q.; Zhang, W.; Zheng, Y.Y.; Wang, D.M.; Xu, Z.Q.; et al. Liu Tumor-associated macrophages correlate with the clinicopathological features and poor outcomes via inducing epithelial to mesenchymal transition in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 12. [Google Scholar] [CrossRef]
- Petty, A.J.; Li, A.; Wang, X.; Dai, R.; Heyman, B.; Hsu, D.; Huang, X.; Yang, Y. Hedgehog signaling promotes tumor-associated macrophage polarization to suppress intratumoral CD8+ T cell recruitment. J. Clin. Investig. 2019, 129, 5151–5162. [Google Scholar] [CrossRef] [PubMed]
- Valverde, L.d.F.; Pereira, T.D.A.; Dias, R.B.; Guimarães, V.S.N.; Ramos, E.A.G.; Santos, J.N.; Rocha, C.A.G. Macrophages and endothelial cells orchestrate tumor-associated angiogenesis in oral cancer via hedgehog pathway activation. Tumor Biol. 2016, 37, 9233–9241. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, T.; Chai, Y.; Chen, F. TGF-β Signaling in Progression of Oral Cancer. Int. J. Mol. Sci. 2023, 24, 10263. [Google Scholar] [CrossRef]
- Sun, H.; Miao, C.; Liu, W.; Qiao, X.; Yang, W.; Li, L. Li TGF-β1/TβRII/Smad3 signaling pathway promotes VEGF expression in oral squamous cell carcinoma tumor-associated macrophages. Biochem. Biophys. Res. Commun. 2018, 497, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Mao, X.H.; Tian, T.; Wang, W.M.; Su, T.; Jiang, C.H.; Hu, C.Y. Role of PFKFB3 and CD163 in Oral Squamous Cell Carcinoma Angiogenesis. Curr. Med. Sci. 2019, 39, 410–414. [Google Scholar] [CrossRef]
- Shi, L.; Pan, H.; Liu, Z.; Xie, J.; Han, W. Roles of PFKFB3 in cancer. Signal Transduct. Target. Ther. 2017, 2, 17044. [Google Scholar] [CrossRef]
- Han, N.; Li, X.; Wang, Y.; Wang, L.; Zhang, C.; Zhang, Z.; Ruan, M.; Zhang, C. Increased tumor-infiltrating plasmacytoid dendritic cells promote cancer cell proliferation and invasion via TNF-α/NF-κB/CXCR-4 pathway in oral squamous cell carcinoma. J. Cancer 2021, 12, 3045–3056. [Google Scholar] [CrossRef]
- Rehman, A.O.; Wang, C.Y. CXCL12/SDF-1α activates NF-κB and promotes oral cancer invasion through the Carma3/Bcl10/Malt1 complex. Int. J. Oral Sci. 2009, 1, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Encarnación-Rosado, J.; Lin, E.Y.; Sohn, A.S.W.; Zhang, H.; Mancias, J.D.; Kimmelman, A.C. Autophagy supports mitochondrial metabolism through the regulation of iron homeostasis in pancreatic cancer. Sci. Adv. 2023, 9, eadf9284. [Google Scholar] [CrossRef] [PubMed]
- Dinić, M.; Pecikoza, U.; Djokić, J.; Stepanović-Petrović, R.; Milenković, M.; Stevanović, M.; Filipović, N.; Begović, J.; Golić, N.; Lukić, J. Exopolysaccharide Produced by Probiotic Strain Lactobacillus paraplantarum BGCG11 Reduces Inflammatory Hyperalgesia in Rats. Front. Pharmacol. 2018, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Pinto, B.; Henriques, A.C.; Silva, P.M.A.; Bousbaa, H. Three-Dimensional Spheroids as In Vitro Preclinical Models for Cancer Research. Pharmaceutics 2020, 12, 1186. [Google Scholar] [CrossRef]
- Pozzi, S.; Scomparin, A.; Dangoor, S.I.; Ajamil, D.R.; Ofek, P.; Neufeld, L.; Krivitsky, A.; Vaskovich-Koubi, D.; Kleiner, R.; Dey, P.; et al. Meet me halfway: Are in vitro 3D cancer models on the way to replace in vivo models for nanomedicine development? Adv. Drug Deliv. Rev. 2021, 175, 113760. [Google Scholar] [CrossRef]
- Abdolahinia, E.D.; Ahmadian, S.; Bohlouli, S.; Gharehbagh, F.J.; Jahandizi, N.G.; Vahed, S.Z.; Saadat, Y.R.; Aghbali, A.; Sharifi, S.; Dizaj, S.M.; et al. Effect of Curcumin on the Head and Neck Squamous Cell Carcinoma Cell Line HN5. Curr. Mol. Pharmacol. 2023, 16, 374–380. [Google Scholar] [PubMed]
- Memar, M.Y.; Abdolahinia, E.D.; Yekani, M.; Kouhsoltani, M.; Sharifi, S.; Dizaj, S.M. Preparation of rutin-loaded mesoporous silica nanoparticles and evaluation of its physicochemical, anticancer, and antibacterial properties. Mol. Biol. Rep. 2023, 50, 203–213. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef]
- Hickman, J.A.; Graeser, R.; de Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; van der Kuip, H. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.E.; Miller, F.R.; Heppner, G.H. Factors affecting growth and drug sensitivity of mouse mammary tumor lines in collagen gel cultures. Cancer Res. 1985, 45, 4200–4205. [Google Scholar] [PubMed]
- Miyamoto, S.; Nishikiori, N.; Sato, T.; Watanabe, M.; Umetsu, A.; Tsugeno, Y.; Hikage, F.; Sasaya, T.; Kato, H.; Ogi, K.; et al. Three-Dimensional Spheroid Configurations and Cellular Metabolic Properties of Oral Squamous Carcinomas Are Possible Pharmacological and Pathological Indicators. Cancers 2023, 15, 2793. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Sato, K.; Nakamura, T.; Yoshida, Y.; Murata, S.; Yoshida, K.; Kanemoto, H.; Umemori, K.; Kawai, H.; Obata, K.; et al. Reproduction of the Antitumor Effect of Cisplatin and Cetuximab Using a Three-dimensional Spheroid Model in Oral Cancer. Int. J. Med. Sci. 2022, 19, 1320–1333. [Google Scholar] [CrossRef]
- Vincent-Chong, V.K.; Seshadri, M. Development and Radiation Response Assessment in A Novel Syngeneic Mouse Model of Tongue Cancer: 2D Culture, 3D Organoids and Orthotopic Allografts. Cancers 2020, 12, 579. [Google Scholar] [CrossRef]
- Howard, D.; Buttery, L.D.; Shakesheff, K.M.; Roberts, S.J. Tissue engineering: Strategies, stem cells and scaffolds. J. Anat. 2008, 213, 66–72. [Google Scholar] [CrossRef]
- Ovsianikov, A.; Khademhosseini, A.; Mironov, V. The synergy of scaffold-based and scaffold-free tissue engineering strategies. Trends Biotechnol. 2018, 36, 348–357. [Google Scholar] [CrossRef]
- Lee, J.; You, J.H.; Shin, D.; Roh, J.L. Ex vivo culture of head and neck cancer explants in cell sheet for testing chemotherapeutic sensitivity. J. Cancer Res. Clin. Oncol. 2020, 146, 2497–2507. [Google Scholar] [CrossRef]
- Sekine, W.; Haraguchi, Y.; Shimizu, T.; Umezawa, A.; Okano, T. Thickness limitation and cell viability of multi-layered cell sheets and overcoming the diffusion limit by a porous-membrane culture insert. J. Biochips Tissue Chips 2011, 1, 2153–2777. [Google Scholar] [CrossRef]
- Sant, S.; Johnston, P.A. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov. Today Technol. 2017, 23, 27–36. [Google Scholar] [CrossRef]
- Rodrigues, T.; Kundu, B.; Silva-Correia, J.; Kundu, S.C.; Oliveira, J.M.; Reis, R.L.; Correlo, V.M. Emerging tumor spheroids technologies for 3D in vitro cancer modeling. Pharmacol. Ther. 2018, 184, 201–211. [Google Scholar] [CrossRef]
- Friedrich, J.; Ebner, R.; Kunz-Schughart, L.A. Experimental anti-tumor therapy in 3-D: Spheroids—Old hat or new challenge? Int. J. Radiat. Biol. 2007, 83, 849–871. [Google Scholar] [CrossRef]
- Chitturi Suryaprakash, R.T.; Kujan, O.; Shearston, K.; Farah, C.S. Three-Dimensional Cell Culture Models to Investigate Oral Carcinogenesis: A Scoping Review. Int. J. Mol. Sci. 2020, 21, 9520. [Google Scholar] [CrossRef]
- Lee, C.; Lee, C.; Atakilit, A.; Siu, A.; Ramos, D.M. Differential spheroid formation by oral cancer cells. Anticancer Res. 2014, 34, 6945–6949. [Google Scholar] [PubMed]
- Singh, S.P.; Sharma, M.; Gupta, P.K. Evaluation of Phototoxic Effects of Curcumin Loaded in Organically Modified Silica Nanoparticles in Tumor Spheroids of Oral Cancer Cells. BioNanoScience 2015, 5, 10–21. [Google Scholar] [CrossRef]
- Laschke, M.W.; Menger, M.D. Life is 3D: Boosting Spheroid Function for Tissue Engineering. Trends Biotechnol. 2017, 35, 133–144. [Google Scholar] [CrossRef]
- Yakavets, I.; Jenard, S.; Francois, A.; Maklygina, Y.; Loschenov, V.; Lassalle, H.-P.; Dolivet, G.; Bezdetnaya, L. Stroma-Rich Co-Culture Multicellular Tumor Spheroids as a Tool for Photoactive Drugs Screening. J. Clin. Med. 2019, 8, 1686. [Google Scholar] [CrossRef]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Demirci, U.; Chen, P. Emerging organoid models: Leaping forward in cancer research. J. Hematol. Oncol. 2019, 12, 142. [Google Scholar] [CrossRef]
- Driehuis, E.; Kolders, S.; Spelier, S.; Lõhmussaar, K.; Willems, S.M.; Devriese, L.A.; de Bree, R.; de Ruiter, E.J.; Korving, J.; Begthel, H. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 2019, 9, 852–871. [Google Scholar] [CrossRef] [PubMed]
- Saglam-Metiner, P.; Gulce-Iz, S.; Biray-Avci, C. Bioengineering-inspired three-dimensional culture systems: Organoids to create tumor microenvironment. Gene 2019, 686, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Sitarski, A.M.; Fairfield, H.; Falank, C.; Reagan, M.R. 3D Tissue Engineered In Vitro Models of Cancer in Bone. ACS Biomater. Sci. Eng. 2018, 4, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Tayebi, L.; Moharamzadeh, K. Biomaterials for Oral and Dental Tissue Engineering; Woodhead Publishing: Sawston, UK, 2017. [Google Scholar]
- Colley, H.E.; Hearnden, V.; Jones, A.V.; Weinreb, P.H.; Violette, S.M.; MacNeil, S.; Thornhill, M.H.; Murdoch, C. Development of tissue-engineered models of oral dysplasia and early invasive oral squamous cell carcinoma. Br. J. Cancer 2011, 105, 1582–1592. [Google Scholar] [CrossRef]
- Costea, D.E.; Kulasekara, K.; Neppelberg, E.; Johannessen, A.C.; Vintermyr, O.K. Species-specific fibroblasts required for triggering invasiveness of partially transformed oral keratinocytes. Am. J. Pathol. 2006, 168, 1889–1897. [Google Scholar] [CrossRef]
- Gaballah, K.; Costea, D.; Hills, A.; Gollin, S.; Harrison, P.; Partridge, M. Tissue engineering of oral dysplasia. J. Pathol. 2008, 215, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Sawant, S.; Dongre, H.; Singh, A.K.; Joshi, S.; Costea, D.E.; Mahadik, S.; Ahire, C.; Makani, V.; Dange, P.; Sharma, S.; et al. Establishment of 3D Co-Culture Models from Different Stages of Human Tongue Tumorigenesis: Utility in Understanding Neoplastic Progression. PLoS ONE 2016, 11, e0160615. [Google Scholar] [CrossRef]
- Brancato, V.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Could 3D models of cancer enhance drug screening? Biomaterials 2020, 232, 119744. [Google Scholar] [CrossRef]
- Al-Samadi, A.; Poor, B.; Tuomainen, K.; Liu, V.; Hyytiäinen, A.; Suleymanova, I.; Mesimaki, K.; Wilkman, T.; Mäkitie, A.; Saavalainen, P.; et al. In vitro humanized 3D microfluidic chip for testing personalized immunotherapeutics for head and neck cancer patients. Exp. Cell Res. 2019, 383, 111508. [Google Scholar] [CrossRef]
- Bower, R.; Green, V.L.; Kuvshinova, E.; Kuvshinov, D.; Karsai, L.; Crank, S.T.; Stafford, N.D.; Greenman, J. Maintenance of head and neck tumor on-chip: Gateway to personalized treatment? Future Sci. OA 2017, 3, FSO174. [Google Scholar] [CrossRef] [PubMed]
- Duzagac, F.; Saorin, G.; Memeo, L.; Canzonieri, V.; Rizzolio, F. Microfluidic Organoids-on-a-Chip: Quantum Leap in Cancer Research. Cancers 2021, 13, 737. [Google Scholar] [CrossRef]
- Hattersley, S.M.; Sylvester, D.C.; Dyer, C.E.; Stafford, N.D.; Haswell, S.J.; Greenman, J. A microfluidic system for testing the responses of head and neck squamous cell carcinoma tissue biopsies to treatment with chemotherapy drugs. Ann. Biomed. Eng. 2012, 40, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Lugo-Cintrón, K.M.; Ayuso, J.M.; Humayun, M.; Gong, M.M.; Kerr, S.C.; Ponik, S.M.; Harari, P.M.; Virumbrales-Muñoz, M.; Beebe, D.J. Primary head and neck tumour-derived fibroblasts promote lymphangiogenesis in a lymphatic organotypic co-culture model. EBioMedicine 2021, 73, 103634. [Google Scholar] [CrossRef]
- Akgönüllü, S.; Bakhshpour, M.; Pişkin, A.K.; Denizli, A. Microfluidic Systems for Cancer Diagnosis and Applications. Micromachines 2021, 12, 1349. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, H. Microfluidic Technologies for Head and Neck Cancer: From Single-Cell Analysis to Tumor-on-a-Chip. In Early Detection and Treatment of Head & Neck Cancers: Practical Applications and Techniques for Detection, Diagnosis, and Treatment; Springer: Berlin/Heidelberg, Germany, 2021; pp. 43–62. [Google Scholar] [CrossRef]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef]
- Irani, S.; Barati, I.; Badiei, M. Periodontitis and oral cancer—Current concepts of the etiopathogenesis. Oncol. Rev. 2020, 14, 465. [Google Scholar] [CrossRef]
- Isola, G.; Giudice, A.L.; Polizzi, A.; Alibrandi, A.; Murabito, P.; Indelicato, F. Identification of the different salivary Interleukin-6 profiles in patients with periodontitis: A cross-sectional study. Arch. Oral Biol. 2021, 122, 104997. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Dewhirst, F.E.; Borisy, G.G. Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis. Annu. Rev. Microbiol. 2019, 73, 335–358. [Google Scholar] [CrossRef]
- Kim, J.Y.; An, C.H.; Kim, J.Y.; Jung, J.K. Experimental Animal Model Systems for Understanding Salivary Secretory Disorders. Int. J. Mol. Sci. 2020, 21, 8423. [Google Scholar] [CrossRef]
- Olek, M.; Machorowska-Pieniążek, A.; Olek, K.; Cieślar, G.; Kawczyk-Krupka, A. Photodynamic therapy in the treatment of oral squamous cell carcinoma—The state of the art in preclinical research on the animal model. Photodiagn. Photodyn. Ther. 2021, 34, 102236. [Google Scholar] [CrossRef] [PubMed]
- Toma, A.I.; Fuller, J.M.; Willett, N.J.; Goudy, S.L. Oral wound healing models and emerging regenerative therapies. Transl. Res. 2021, 236, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Johnston, W.; Delaney, C.; Rajendran, R.; Butcher, J.; Khan, S.; Bradshaw, D.; Ramage, G.; Culshaw, S. Biofilm-stimulated epithelium modulates the inflammatory responses in co-cultured immune cells. Sci. Rep. 2019, 9, 15779. [Google Scholar] [CrossRef] [PubMed]
- Dongari-Bagtzoglou, A. Pathogenesis of mucosal biofilm infections: Challenges and progress. Exp. Rev. Anti-Infect. Ther. 2008, 6, 201–208. [Google Scholar] [CrossRef]
- Valm, A.M. The Structure of Dental Plaque Microbial Communities in the Transition from Health to Dental Caries and Periodontal Disease. J. Mol. Biol. 2019, 431, 2957–2969. [Google Scholar] [CrossRef]
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40. [Google Scholar] [CrossRef]
- Ohnemus, U.; Willers, C.; Bubenheim, M.; Horstkotte, M.A.; Houdek, P.; Fischer, F.; Schmage, P.; Moll, I.; Brandner, J.M. An ex-vivo oral mucosa infection model for the evaluation of the topical activity of antifungal agents. Mycoses 2008, 51, 21–29. [Google Scholar] [CrossRef]
- Sacks, P.G. Cell, tissue and organ culture as in vitro models to study the biology of squamous cell carcinomas of the head and neck. Cancer Metastasis Rev. 1996, 15, 27–51. [Google Scholar] [CrossRef]
- Gibbs, S.; Roffel, S.; Meyer, M.; Gasser, A. Biology of soft tissue repair: Gingival epithelium in wound healing and attachment to the tooth and abutment surface. Eur. Cells Mater. 2019, 38, 63–78. [Google Scholar] [CrossRef]
- Moutsopoulos, N.M.; Konkel, J.E. Tissue-Specific Immunity at the Oral Mucosal Barrier. Trends Immunol. 2018, 39, 276–287. [Google Scholar] [CrossRef]
- Takahashi, N.; Sulijaya, B.; Yamada-Hara, M.; Tsuzuno, T.; Tabeta, K.; Yamazaki, K. Gingival epithelial barrier: Regulation by beneficial and harmful microbes. Tissue Barriers 2019, 7, e1651158. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.J.; Wilson, M.J.; Wei, X.; Lewis, M.A.O.; Bradshaw, D.J.; Murdoch, C.; Williams, D.W. Denture-associated biofilm infection in three-dimensional oral mucosal tissue models. J. Med. Microbiol. 2018, 67, 364–375. [Google Scholar] [CrossRef]
- Souza, J.G.S.; Bertolini, M.; Thompson, A.; Barão, V.A.R.; Dongari-Bagtzoglou, A. Biofilm Interactions of Candida albicans and Mitis Group Streptococci in a Titanium-Mucosal Interface Model. Appl. Environ. Microbiol. 2020, 86, e02950-19. [Google Scholar] [CrossRef]
- Beklen, A.; Torittu, A.; Ihalin, R.; Pöllänen, M. Aggregatibacter actinomycetemcomitans Biofilm Reduces Gingival Epithelial Cell Keratin Expression in an Organotypic Gingival Tissue Culture Model. Pathogens 2019, 8, 278. [Google Scholar] [CrossRef]
- Ingendoh-Tsakmakidis, A.; Mikolai, C.; Winkel, A.; Szafrański, S.P.; Falk, C.S.; Rossi, A.; Walles, H.; Stiesch, M. Commensal and pathogenic biofilms differently modulate peri-implant oral mucosa in an organotypic model. Cell. Microbiol. 2019, 21, e13078. [Google Scholar] [CrossRef] [PubMed]
- Bugueno, I.M.; Batool, F.; Keller, L.; Kuchler-Bopp, S.; Benkirane-Jessel, N.; Huck, O. Porphyromonas gingivalis bypasses epithelial barrier and modulates fibroblastic inflammatory response in an in vitro 3D spheroid model. Sci. Rep. 2018, 8, 14914. [Google Scholar] [CrossRef]
- Dongari-Bagtzoglou, A.; Kashleva, H. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat. Protoc. 2006, 1, 2012–2018. [Google Scholar] [CrossRef]
- Pinnock, A.; Murdoch, C.; Moharamzadeh, K.; Whawell, S.; Douglas, C.W. Characterisation and optimisation of organotypic oral mucosal models to study Porphyromonas gingivalis invasion. Microbes Infect. 2014, 16, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Iannelli, F.; Zotti, A.I.; Roca, M.S.; Grumetti, L.; Lombardi, R.; Moccia, T.; Vitagliano, C.; Milone, M.R.; Ciardiello, C.; Bruzzese, F.; et al. Valproic Acid Synergizes With Cisplatin and Cetuximab in vitro and in vivo in Head and Neck Cancer by Targeting the Mechanisms of Resistance. Front. Cell Dev. Biol. 2020, 8, 732. [Google Scholar] [CrossRef]
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019, 19, 16. [Google Scholar] [CrossRef]
- Van Vuuren, R.J.; Botes, M.; Jurgens, T.; Joubert, A.M.; van den Bout, I. Novel sulphamoylated 2-methoxy estradiol derivatives inhibit breast cancer migration by disrupting microtubule turnover and organization. Cancer Cell Int. 2019, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Azharuddin, M.; Roberg, K.; Dhara, A.K.; Jain, M.V.; Darcy, P.; Hinkula, J.; Slater, N.K.H.; Patra, H.K. Dissecting multi drug resistance in head and neck cancer cells using multicellular tumor spheroids. Sci. Rep. 2019, 9, 20066. [Google Scholar] [CrossRef] [PubMed]
- Perche, F.; Torchilin, V.P. Cancer cell spheroids as a model to evaluate chemotherapy protocols. Cancer Biol. Ther. 2012, 13, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.G.; da Rocha, G.O.; de Andrade, J.B. Occurrence of the potent mutagens 2-nitrobenzanthrone and 3-nitrobenzanthrone in fine airborne particles. Sci. Rep. 2019, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, J.; Jacobi, C.; Gstoettner, S.; Welz, C.; Schwenk-Zieger, S.; Stauber, R.; Strieth, S.; Kuenzel, J.; Baumeister, P.; Becker, S. Therapy testing in a spheroid-based 3D cell culture model for head and neck squamous cell carcinoma. J. Vis. Exp 2018, 134, e57012. [Google Scholar] [CrossRef]
- Hagemann, J.; Jacobi, C.; Hahn, M.; Schmid, V.; Welz, C.; Schwenk-Zieger, S.; Stauber, R.; Baumeister, P.; Becker, S. Spheroid-based 3D Cell Cultures Enable Personalized Therapy Testing and Drug Discovery in Head and Neck Cancer. Anticancer Res. 2017, 37, 2201–2210. [Google Scholar] [CrossRef]
- Magan, M.; Wiechec, E.; Roberg, K. CAFs affect the proliferation and treatment response of head and neck cancer spheroids during co-culturing in a unique in vitro model. Cancer Cell Int. 2020, 20, 599. [Google Scholar] [CrossRef]
- Lin, K.-C.; Ting, L.-L.; Chang, C.-L.; Lu, L.-S.; Lee, H.-L.; Hsu, F.-C.; Chiou, J.-F.; Wang, P.-Y.; Burnouf, T.; Ho, D.C.-Y.; et al. Ex Vivo Expanded Circulating Tumor Cells for Clinical Anti-Cancer Drug Prediction in Patients with Head and Neck Cancer. Cancers 2021, 13, 6076. [Google Scholar] [CrossRef]
- Tanaka, N.; Osman, A.A.; Takahashi, Y.; Lindemann, A.; Patel, A.A.; Zhao, M.; Takahashi, H.; Myers, J.N. Head and neck cancer organoids established by modification of the CTOS method can be used to predict in vivo drug sensitivity. Oral Oncol. 2018, 87, 49–57. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, L.; Yu, S.; Zheng, J.; Wang, H.; Zhang, Y. Decellularized tongue tissue as an in vitro model for studying tongue cancer and tongue regeneration. Acta Biomater. 2017, 58, 122–135. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Vitek, R.; Swick, A.D.; Skala, M.C.; Wisinski, K.B.; Kimple, R.J.; Lambert, P.F.; Beebe, D.J. Effects of culture method on response to EGFR therapy in head and neck squamous cell carcinoma cells. Sci. Rep. 2019, 9, 12480. [Google Scholar] [CrossRef]
- Tuomainen, K.; Al-Samadi, A.; Potdar, S.; Turunen, L.; Turunen, M.; Karhemo, P.-R.; Bergman, P.; Risteli, M.; Åström, P.; Tiikkaja, R.; et al. Human Tumor–Derived Matrix Improves the Predictability of Head and Neck Cancer Drug Testing. Cancers 2020, 12, 92. [Google Scholar] [CrossRef]
- Jin, D.; Ma, X.; Luo, Y.; Fang, S.; Xie, Z.; Li, X.; Qi, D.; Zhang, F.; Kong, J.; Li, J. Application of a microfluidic-based perivascular tumor model for testing drug sensitivity in head and neck cancers and toxicity in endothelium. RSC Adv. 2016, 6, 29598–29607. [Google Scholar] [CrossRef]
- Wahbi, W.; Korelin, K.; Sieviläinen, M.; Karihtala, P.; Wilkman, T.; Tarkkanen, J.; Salo, T.; Al-Samadi, A. Evaluation of in vitro and in vivo personalized cancer treatment assays for oral squamous cell carcinoma. Transl. Oncol. 2023, 33, 101677. [Google Scholar] [CrossRef]
- Sieviläinen, M.; Saavalainen, J.; Adnan-Awad, S.; Salo, T.; Al-Samadi, A. IDO1 Inhibition Reduces Immune Cell Exclusion Through Inducing Cell Migration While PD-1 Blockage Increases IL-6 and -8 Secretion From T Cells in Head and Neck Cancer. Front. Immunol. 2022, 13, 812822. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Micalet, A.; Moeendarbary, E.; Cheema, U. 3D In Vitro Models for Investigating the Role of Stiffness in Cancer Invasion. ACS Biomater. Sci. Eng. 2021, 9, 3729–3741. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Heimdal, J.; Aarstad, H.J.; Olofsson, J. Monocytes secrete interleukin-6 when co-cultured in vitro with benign or malignant autologous fragment spheroids from squamous cell carcinoma patients. Scand. J. Immunol. 2000, 51, 271–278. [Google Scholar] [CrossRef]
- Heimdal, J.H.; Olsnes, C.; Olofsson, J.; Aarstad, H.J. Monocyte and monocyte-derived macrophage secretion of MCP-1 in co-culture with autologous malignant and benign control fragment spheroids. Cancer Immunol. Immunother. 2001, 50, 300–306. [Google Scholar] [CrossRef]
- Kross, K.W.; Heimdal, J.H.; Aarstad, H.J. Mononuclear phagocytes in head and neck squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2010, 267, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Kross, K.W.; Heimdal, J.H.; Olsnes, C.; Olofsson, J.; Aarstad, H.J. Co-culture of head and neck squamous cell carcinoma spheroids with autologous monocytes predicts prognosis. Scand. J. Immunol. 2008, 67, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.K.; Schirlau, K.; Sonkoly, E.; Brandau, S.; Lang, S.; Pivarcsi, A.; Balz, V.; Müller, A.; Homey, B.; Boelke, E.; et al. A novel mechanism for anti-EGFR antibody action involves chemokine-mediated leukocyte infiltration. Int. J. Cancer 2009, 124, 2589–2596. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Leidig, T.; Rodday, B.; Lindemann, C.; Mueller-Klieser, W. Test System for Trifunctional Antibodies in 3D MCTS Culture. J. Biomol. Screen. 2009, 14, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Q.; Kuo, J.-C.; Wei, M.-T.; Wu, M.-C.; Yang, M.-H.; Chiou, A. Fibroblast promotes head and neck squamous cell carcinoma cell invasion through mechanical barriers in 3D collagen microenvironments. ACS Appl. Bio Mater. 2020, 3, 6419–6429. [Google Scholar] [CrossRef]
- Gomez, K.E.; Wu, F.; Keysar, S.B.; Morton, J.J.; Miller, B.; Chimed, T.S.; Le, P.N.; Nieto, C.; Chowdhury, F.N.; Tyagi, A.; et al. Cancer Cell CD44 Mediates Macrophage/Monocyte-Driven Regulation of Head and Neck Cancer Stem Cells. Cancer Res. 2020, 80, 4185–4198. [Google Scholar] [CrossRef]
- Huang, C.; Yoon, C.; Zhou, X.H.; Zhou, Y.C.; Zhou, W.W.; Liu, H.; Yang, X.; Lu, J.; Lee, S.Y.; Huang, K. ERK1/2-Nanog signaling pathway enhances CD44+ cancer stem-like cell phenotypes and epithelial-to-mesenchymal transition in head and neck squamous cell carcinomas. Cell Death Dis. 2020, 11, 266. [Google Scholar] [CrossRef]
- Engelmann, L.; Thierauf, J.; Laureano, N.K.; Stark, H.-J.; Prigge, E.-S.; Horn, D.; Freier, K.; Grabe, N.; Rong, C.; Federspil, P. Organotypic co-cultures as a novel 3D model for head and neck squamous cell carcinoma. Cancers 2020, 12, 2330. [Google Scholar] [CrossRef]
- Kochanek, S.J.; Close, D.A.; Johnston, P.A. High Content Screening Characterization of Head and Neck Squamous Cell Carcinoma Multicellular Tumor Spheroid Cultures Generated in 384-Well Ultra-Low Attachment Plates to Screen for Better Cancer Drug Leads. Assay Drug Dev. Technol. 2019, 17, 17–36. [Google Scholar] [CrossRef]
- Braunholz, D.; Saki, M.; Niehr, F.; Öztürk, M.; Puértolas, B.B.; Konschak, R.; Budach, V.; Tinhofer, I. Spheroid Culture of Head and Neck Cancer Cells Reveals an Important Role of EGFR Signalling in Anchorage Independent Survival. PLoS ONE 2016, 11, e0163149. [Google Scholar] [CrossRef]
- He, C.K.; Chen, Y.W.; Wang, S.H.; Hsu, C.H. Hydrodynamic shuttling for deterministic high-efficiency multiple single-cell capture in a microfluidic chip. Lab Chip 2019, 19, 1370–1377. [Google Scholar] [CrossRef]
- Byun, J.-Y.; Huang, K.; Lee, J.S.; Huang, W.; Hu, L.; Zhen, X.; Tang, X.; Li, F.; Jo, D.-G.; Song, X.; et al. Targeting HIF-1α/NOTCH1 pathway eliminates CD44+ cancer stem-like cell phenotypes, malignancy, and resistance to therapy in head and neck squamous cell carcinoma. Oncogene 2022, 41, 1352–1363. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, R.; Chen, Y.; Yang, X.; Shang, Z. Stromal nicotinamide N-methyltransferase orchestrates the crosstalk between fibroblasts and tumour cells in oral squamous cell carcinoma: Evidence from patient-derived assembled organoids. Oncogene 2023, 42, 1166–1180. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-R.; Brown, J.; Yu, Y.; Lee, D.; Zhou, K.; Dunn, Z.S.; Hon, R.; Wilson, M.; Kramer, A.; Zhu, Y.; et al. Targeting Immunosuppressive Tumor-Associated Macrophages Using Innate T Cells for Enhanced Antitumor Reactivity. Cancers 2022, 14, 2749. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef] [PubMed]
- Doedens, A.L.; Stockmann, C.; Rubinstein, M.P.; Liao, D.; Zhang, N.; DeNardo, D.G.; Coussens, L.M.; Karin, M.; Goldrath, A.W.; Johnson, R.S. Macrophage expression of hypoxia-inducible factor-1α suppresses T-cell function and promotes tumor progression. Cancer Res. 2010, 70, 7465–7475. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Song, B.; Kim, Y.-S.; Kim, E.-K.; Lee, J.-M.; Lee, G.-E.; Kim, J.-O.; Kim, Y.-J.; Chang, W.-S.; Kang, C.-Y. Tumor Microenvironmental Conversion of Natural Killer Cells into Myeloid-Derived Suppressor CellsNK Cell Conversion into MDSCs. Cancer Res. 2013, 73, 5669–5681. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Gutschalk, C.M.; Herold-Mende, C.C.; Fusenig, N.E.; Mueller, M.M. Granulocyte Colony-Stimulating Factor and Granulocyte-Macrophage Colony-Stimulating Factor Promote Malignant Growth of Cells from Head and Neck Squamous Cell Carcinomas In vivo. Cancer Res. 2006, 66, 8026–8036. [Google Scholar] [CrossRef]
- Linde, N.; Gutschalk, C.M.; Hoffmann, C.; Yilmaz, D.; Mueller, M.M. Integrating macrophages into organotypic co-cultures: A 3D in vitro model to study tumor-associated macrophages. PLoS ONE 2012, 7, e40058. [Google Scholar] [CrossRef]
- Lu, Y.; Li, S.; Ma, L.; Li, Y.; Zhang, X.; Peng, Q.; Mo, C.; Huang, L.; Qin, X.; Liu, Y. Type conversion of secretomes in a 3D TAM2 and HCC cell co-culture system and functional importance of CXCL2 in HCC. Sci. Rep. 2016, 6, 24558. [Google Scholar] [CrossRef]
- Chávez-Galán, L.; Olleros, M.L.; Vesin, D.; Garcia, I. Much More than M1 and M2 Macrophages, There are also CD169+ and TCR+ Macrophages. Front. Immunol. 2015, 6, 263. [Google Scholar] [CrossRef]
- Laoui, D.; Van Overmeire, E.; Movahedi, K.; Van den Bossche, J.; Schouppe, E.; Mommer, C.; Nikolaou, A.; Morias, Y.; De Baetselier, P.; Van Ginderachter, J.A. Mononuclear phagocyte heterogeneity in cancer: Different subsets and activation states reaching out at the tumor site. Immunobiology 2011, 216, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Kulawiec, D.; Jazwinska, D.; Zervantonakis, I. Paracrine CSF1 signaling regulates macrophage migration dynamics towards ovarian cancer cells in a 3D microfluidic model that recapitulates in vivo infiltration patterns in patient-derived xenograft models. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lewis, C.E.; Pollard, J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006, 66, 605–612. [Google Scholar] [CrossRef]
- Wyckoff, J.B.; Wang, Y.; Lin, E.Y.; Li, J.-F.; Goswami, S.; Stanley, E.R.; Segall, J.E.; Pollard, J.W.; Condeelis, J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007, 67, 2649–2656. [Google Scholar] [CrossRef]
- Heldin, C.H.; Rubin, K.; Pietras, K.; Ostman, A. High interstitial fluid pressure—An obstacle in cancer therapy. Nat. Rev. Cancer 2004, 4, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Hompland, T.; Ellingsen, C.; Øvrebø, K.M.; Rofstad, E.K. Interstitial fluid pressure and associated lymph node metastasis revealed in tumors by dynamic contrast-enhanced MRI. Cancer Res. 2012, 72, 4899–4908. [Google Scholar] [CrossRef] [PubMed]
- Penny, H.L.; Sieow, J.L.; Adriani, G.; Yeap, W.H.; Ee, P.S.C.; Luis, B.S.; Lee, B.; Lee, T.; Mak, S.Y.; Ho, Y.S. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 2016, 5, e1191731. [Google Scholar] [CrossRef] [PubMed]
- Battat, S.; Weitz, D.A.; Whitesides, G.M. An outlook on microfluidics: The promise and the challenge. Lab Chip 2022, 22, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.L.; Seager, R.J.; Litvak, F.; Spill, F.; Sieow, J.L.; Leong, P.H.; Kumar, D.; Tan, A.S.M.; Wong, S.C.; Adriani, G.; et al. Integrated in silico and 3D in vitro model of macrophage migration in response to physical and chemical factors in the tumor microenvironment. Integr. Biol. 2020, 12, 90–108. [Google Scholar] [CrossRef]

| Type | Secretory Cell | Markers | References |
|---|---|---|---|
| Immune Cell | M1 TAMs | CD11c, CD80, HLA-DR 1 | [41,42,43] |
| M2 TAMs | CD163, CD11b, CD206, MRC1 2 | [43,44,45,46] | |
| DC | S100, CD1a, CD83, CD207, CD208, CD80, CD11c, CD86, HLA-DR CLEC9A 3 | [47,48,49] | |
| NK cells | CD57 | [50,51,52] | |
| pan T cell | CD3 | [12,53,54] | |
| cytotoxic T cell | CD8 | [12,55,56] | |
| T helper cell | CD4 | [12,57,58] | |
| Pan B cell | CD19, CD20 | [12,59,60] | |
| MDSCs | CD33, CD11b | [61,62,63] | |
| CAFs | α SMA 4 | [64,65,66] | |
| Treg | FOXP3+ CD4+ T cells or CD4+ CD25+ CD127low | [39,67,68] | |
| Non-immune cell | endothelial cells | CD34 | [69,70,71] |
| Salivary biomarkers | L-phenylalanine Sphinganine Phytosphingosine S-carboxymethyl-L-cysteine | [72,73,74] | |
| Genomic biomarkers | ITGA3 5, ITGB4 expression | [75,76,77] | |
| Oral cancer cell | CCR7 6 | [78,79,80] | |
| Oral cancer cell | MYO1B 7 | [81,82,83] |
| Stromal Cells | Mechanism | Function | References |
|---|---|---|---|
| CAFs | Production of numerous ECM proteins such as HAS2 1 expression by CAFs | Tumor cell invasion by increasing ECM-degrading MMPs 2 and decreasing TIMPs 3 | [95,96] |
| high expression of α-SMA in CAFs | OSCC invasion into the bone by increasing expression of RANKL 4 and OPG 5 | [97,98,99,100] | |
| High secretion of IL-1α by OSCC upregulated expression of secretory cytokines, including CCL7 6, CXCL1 7, and IL-8 | Tumor cell proliferation | [101,102,103] | |
| IGF-1 overexpression in CAF and activation of PI3K-AKT and Hedgehog signaling pathways | Tumor cell proliferation, migration invasion, tumorsphere formation angiogenesis. | [104,105,106] | |
| Overexpression of NOTCH-1 in CAFs | Increasing tumor volume angiogenesis in OSCC | [107,108,109] | |
| Overexpression of IL-6 in CAFs | Expression of VGEF in CAFs and OSCC angiogenesis in OSCC | [110,111,112] | |
| Multiple factors derived from CAFs, such as CXCL12 and MCP-1 attract macrophages to tumors and induce the M2 phenotype | A mediator for T-cell suppression | [113,114] | |
| IL-1α secreted from OSCC cells induces the chemokine CCL7 in co-cultured CAF | OSCC invasion and progression | [115,116] | |
| TAMs | increased expression of arginase I, IL-10 and TGF-β | Suppressive effect on T cells and invasion and metastasis of OSCC | [117,118] |
| PDL-1 and IL-10 production in TAMs | Immune escape of OSCC cells | [119,120] | |
| the secretion of EGFs 8 and the management of collagen production by TAMs | OSCC invasion and progression | [121,122] | |
| EMT 9 induced by TAMs decreased E-mucin and E-cadherin and increased-vimentin protein in OSCC cells | OSCC invasion and progression | [123,124,125] | |
| activated the Hh 10 signaling pathway by TAMs | Angiogenesis in OSCC | [126,127] | |
| Activation of TGF-β1/TβRII/Smad3 signaling pathway in TAMs | VEGF secretions in OSCC | [128,129] | |
| TAM number modulation by PFKFB3 11 | Angiogenesis in OSCC | [130,131] | |
| DCs | activated the TNF-α/NF-κB/CXCR-4 pathway by pDCs 12 | Oral cancer proliferation and invasion | [132,133] |
| 2D Model | 3D Model | Animal Model | |
|---|---|---|---|
| Modeling human development and disease | - | + | + |
| High costs, high personal, and work effort | - | - | + |
| High-throughput screening | + | + | - |
| Personalized medicine | - | + | - |
| Vascularization and immune system | - | - | + |
| Architecture | - | + | + |
| 3D Models | Drug | Application | Reference |
|---|---|---|---|
| Spheroid | Cetuximab Cisplatin | To assess the impact of both 2D and 3D culture techniques on gene expression related to cell junctions, cell adhesions, cell cycle, and metabolism. To verify the feasibility and practicality of this novel 3D culture approach for oral cancer research. | [147,202,203,204] |
| Cisplatin Doxorubicin Methotrexate | To evaluate the differences in chemoresistance between 2D and 3D culture methods. | [205,206,207] | |
| Cisplatin 5-FU 2-Gy Radiation | To assess and compare the efficacy of 2D and 3D methods as platforms for chemotherapy and radiotherapy testing. | [208,209] | |
| Cetuximab | To develop a biologically significant in vitro model of HNSCC that accurately replicates the tumor environment by incorporating both tumor cells and CAFs in a 3D culture system. | [210] | |
| Organotypic models | Cisplatin Docetaxel ± 5-FU | To evaluate and compare the suitability of 2D and 3D methods as platforms for assessing chemotherapy sensitivity. | [151,211,212] |
| Cisplatin ± (carboplatin, cetuximab, radiotherapy) | To assess and compare the effectiveness of 2D and 3D methods as platforms for chemotherapy screening and regenerative purposes. | [163,213] | |
| Cetuximab mTOR inhibitor Canertinib Dactolisib PF-04691502 Apitolisib Omipalisib Refametinib binimetinib trametinib pimasertib trametinib | To evaluate and compare the efficacy of 2D and 3D methods as platforms for dual drug screening. | [214,215] | |
| Microfluidic platforms | 5-FU Cisplatin ± (Paclitaxel, Cetuximab, Carboplatin) | The use of a dynamic culture method as a platform for chemotherapy screening. | [175,216,217] |
| IDO1 inhibitor ± (PDL1 antibody, Nivolumab) | [172,218] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalir Abdolahinia, E.; Han, X. The Three-Dimensional In Vitro Cell Culture Models in the Study of Oral Cancer Immune Microenvironment. Cancers 2023, 15, 4266. https://doi.org/10.3390/cancers15174266
Dalir Abdolahinia E, Han X. The Three-Dimensional In Vitro Cell Culture Models in the Study of Oral Cancer Immune Microenvironment. Cancers. 2023; 15(17):4266. https://doi.org/10.3390/cancers15174266
Chicago/Turabian StyleDalir Abdolahinia, Elaheh, and Xiaozhe Han. 2023. "The Three-Dimensional In Vitro Cell Culture Models in the Study of Oral Cancer Immune Microenvironment" Cancers 15, no. 17: 4266. https://doi.org/10.3390/cancers15174266






