Analysis of Artificial Intelligence-Based Approaches Applied to Non-Invasive Imaging for Early Detection of Melanoma: A Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Study Eligibility and Selection
2.3. Study Analysis and Performance Metrics
3. Results
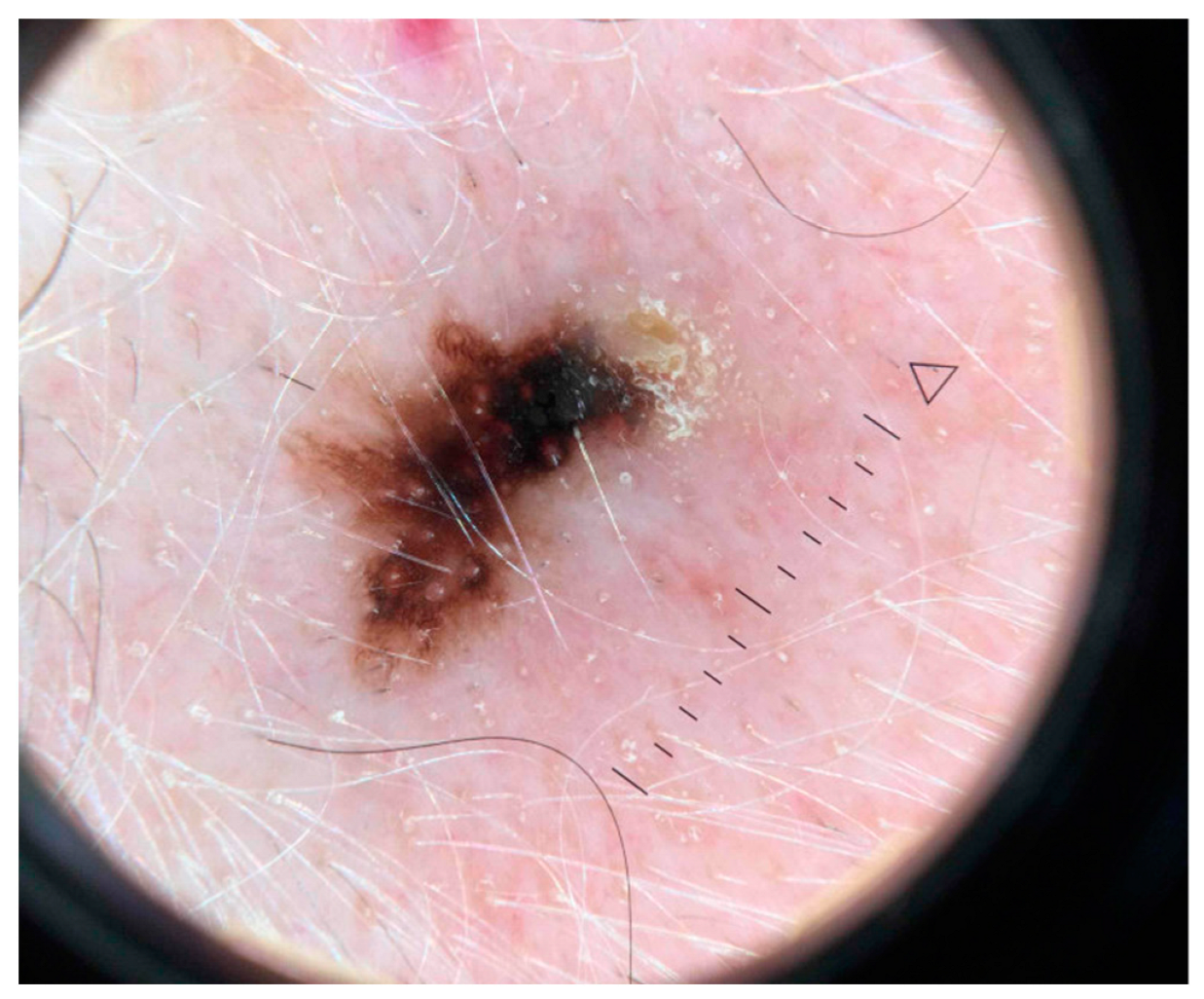
3.1. Artificial Intelligence-Based Approaches Applied to Dermoscopic Images
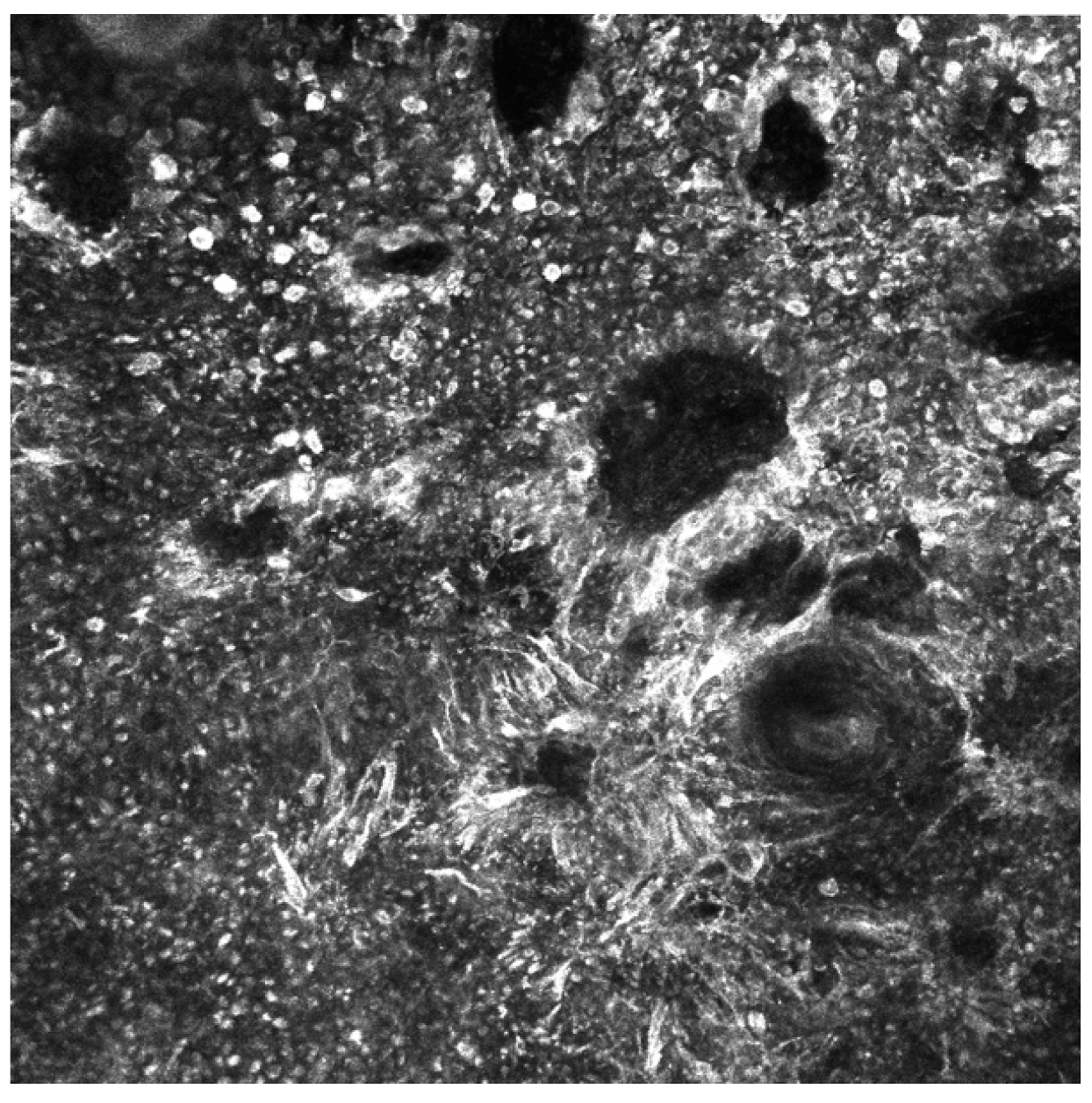
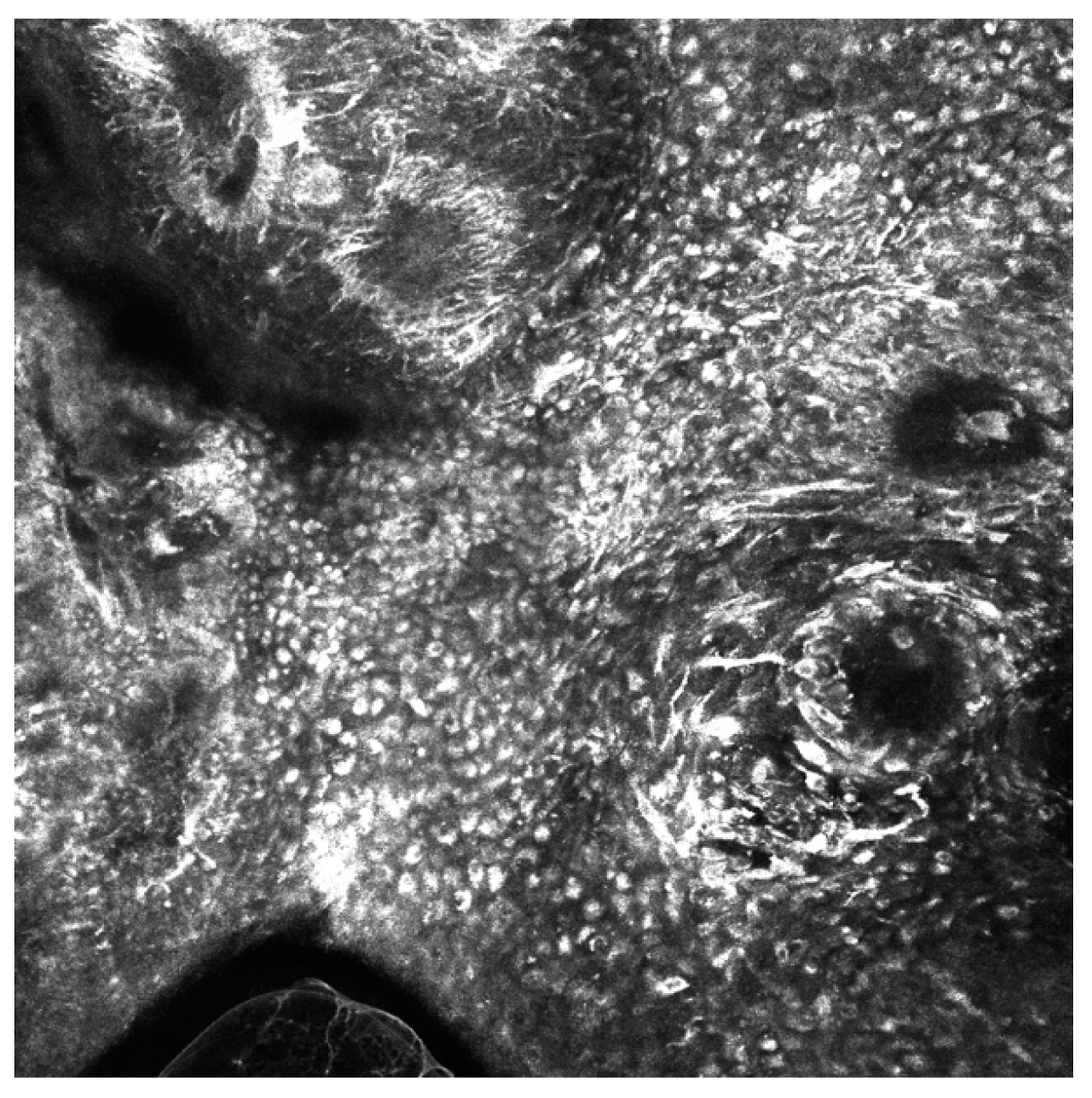
3.2. Artificial Intelligence-Based Approaches to Analysis of Reflectance Confocal Microscopy (RCM) Images
3.3. Artificial Intelligence-Based Approaches Applied to Optical Coherence Tomography (OCT) Images
4. Discussion
4.1. Clinical Utility and Perceptions of AI in Dermatology
4.2. Ethical Implications
4.3. Limitations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Johansson, M.; Brodersen, J.; Gøtzsche, P.C.; Jørgensen, K.J. Screening for reducing morbidity and mortality in malignant melanoma. Cochrane Database Syst. Rev. 2019, 6, CD012352. [Google Scholar] [CrossRef]
- Franceschini, C.; Persechino, F.; Ardigò, M. In Vivo Reflectance Confocal Microscopy in General Dermatology: How to Choose the Right Indication. Dermatol. Pract. Concept. 2020, 10, e2020032. [Google Scholar] [CrossRef]
- di Ruffano, L.F.; Dinnes, J.; Deeks, J.J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Takwoingi, Y.; Godfrey, K.; O’Sullivan, C.; Matin, R.N.; et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 12, CD013189. [Google Scholar] [CrossRef]
- Sonthalia, S.; Yumeen, S.; Kaliyadan, F. Dermoscopy Overview and Extradiagnostic Applications. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK537131/ (accessed on 8 August 2022).
- Maron, R.C.; Utikal, J.S.; Hekler, A.; Hauschild, A.; Sattler, E.; Sondermann, W.; Haferkamp, S.; Schilling, B.; Heppt, M.V.; Jansen, P.; et al. Artificial Intelligence and Its Effect on Dermatologists’ Accuracy in Dermoscopic Melanoma Image Classification: Web-Based Survey Study. J. Med. Internet Res. 2020, 22, e18091. [Google Scholar] [CrossRef]
- Ahmad, Z.; Rahim, S.; Zubair, M.; Abdul-Ghafar, J. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: Present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn. Pathol. 2021, 16, 24. [Google Scholar] [CrossRef]
- Jayakumar, S.; Sounderajah, V.; Normahani, P.; Harling, L.; Markar, S.R.; Ashrafian, H.; Darzi, A. Quality assessment standards in artificial intelligence diagnostic accuracy systematic reviews: A meta-research study. npj Digit. Med. 2022, 5, 11. [Google Scholar] [CrossRef]
- Rezk, E.; Eltorki, M.; El-Dakhakhni, W. Leveraging Artificial Intelligence to Improve the Diversity of Dermatological Skin Color Pathology: Protocol for an Algorithm Development and Validation Study. JMIR Res. Protoc. 2022, 11, e34896. [Google Scholar] [CrossRef]
- Daneshjou, R.; Vodrahalli, K.; Novoa, R.A.; Jenkins, M.; Liang, W.; Rotemberg, V.; Ko, J.; Swetter, S.M.; Bailey, E.E.; Gevaert, O.; et al. Disparities in dermatology AI performance on a diverse, curated clinical image set. Sci. Adv. 2022, 8, eabq6147. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Damaševičius, R.; Eltoukhy, M.M. Machine Learning and Deep Learning Methods for Skin Lesion Classification and Diagnosis: A Systematic Review. Diagnostics 2021, 11, 1390. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Gouabou, A.C.F.; Collenne, J.; Monnier, J.; Iguernaissi, R.; Damoiseaux, J.-L.; Moudafi, A.; Merad, D. Computer Aided Diagnosis of Melanoma Using Deep Neural Networks and Game Theory: Application on Dermoscopic Images of Skin Lesions. Int. J. Mol. Sci. 2022, 23, 13838. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Codella, N.C.; Dusza, S.W.; Gutman, D.A.; Helba, B.; Kalloo, A.; Mishra, N.; Carrera, C.; Celebi, M.E.; DeFazio, J.L.; et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol. 2018, 78, 270–277.e1. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Liopyris, K.; Dusza, S.W.; Codella, N.C.; Gutman, D.A.; Helba, B.; Kalloo, A.; Halpern, A.C.; Soyer, H.P.; Curiel-Lewandrowski, C.; et al. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J. Am. Acad. Dermatol. 2020, 82, 622–627. [Google Scholar] [CrossRef]
- Xia, M.; Kheterpal, M.K.; Wong, S.C.; Park, C.; Ratliff, W.; Carin, L.; Henao, R. Lesion identification and malignancy prediction from clinical dermatological images. Sci. Rep. 2022, 12, 15836. [Google Scholar] [CrossRef]
- Xin, C.; Liu, Z.; Zhao, K.; Miao, L.; Ma, Y.; Zhu, X.; Zhou, Q.; Wang, S.; Li, L.; Yang, F.; et al. An improved transformer network for skin cancer classification. Comput. Biol. Med. 2022, 149, 105939. [Google Scholar] [CrossRef]
- Singh, S.K.; Abolghasemi, V.; Anisi, M.H. Skin Cancer Diagnosis Based on Neutrosophic Features with a Deep Neural Network. Sensors 2022, 22, 6261. [Google Scholar] [CrossRef]
- Naeem, A.; Anees, T.; Fiza, M.; Naqvi, R.A.; Lee, S.-W. SCDNet: A Deep Learning-Based Framework for the Multiclassification of Skin Cancer Using Dermoscopy Images. Sensors 2022, 22, 5652. [Google Scholar] [CrossRef]
- Lee, J.R.H.; Pavlova, M.; Famouri, M.; Wong, A. Cancer-Net SCa: Tailored deep neural network designs for detection of skin cancer from dermoscopy images. BMC Med. Imaging 2022, 22, 143. [Google Scholar] [CrossRef]
- Fraiwan, M.; Faouri, E. On the Automatic Detection and Classification of Skin Cancer Using Deep Transfer Learning. Sensors 2022, 22, 4963. [Google Scholar] [CrossRef]
- Vaiyapuri, T.; Balaji, P.; Alaskar, H.; Sbai, Z. Computational Intelligence-Based Melanoma Detection and Classification Using Dermoscopic Images. Comput. Intell. Neurosci. 2022, 2022, 2370190. [Google Scholar] [CrossRef]
- Martin-Gonzalez, M.; Azcarraga, C.; Martin-Gil, A.; Carpena-Torres, C.; Jaen, P. Efficacy of a Deep Learning Convolutional Neural Network System for Melanoma Diagnosis in a Hospital Population. Int. J. Environ. Res. Public Health 2022, 19, 3892. [Google Scholar] [CrossRef]
- Lu, X.; Zadeh, Y.A.F.A. Deep Learning-Based Classification for Melanoma Detection Using XceptionNet. J. Health Eng. 2022, 2022, 2196096. [Google Scholar] [CrossRef]
- Kaur, R.; GholamHosseini, H.; Sinha, R.; Lindén, M. Melanoma Classification Using a Novel Deep Convolutional Neural Network with Dermoscopic Images. Sensors 2022, 22, 1134. [Google Scholar] [CrossRef]
- Arshad, M.; Khan, M.A.; Tariq, U.; Armghan, A.; Alenezi, F.; Javed, M.Y.; Aslam, S.M.; Kadry, S. A Computer-Aided Diagnosis System Using Deep Learning for Multiclass Skin Lesion Classification. Comput. Intell. Neurosci. 2021, 2021, 9619079. [Google Scholar] [CrossRef]
- Xing, X.; Song, P.; Zhang, K.; Yang, F.; Dong, Y. ZooME: Efficient Melanoma Detection Using Zoom-in Attention and Metadata Embedding Deep Neural Network. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 4041–4044. [Google Scholar] [CrossRef]
- Pham, T.-C.; Luong, C.-M.; Hoang, V.-D.; Doucet, A. AI outperformed every dermatologist in dermoscopic melanoma diagnosis, using an optimized deep-CNN architecture with custom mini-batch logic and loss function. Sci. Rep. 2021, 11, 17485. [Google Scholar] [CrossRef]
- Kim, C.-I.; Hwang, S.-M.; Park, E.-B.; Won, C.-H.; Lee, J.-H. Computer-Aided Diagnosis Algorithm for Classification of Malignant Melanoma Using Deep Neural Networks. Sensors 2021, 21, 5551. [Google Scholar] [CrossRef]
- Nawaz, M.; Mehmood, Z.; Nazir, T.; Naqvi, R.A.; Rehman, A.; Iqbal, M.; Saba, T. Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering. Microsc. Res. Tech. 2022, 85, 339–351. [Google Scholar] [CrossRef]
- Sayed, G.I.; Soliman, M.M.; Hassanien, A.E. A novel melanoma prediction model for imbalanced data using optimized SqueezeNet by bald eagle search optimization. Comput. Biol. Med. 2021, 136, 104712. [Google Scholar] [CrossRef]
- Gouabou, A.C.F.; Damoiseaux, J.-L.; Monnier, J.; Iguernaissi, R.; Moudafi, A.; Merad, D. Ensemble Method of Convolutional Neural Networks with Directed Acyclic Graph Using Dermoscopic Images: Melanoma Detection Application. Sensors 2021, 21, 3999. [Google Scholar] [CrossRef]
- Alsaade, F.W.; Aldhyani, T.H.H.; Al-Adhaileh, M.H. Developing a Recognition System for Diagnosing Melanoma Skin Lesions Using Artificial Intelligence Algorithms. Comput. Math. Methods Med. 2021, 2021, 9998379. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, I.; Younus, M.; Walayat, K.; Kakar, M.U.; Ma, J. Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput. Med. Imaging Graph. 2021, 88, 101843. [Google Scholar] [CrossRef]
- Jojoa Acosta, M.F.; Caballero Tovar, L.Y.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef]
- Tognetti, L.; Bonechi, S.; Andreini, P.; Bianchini, M.; Scarselli, F.; Cevenini, G.; Moscarella, E.; Farnetani, F.; Longo, C.; Lallas, A.; et al. A new deep learning approach integrated with clinical data for the dermoscopic differentiation of early melanomas from atypical nevi. J. Dermatol. Sci. 2021, 101, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Gareau, D.S.; Browning, J.; Da Rosa, J.C.; Suarez-Farinas, M.; Lish, S.; Zong, A.M.; Firester, B.; Vrattos, C.; Renert-Yuval, Y.; Gamboa, M.; et al. Deep learning-level melanoma detection by interpretable machine learning and imaging biomarker cues. J. Biomed. Opt. 2020, 25, 112906. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xie, G.; Xu, X.; Ren, J. Effective Melanoma Recognition Using Deep Convolutional Neural Network with Covariance Discriminant Loss. Sensors 2020, 20, 5786. [Google Scholar] [CrossRef]
- Kaur, R.; GholamHosseini, H.; Sinha, R. Deep Convolutional Neural Network for Melanoma Detection using Dermoscopy Images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2020, 2020, 1524–1527. [Google Scholar] [CrossRef]
- Minagawa, A.; Koga, H.; Sano, T.; Matsunaga, K.; Teshima, Y.; Hamada, A.; Houjou, Y.; Okuyama, R. Dermoscopic diagnostic performance of Japanese dermatologists for skin tumors differs by patient origin: A deep learning convolutional neural network closes the gap. J. Dermatol. 2021, 48, 232–236. [Google Scholar] [CrossRef]
- Nasiri, S.; Helsper, J.; Jung, M.; Fathi, M. DePicT Melanoma Deep-CLASS: A deep convolutional neural networks approach to classify skin lesion images. BMC Bioinform. 2020, 21, 84. [Google Scholar] [CrossRef]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; et al. Melanoma recognition by a deep learning convolutional neural network—Performance in different melanoma subtypes and localisations. Eur. J. Cancer 2020, 127, 21–29. [Google Scholar] [CrossRef]
- Phillips, M.; Greenhalgh, J.; Marsden, H.; Palamaras, I. Detection of Malignant Melanoma Using Artificial Intelligence: An Observational Study of Diagnostic Accuracy. Dermatol. Pract. Concept. 2019, 10, e2020011. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 2019, 119, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Hagerty, J.R.; Stanley, R.J.; Almubarak, H.A.; Lama, N.; Kasmi, R.; Guo, P.; Drugge, R.J.; Rabinovitz, H.S.; Oliviero, M.; Stoecker, W.V. Deep Learning and Handcrafted Method Fusion: Higher Diagnostic Accuracy for Melanoma Dermoscopy Images. IEEE J. Biomed. Health Inform. 2019, 23, 1385–1391. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Li, Y.; Shen, L. Skin Lesion Analysis towards Melanoma Detection Using Deep Learning Network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef]
- Silver, F.H.; Mesica, A.; Gonzalez-Mercedes, M.; Deshmukh, T. Identification of Cancerous Skin Lesions Using Vibrational Optical Coherence Tomography (VOCT): Use of VOCT in Conjunction with Machine Learning to Diagnose Skin Cancer Remotely Using Telemedicine. Cancers 2022, 15, 156. [Google Scholar] [CrossRef]
- Wodzinski, M.; Skalski, A.; Witkowski, A.; Pellacani, G.; Ludzik, J. Convolutional Neural Network Approach to Classify Skin Lesions Using Reflectance Confocal Microscopy. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 4754–4757. [Google Scholar] [CrossRef]
- D’alonzo, M.; Bozkurt, A.; Alessi-Fox, C.; Gill, M.; Brooks, D.H.; Rajadhyaksha, M.; Kose, K.; Dy, J.G. Semantic segmentation of reflectance confocal microscopy mosaics of pigmented lesions using weak labels. Sci. Rep. 2021, 11, 3679. [Google Scholar] [CrossRef]
- Phillips, M.; Marsden, H.; Jaffe, W.; Matin, R.N.; Wali, G.N.; Greenhalgh, J.; McGrath, E.; James, R.; Ladoyanni, E.; Bewley, A.; et al. Assessment of Accuracy of an Artificial Intelligence Algorithm to Detect Melanoma in Images of Skin Lesions. JAMA Netw. Open 2019, 2, e1913436. [Google Scholar] [CrossRef]
- Levine, A.; Markowitz, O. Introduction to reflectance confocal microscopy and its use in clinical practice. JAAD Case Rep. 2018, 4, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- González, S.; Gilaberte-Calzada, Y. In vivo reflectance-mode confocal microscopy in clinical dermatology and cosmetology. Int. J. Cosmet. Sci. 2008, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Narayanamurthy, V.; Padmapriya, P.; Noorasafrin, A.; Pooja, B.; Hema, K.; Khan, A.Y.F.; Nithyakalyani, K.; Samsuri, F. Skin cancer detection using non-invasive techniques. RSC Adv. 2018, 8, 28095–28130. [Google Scholar] [CrossRef] [PubMed]
- Sattler, E.; Kästle, R.; Welzel, J. Optical coherence tomography in dermatology. J. Biomed. Opt. 2013, 18, 061224. [Google Scholar] [CrossRef]
- Mogensen, M.; Thrane, L.; Jørgensen, T.M.; Andersen, P.E.; Jemec, G.B.E. OCT imaging of skin cancer and other dermatological diseases. J. Biophotonics 2009, 2, 442–451. [Google Scholar] [CrossRef]
- Fujimoto, J.G.; Pitris, C.; Boppart, S.A.; Brezinski, M.E. Optical Coherence Tomography: An Emerging Technology for Biomedical Imaging and Optical Biopsy. Neoplasia 2000, 2, 9–25. [Google Scholar] [CrossRef]
- Wan, B.; Ganier, C.; Du-Harpur, X.; Harun, N.; Watt, F.; Patalay, R.; Lynch, M.D. Applications and future directions for optical coherence tomography in dermatology. Br. J. Dermatol. 2021, 184, 1014–1022. [Google Scholar] [CrossRef]
- Xiong, Y.-Q.; Mo, Y.; Wen, Y.-Q.; Cheng, M.-J.; Huo, S.-T.; Chen, X.-J.; Chen, Q. Optical coherence tomography for the diagnosis of malignant skin tumors: A meta-analysis. J. Biomed. Opt. 2018, 23, 1–10. [Google Scholar] [CrossRef]
- Schuh, S.; Holmes, J.; Ulrich, M.; Themstrup, L.; Jemec, G.B.E.; De Carvalho, N.; Pellacani, G.; Welzel, J. Imaging Blood Vessel Morphology in Skin: Dynamic Optical Coherence Tomography as a Novel Potential Diagnostic Tool in Dermatology. Dermatol. Ther. 2017, 7, 187–202. [Google Scholar] [CrossRef]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef]
- Nelson, C.A.; Pérez-Chada, L.M.; Creadore, A.; Li, S.J.; Lo, K.; Manjaly, P.; Pournamdari, A.B.; Tkachenko, E.; Barbieri, J.S.; Ko, J.M.; et al. Patient Perspectives on the Use of Artificial Intelligence for Skin Cancer Screening: A Qualitative Study. JAMA Dermatol. 2020, 156, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Willingham, M.L.J.; Spencer, S.Y.; Lum, C.A.; Sanchez, J.M.N.; Burnett, T.; Shepherd, J.; Cassel, K. The potential of using artificial intelligence to improve skin cancer diagnoses in Hawai‘i’s multiethnic population. Melanoma Res. 2021, 31, 504–514. [Google Scholar] [CrossRef]
- Shellenberger, R.A.; Johnson, T.M.; Fayyaz, F.; Swamy, B.; Albright, J.; Geller, A.C. Disparities in melanoma incidence and mortality in rural versus urban Michigan. Cancer Rep. 2023, 6, e1713. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Sarda, A.; Gupta, S.; Das, S. Use of artificial intelligence in dermatology. Indian J. Dermatol. 2020, 65, 352–357. [Google Scholar] [CrossRef] [PubMed]

| Paper Title | Authors | Objective | AI Technique | Dataset | Diagnostic Accuracy Rate |
|---|---|---|---|---|---|
| Computer Aided Diagnosis of Melanoma Using Deep Neural Networks and Game Theory: Application on Dermoscopic Images of Skin Lesions | Foahom Gouabou et al. [12] | Development of a novel deep learning ensemble method for computer aided diagnosis of melanoma | Deep Learning Ensemble Method | ISIC 2018 | AUROC = 0.93 for melanoma, BACC = 86% |
| Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images | Marchetti et al. [13] | Comparison of melanoma diagnostic accuracy of computer algorithms to dermatologists using dermoscopic images | Deep Learning | ISIC 2016 | ROC of the top fusion algorithm = 0.86, mean ROC of dermatologists = 0.71. (p = 0.001) |
| Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017 | Marchetti et al. [14] | Potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma | Deep Learning | ISIC 2017 | ROC of top-ranked computer algorithm = 0.87, ROC of dermatologists and residents = 0.66 (p < 0.001), dermatologists overall sensitivity = 76.0%, algorithm had superior specificity (85.0% vs. 72.6%, p = 0.001) |
| Lesion identification and malignancy prediction from clinical dermatological images | Xia et al. [15] | Two-stage approach that identifies all lesions in an image, estimates their likelihood of malignancy, and generates an image-level likelihood for high-level screening | Deep Learning | ISIC 2018 | AUC of 0.959 based on dermoscopic images, which was augmented to an AUC of 0.961 using ISIC2018 |
| An improved transformer network for skin cancer classification | Xin et al. [16] | Establishment of an improved transformer network named SkinTrans | Vision Transformer (VIT) | HAM10000 | Accuracy = 94.3% |
| Skin Cancer Diagnosis Based on Neutrosophic Features with a Deep Neural Network | Singh et al. [17] | Computer-aided diagnosis system for the classification of a malignant lesion, where the acquired image is primarily pre-processed using novel methods | Deep Learning | PH2, ISIC 2017, ISIC 2018, and ISIC 2019 | PH2 = 99.50%, ISIC 2017 = 99.33%, ISIC 2018 = 98.56%, and ISIC 2019 = 98.04% |
| SCDNet: A Deep Learning-Based Framework for the Multiclassification of Skin Cancer Using Dermoscopy Images | Naeem et al. [18] | Novel framework for the multiclassification of skin cancer types such as melanoma, melanocytic nevi, basal cell carcinoma, and benign keratosis | Deep Learning | ISIC 2019 | Accuracy = 92.18% |
| Cancer-Net SCa: tailored deep neural network designs for detection of skin cancer from dermoscopy images | Lee et al. [19] | Deep neural network designs tailored for melanoma detection from dermoscopy images | Deep Learning | ISIC 2018 | Sensitivity of 92.8%, PPV of 78.5%, and NPV of 91.2% |
| On the Automatic Detection and Classification of Skin Cancer Using Deep Transfer Learning | Fraiwan et al. [20] | Applicability of raw deep transfer learning in classifying images of skin lesions into seven possible categories | Deep Learning | HAM10000 | 71% sensitivity (i.e., recall) but 43.1% precision. Highest reported accuracy rate = 76.7% |
| Computational Intelligence-Based Melanoma Detection and Classification Using Dermoscopic Images | Vaiyapuri et al. [21] | Develops a novel computational intelligence-based melanoma detection and classification technique using dermoscopic images (CIMDC-DIs) | Computational Intelligence (CI) and Deep Learning (DL) | ISIC 2016, 2017, 2020 | Maximum accuracy of 97.50% |
| Efficacy of a Deep Learning Convolutional Neural Network System for Melanoma Diagnosis in a Hospital Population | Martin-Gonzalez et al. [22] | Deep learning-based tool to differentiate between benign skin lesions versus melanoma in the hospital setting | Deep Learning | 232 dermoscopic images | AUC = 0.813, sensitivity of 0.691, specificity of 0.802, and accuracy of 0.776 |
| Deep Learning-Based Classification for Melanoma Detection Using XceptionNet | Lu et al. [23] | Automatic method for diagnosis of skin cancer from dermoscopy images | Deep Learning | HAM10000 | Accuracy rate of 100% for the detection of melanoma, sensitivity rate of 94.05%, and precision rate of 97.07% |
| Melanoma Classification Using a Novel Deep Convolutional Neural Network with Dermoscopic Images | Kaur et al. [24] | Proposed a lightweight and less complex DCNN than other state-of-the-art methods to classify melanoma skin cancer with high efficiency | Deep Learning | ISIC 2016, ISIC 2017, and ISIC 2020 | ISIC 2016 = 81.41%, ISIC 2017 = 88.23%, and ISIC 2020 = 90.42% |
| A Computer-Aided Diagnosis System Using Deep Learning for Multiclass Skin Lesion Classification | Arshad et al. [25] | Series-based new automated framework for multiclass skin lesion classification | Deep Learning | HAM10000 | Accuracy = 91.7% |
| ZooME: Efficient Melanoma Detection Using Zoom-in Attention and Metadata Embedding Deep Neural Network | Xing et al. [26] | Zoom-in Attention and Metadata Embedding (ZooME) melanoma detection network | Deep Learning | ISIC 2020 | 92.23% AUC score, 84.59% accuracy, 85.95% sensitivity, and 84.63% specificity |
| AI outperformed every dermatologist in dermoscopic melanoma diagnosis, using an optimized deep-CNN architecture with custom mini-batch logic and loss function | Pham et al. [27] | Method for improving melanoma prediction on an imbalanced dataset by reconstructed appropriate CNN architecture and optimized algorithms | Deep Learning | 17,302 dermoscopic images | AUC of 94.4%, sensitivity of 85.0%, and specificity of 95.0% |
| Computer-Aided Diagnosis Algorithm for Classification of Malignant Melanoma Using Deep Neural Networks | Kim et al. [28] | Tumor lesion segmentation model and a classification model of malignant melanoma | Deep Learning | ISIC 2017 | Classification accuracy of malignant melanoma reached 80.06% |
| Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering | Nawaz et al. [29] | Fully automated method for segmenting skin melanoma at its earliest stage by employing a deep learning-based approach, namely faster region-based convolutional neural networks (RCNN) along with fuzzy k-means clustering (FKM) | DL-based approach | ISIC 2016, ISIC 2017, and PH2 | ISIC 2016 = 95.40, ISIC 2017 = 93.1, and PH2 = 95.6% |
| A novel melanoma prediction model for imbalanced data using optimized SqueezeNet by bald eagle search optimization | Sayed et al. [30] | New model for the classification of skin lesions as either normal or melanoma | Deep Learning | ISIC 2020 | Accuracy of 98.37%, specificity of 96.47%, sensitivity of 100%, f-score of 98.40%, and area under the curve of 99% |
| Ensemble Method of Convolutional Neural Networks with Directed Acyclic Graph Using Dermoscopic Images: Melanoma Detection Application | Foahom Gouabou et al. [31] | Novel ensemble scheme of convolutional neural networks (CNNs), inspired by decomposition and ensemble methods, to improve the performance of the CAD system | CAD/CNN | ISIC 2018 | Best balanced accuracy = (76.6%) |
| Developing a Recognition System for Diagnosing Melanoma Skin Lesions Using Artificial Intelligence Algorithms | Alsaade et al. [32] | Development of feature-based and deep learning-based systems for melanoma classification | Deep learning and traditional artificial intelligence machine learning algorithms | PH2, ISIC 2018 | Accuracy = PH2 (97.50%) and ISIC 2018 (98.35%) |
| Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images | Iqbal et al. [33] | Develop, implement, and calibrate an advanced deep learning model in the context of automated multiclass classification of skin lesions | Deep Convolutional Neural Network (DCNN) Model | ISIC-2017, ISIC-2018, and ISIC-2019 | AUROC = 0.964, 94% precision, 93% sensitivity, and 91% specificity in ISIC-17 |
| Melanoma diagnosis using deep learning techniques on dermatoscopic images | Jojoa Acosta et al. [34] | Two-stage DL-based method | Deep Learning | ISIC 2017 | Overall accuracy (0.904), sensitivity (0.820), and specificity (0.925) |
| A new deep learning approach integrated with clinical data for the dermoscopic differentiation of early melanomas from atypical nevi | Tognetti et al. [35] | Deep convolutional neural network (DCNN) model able to support dermatologists in the classification and management of atypical melanocytic skin lesions (aMSL) | Deep Convolutional Neural Network (DCNN) | 630 dermoscopic images | AUC = 90.3%, SE = 86.5%, and SP = 73.6% |
| Deep learning-level melanoma detection by interpretable machine learning and imaging biomarker cues | Gareau et al. [36] | Transparent machine learning technology (i.e., not deep learning) to discriminate melanomas from nevi in dermoscopy images and an interface for sensory cue integration | Transparent Machine Learning | 349 images | AUROC was 0.71 ± 0.07, Sens = 98%, Spec = 36% |
| Effective Melanoma Recognition Using Deep Convolutional Neural Network with Covariance Discriminant Loss | Guo et al. [37] | Melanoma recognition method using deep convolutional neural network with covariance discriminant loss in dermoscopy images | Deep Learning | ISIC 2018 | Sensitivity = 0.942 and 0.917 |
| Deep Convolutional Neural Network for Melanoma Detection using Dermoscopy Images | R K et al. [38] | Propose a deep convolutional neural network for automated melanoma detection that is scalable to accommodate a variety of hardware and software constraints | Deep Learning | 2150 dermosocpic images | Accuracy = 82.95%, sensitivity = 82.99%, and specificity = 83.89% |
| Dermoscopic diagnostic performance of Japanese dermatologists for skin tumors differs by patient origin: A deep learning convolutional nmeural network closes the gap | Minagawa et al. [39] | Compared the performance of 30 Japanese dermatologists with that of a DNN for the dermoscopic diagnosis of International Skin Imaging Collaboration (ISIC) and Shinshu (Japanese only) datasets to classify malignant melanoma, melanocytic nevus, basal cell carcinoma, and benign keratosis on non-volar skin | Deep Learning | ISIC and Shinsu (Japanese) dataset | Specificity of the DNN at the dermatologists’ mean sensitivity value = 0.962; Shinshu set = 1.00 |
| DePicT Melanoma Deep-CLASS: a deep convolutional neural networks approach to classify skin lesion images | Nasiri et al. [40] | Approach to classify skin lesions using deep learning for early detection of melanoma in a case-based reasoning (CBR) system | Deep Learning | ISIC Archive | Accuracy = 0.77 |
| Melanoma recognition by a deep learning convolutional neural network—Performance in different melanoma subtypes and localisations | Winkler et al. [41] | Investigated the diagnostic performance of a CNN with approval for the European market across different melanoma localizations and subtypes | Deep Learning | 6 dermoscopic image sets (each set included 30 melanomas and 100 benign lesions of related localisations and morphology) | Sensitivities > 93.3%, specificities > 65%, receiver operating characteristic–area under the curve (ROC-AUC) > 0.926 |
| Detection of Malignant Melanoma Using Artificial Intelligence: An Observational Study of Diagnostic Accuracy | Phillips et al. [42] | Evaluated the accuracy of an AI neural network (Deep Ensemble for Recognition of Melanoma (DERM)) to identify malignant melanoma from dermoscopic images | DERM | 7102 dermoscopic images | ROC (AUC) of 0.93 (95% confidence interval: 0.92–0.94), and sensitivity and specificity of 85.0% and 85.3% |
| Deep neural networks are superior to dermatologists in melanoma image classification | Brinker et al. [43] | Automated dermoscopic melanoma image classification compared with dermatologists | Deep Learning | 4204 biopsy-proven images of melanoma and nevi | Trained CNN achieved higher sensitivity of 82.3% (95% CI: 78.3–85.7%) and higher specificity of 77.9% (95% CI: 73.8–81.8%) |
| Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task | Brinker et al. [44] | Performance of a deep learning algorithm trained by open-source images exclusively is compared to a large number of dermatologists covering all levels within the clinical hierarchy | Deep Learning | 12,378 open-source dermoscopic images (training set), 100 images for comparison | At mean sensitivity of 74.1%, the CNN exhibited mean specificity of 86.5% (range 70.8–91.3%). At mean specificity of 60%, mean sensitivity of 87.5% (range 80–95%) was achieved by our algorithm |
| Deep Learning and Handcrafted Method Fusion: Higher Diagnostic Accuracy for Melanoma Dermoscopy Images | Hagerty et al. [45] | Approach that combines conventional image processing with deep learning by fusing the features from the individual techniques | Deep Learning | ISIC | AUC of 0.87, classification accuracy of 0.94 |
| Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists | Haenssle et al. [46] | Compared a CNN’s diagnostic performance with a large international group of 58 dermatologists | Deep Learning | 100 dermoscopic images | CNN ROC AUC was greater than the mean ROC area of dermatologists (0.86 versus 0.79, p < 0.01) |
| Skin Lesion Analysis towards Melanoma Detection Using Deep Learning Network | Li et al. [47] | Proposed two deep learning methods to address three main tasks emerging in the area of skin lesion image processing | Deep Learning | ISIC 2017 | Highest accuracy = 0.912 |
| Paper Title | Authors | Objective | AI Technique | Dataset | Diagnostic Accuracy Rate |
|---|---|---|---|---|---|
| Identification of Cancerous Skin Lesions Using Vibrational Optical Coherence Tomography (VOCT): Use of VOCT in Conjunction with Machine Learning to Diagnose Skin Cancer Remotely Using Telemedicine | Silver FH et al. [48] | Used VOCT and machine learning to evaluate the specificity and sensitivity of differentiating normal skin from skin cancers | Conventional Machine Learning | 80 images | Sensitivity = 83.3%, specificity = 77.8% |
| Paper Title | Authors | Objective | AI Technique | Dataset | Diagnostic Accuracy Rate |
|---|---|---|---|---|---|
| Convolutional Neural Network Approach to Classify Skin Lesions Using Reflectance Confocal Microscopy | Wodzinski et al. [49] | CNN-based approach to classify skin lesions using the reflectance confocal microscopy (RCM) mosaics | CNN | 429 RCM mosaics | Test set classification accuracy = 87% |
| Semantic segmentation of reflectance confocal microscopy mosaics of pigmented lesions using weak labels | D’Alonzo et al. [50] | Development of a weakly supervised machine learning model to perform semantic segmentation of architectural patterns encountered in RCM mosaics | Deep Learning | 157 RCM mosaics | Trained DNN achieved an average AUC of 0.969; Dice coefficient = 0.778 |
| Dataset | Total Dermoscopic Images |
|---|---|
| International Skin Imaging Collaboration (ISIC 2016) | 1279 |
| International Skin Imaging Collaboration (ISIC 2017) | 2600 |
| International Skin Imaging Collaboration (ISIC 2018) | 11,527 |
| International Skin Imaging Collaboration (ISIC 2019) | 33,569 |
| International Skin Imaging Collaboration (ISIC 2020) | 44,108 |
| PH2 | 200 |
| Human Against Machine with 10,000 training images (HAM10000) | 10,015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, R.H.; Foltz, E.A.; Witkowski, A.; Ludzik, J. Analysis of Artificial Intelligence-Based Approaches Applied to Non-Invasive Imaging for Early Detection of Melanoma: A Systematic Review. Cancers 2023, 15, 4694. https://doi.org/10.3390/cancers15194694
Patel RH, Foltz EA, Witkowski A, Ludzik J. Analysis of Artificial Intelligence-Based Approaches Applied to Non-Invasive Imaging for Early Detection of Melanoma: A Systematic Review. Cancers. 2023; 15(19):4694. https://doi.org/10.3390/cancers15194694
Chicago/Turabian StylePatel, Raj H., Emilie A. Foltz, Alexander Witkowski, and Joanna Ludzik. 2023. "Analysis of Artificial Intelligence-Based Approaches Applied to Non-Invasive Imaging for Early Detection of Melanoma: A Systematic Review" Cancers 15, no. 19: 4694. https://doi.org/10.3390/cancers15194694





