Pancreatic Cancer-Secreted Proteins: Targeting Their Functions in Tumor Microenvironment
Abstract
:Simple Summary
Abstract
1. Pancreatic Cancer Fact Sheet
2. Proteins Secreted by Pancreatic Cancer Cells: Messages Sent to the Neighborhood
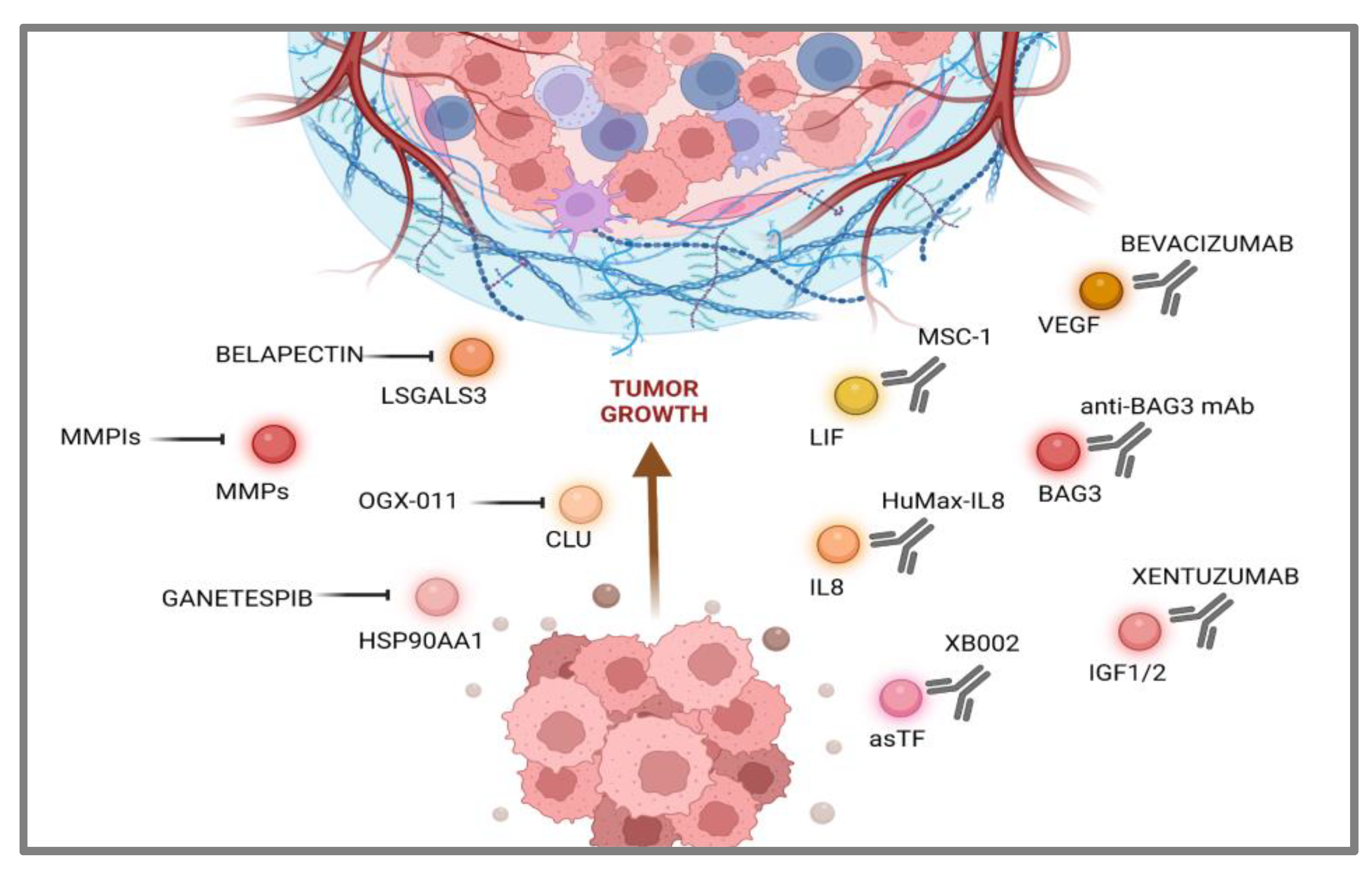
3. Targeting Pancreatic Cancer-Secreted Proteins
3.1. Communications Breakdown Operated by Small Molecule Drugs
3.2. Communications Breakdown Operated by Monoclonal Antibodies
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khalaf, N.; El-Serag, H.B.; Abrams, H.R.; Thrift, A.P. Burden of Pancreatic Cancer: From Epidemiology to Practice. Clin. Gastroenterol. Hepatol. 2021, 19, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer. JAMA 2021, 326, 851. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Mattiuzzi, C. The global burden of pancreatic cancer. Arch. Med. Sci. 2020, 16, 820–824. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA: A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Rahib, L.; Wehner, M.R.; Matrisian, L.M.; Nead, K.T. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw. Open 2021, 4, e214708. [Google Scholar] [CrossRef]
- Kindler, H.L. A Glimmer of Hope for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2463–2464. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Schepis, T.; De Lucia, S.S.; Pellegrino, A.; Del Gaudio, A.; Maresca, R.; Coppola, G.; Chiappetta, M.F.; Gasbarrini, A.; Franceschi, F. State-of-the-Art and Upcoming Innovations in Pancreatic Cancer Care: A Step Forward to Precision Medicine. Cancers 2023, 15, 3423. [Google Scholar] [CrossRef]
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; Benson, A.B., III; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. J. Clin. Oncol. 2023, 41, 678–700. [Google Scholar] [CrossRef]
- Shah, M.A.; Kennedy, E.B.; Alarcon-Rozas, A.E.; Alcindor, T.; Bartley, A.N.; Malowany, A.B.; Bhadkamkar, N.A.; Deighton, D.C.; Janjigian, Y.; Karippot, A.; et al. Immunotherapy and Targeted Therapy for Advanced Gastroesophageal Cancer: ASCO Guideline. J. Clin. Oncol. 2023, 41, 1470–1491. [Google Scholar] [CrossRef]
- Moy, B.; Rumble, R.B.; Come, S.E.; Davidson, N.E.; Di Leo, A.; Gralow, J.R.; Hortobagyi, G.N.; Yee, D.; Smith, I.E.; Chavez-MacGregor, M.; et al. Chemotherapy and Targeted Therapy for Patients with Human Epidermal Growth Factor Receptor 2–Negative Metastatic Breast Cancer That is Either Endocrine-Pretreated or Hormone Receptor–Negative: ASCO Guideline Update. J. Clin. Oncol. 2021, 39, 3938–3958. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, E.; Ismaila, N.; Bauman, J.E.; Dabney, R.; Gan, G.; Jordan, R.; Kaufman, M.; Kirtane, K.; McBride, S.M.; Old, M.O.; et al. Immunotherapy and Biomarker Testing in Recurrent and Metastatic Head and Neck Cancers: ASCO Guideline. J. Clin. Oncol. 2023, 41, 1132–1146. [Google Scholar] [CrossRef] [PubMed]
- Mamdani, H.; Matosevic, S.; Khalid, A.B.; Durm, G.; Jalal, S.I. Immunotherapy in Lung Cancer: Current Landscape and Future Directions. Front. Immunol. 2022, 13, 823618. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, F.; Di Costanzo, F.; Antonuzzo, L.; Mazza, E.; Giommoni, E. Optimizing First-Line Chemotherapy in Metastatic Pancreatic Cancer: Efficacy of FOLFIRINOX versus Nab-Paclitaxel Plus Gemcitabine. Cancers 2023, 15, 416. [Google Scholar] [CrossRef] [PubMed]
- Principe, D.R.; Underwood, P.W.; Korc, M.; Trevino, J.G.; Munshi, H.G.; Rana, A. The Current Treatment Paradigm for Pancreatic Ductal Adenocarcinoma and Barriers to Therapeutic Efficacy. Front. Oncol. 2021, 11, 688377. [Google Scholar] [CrossRef] [PubMed]
- Di Federico, A.; Mosca, M.; Pagani, R.R.; Carloni, R.; Frega, G.; De Giglio, A.; Rizzo, A.; Ricci, D.; Tavolari, S.; Di Marco, M.; et al. Immunotherapy in Pancreatic Cancer: Why Do We Keep Failing? A Focus on Tumor Immune Microenvironment, Predictive Biomarkers and Treatment Outcomes. Cancers 2022, 14, 2429. [Google Scholar] [CrossRef]
- Singhi, A.D.; George, B.; Greenbowe, J.R.; Chung, J.; Suh, J.; Maitra, A.; Klempner, S.J.; Hendifar, A.; Milind, J.M.; Golan, T.; et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted with Existing Drugs or Used as Biomarkers. Gastroenterology 2019, 156, 2242–2253.e4. [Google Scholar] [CrossRef]
- Ben-Ammar, I.; Tarabay, A.; Hollebecque, A.; Tanguy, V.; Boige, D.; Malka, C.; Smolenschi, M.; Ducreux, A.; Boileve, A. SO-1 Clinical and molecular features of patients with KRAS wild-type pancreatic adenocarcinoma. Ann. Oncol. 2022, 33, S356. [Google Scholar] [CrossRef]
- Huffman, B.M.; Ellis, H.; Jordan, A.C.; Freed-Pastor, W.A.; Perez, K.; Rubinson, D.A.; Sethi, N.; Singh, H.; Surana, R.; Wolpin, B.M.; et al. Emerging Role of Targeted Therapy in Metastatic Pancreatic Adenocarcinoma. Cancers 2022, 14, 6223. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Lyssiotis, C.A.; Pasca di Magliano, M.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef]
- Mahalingam, D.; Burns, M.C.; Kalyan, A.; Kircher, S.M.; Kocherginsky, M.; Mulcahy, M.F.; Benson, A.B. A phase Ib/II study of sotorasib combined with chemotherapy for second-line treatment of KRAS p. G12C–mutated advanced pancreatic cancer. J. Clin. Oncol. 2022, 40 (Suppl. 16), TPS4194. [Google Scholar] [CrossRef]
- Tsai, Y.S.; Woodcock, M.G.; Azam, S.H.; Thorne, L.B.; Kanchi, K.L.; Parker, J.S.; Vincent, B.G.; Pecot, C.V. Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J. Clin. Investig. 2022, 132, e155523. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, T.; Inamura, K.; Nishizono, Y.; Suzuki, A.; Tanaka, H.; Yoshinari, T.; Yamanaka, Y. ASP3082, a First-in-class novel KRAS G12D degrader, exhibits remarkable anti-tumor activity in KRAS G12D mutated cancer models. Eur. J. Cancer 2022, 174, S30. [Google Scholar] [CrossRef]
- Evan, T.; Wang, V.M.Y.; Behrens, A. The roles of intratumour heterogeneity in the biology and treatment of pancreatic ductal adenocarcinoma. Oncogene 2022, 41, 4686–4695. [Google Scholar] [CrossRef]
- Shaya, J.; Kato, S.; Adashek, J.J.; Patel, H.; Fanta, P.T.; Botta, G.P.; Sicklick, J.K.; Kurzrock, R. Personalized matched targeted therapy in advanced pancreatic cancer: A pilot cohort analysis. NPJ Genom. Med. 2023, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, S.; Pan, W.; Wang, J.; Zhan, H.; Liu, S. Targeting tumor immunosuppressive microenvironment for pancreatic cancer immunotherapy: Current research and future perspective. Front. Oncol. 2023, 13, 1166860. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.H.; Beatty, G.L. Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance. Annu. Rev. Pathol. Mech. Dis. 2023, 18, 123–148. [Google Scholar] [CrossRef]
- Herpels, M.; Ishihara, J.; Sadanandam, A. The clinical terrain of immunotherapies in heterogeneous pancreatic cancer: Unravelling challenges and opportunities. J. Pathol. 2023, 260, 533–550. [Google Scholar] [CrossRef]
- Robinson, J.L.; Feizi, A.; Uhlén, M.; Nielsen, J. A Systematic Investigation of the Malignant Functions and Diagnostic Potential of the Cancer Secretome. Cell Rep. 2019, 26, 2622–2635.e5. [Google Scholar] [CrossRef]
- De Morais, J.A.; Zelanis, A. Bioinformatic reanalysis of public proteomics data reveals that nuclear proteins are recurrent in cancer secretomes. Traffic 2021, 23, 98–108. [Google Scholar] [CrossRef]
- Gautam, S.K.; Batra, S.K.; Jain, M. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol. Cancer 2023, 22, 118. [Google Scholar] [CrossRef] [PubMed]
- Padgaonkar, M.; Shendre, S.; Chatterjee, P.; Banerjee, S. Cancer secretome: Finding out hidden messages in extracellular secretions. Clin. Transl. Oncol. 2022, 25, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Espinet, E.; Klein, L.; Puré, E.; Singh, S.K. Mechanisms of PDAC subtype heterogeneity and therapy response. Trends in Cancer. 2022, 8, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.C.; Oh, M.J.; Choi, S.H.; Bae, C.D. Proteomic analysis to identify biomarker proteins in pancreatic ductal adenocarcinoma. ANZ J. Surg. 2008, 78, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Brandi, J.; Pozza, E.D.; Dando, I.; Biondani, G.; Robotti, E.; Jenkins, R.; Elliott, V.; Park, K.; Marengo, E.; Costello, E.; et al. Secretome protein signature of human pancreatic cancer stem-like cells. J. Proteom. 2016, 136, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, H.; Dun, M.D.; Faulkner, S.; Liu, X.; Jiang, C.C.; Hondermarck, H. Proteome and secretome analysis of pancreatic cancer cells. Proteomics 2022, 22, 2100320. [Google Scholar] [CrossRef]
- Liu, P.; Kong, L.; Jin, H.; Wu, Y.; Tan, X.; Song, B. Differential secretome of pancreatic cancer cells in serum-containing conditioned medium reveals CCT8 as a new biomarker of pancreatic cancer invasion and metastasis. Cancer Cell Int. 2019, 19, 262. [Google Scholar] [CrossRef]
- Liu, P.; Weng, Y.; Sui, Z.; Wu, Y.; Tan, X.; Song, B. Quantitative secretomic analysis of pancreatic cancer cells in serum-containing conditioned medium. Sci. Rep. 2016, 6, 37606. [Google Scholar] [CrossRef]
- Jo, J.H.; Kim, S.A.; Lee, J.H.; Park, Y.R.; Kim, C.; Park, S.B.; Jung, D.E.; Lee, H.S.; Chung, M.J.; Song, S.Y. GLRX3, a novel cancer stem cell-related secretory biomarker of pancreatic ductal adenocarcinoma. BMC Cancer 2021, 21, 1241. [Google Scholar] [CrossRef]
- Kelly, K.A.; Bardeesy, N.; Anbazhagan, R.; Park, Y.R.; Kim, C.; Park, S.B.; Jung, D.E.; Lee, H.S.; Chung, M.J.; Song, S.Y. Targeted Nanoparticles for Imaging Incipient Pancreatic Ductal Adenocarcinoma. PLoS Med. 2008, 5, e85. [Google Scholar] [CrossRef]
- Liu, P.; Kong, L.; Liang, K.; Wu, Y.; Tan, X.; Song, B. Identification of dissociation factors in pancreatic Cancer using a mass spectrometry-based proteomic approach. BMC Cancer 2020, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Guo, M.; Liu, Y.; Zheng, J. Progress in Animal Models of Pancreatic Ductal Adenocarcinoma. J. Cancer 2020, 11, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Levink, I.J.M.; Visser, I.J.; Koopmann, B.D.M.; van Driel, L.M.J.W.; Poley, J.W.; Cahen, D.L.; Bruno, M.J.; Fuhler, G.M. Protein biomarkers in pancreatic juice and serum for identification of pancreatic cancer. Gastrointest. Endosc. 2022, 96, 801–813.e2. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.T.; Wu, C.C.; Shyr, Y.M.; Chen, T.-C.; Hwang, T.-L.; Yeh, T.-S.; Chang, K.-P.; Liu, H.-P.; Liu, Y.-L.; Tsai, M.-H.; et al. Secretome-Based Identification of ULBP2 as a Novel Serum Marker for Pancreatic Cancer Detection. PLoS ONE 2011, 6, e20029. [Google Scholar] [CrossRef] [PubMed]
- Firpo, M.A.; Boucher, K.M.; Bleicher, J.; Khanderao, G.D.; Rosati, A.; Poruk, K.E.; Kamal, S.; Marzullo, L.; De Marco, M.; Falco, A.; et al. Multianalyte Serum Biomarker Panel for Early Detection of Pancreatic Adenocarcinoma. JCO Clin. Cancer Inform. 2023, 7, e2200160. [Google Scholar] [CrossRef] [PubMed]
- Kapszewicz, M.; Małecka-Wojciesko, E. Simple Serum Pancreat. Ductal Adenocarcinoma (PDAC) Protein Biomark—Is There Anything Sight? J. Clin. Med. 2021, 10, 5463. [Google Scholar] [CrossRef]
- de Oliveira, G.; Paccielli Freire, P.; Santiloni Cury, S.; de Moraes, D.; Santos Oliveira, J.; Dal-Pai-Silva, M.; do Reis, P.P.; Francisco Carvalho, R. An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 716. [Google Scholar] [CrossRef]
- Xu, T.; Xu, X.; Liu, P.C.; Mao, H.; Ju, S. Transcriptomic Analyses and Potential Therapeutic Targets of Pancreatic Cancer with Concomitant Diabetes. Front. Oncol. 2020, 10, 563527. [Google Scholar] [CrossRef]
- Slapak, E.J.; Duitman, J.; Tekin, C.; Bijlsma, M.F.; Spek, C.A. Matrix Metalloproteases in Pancreatic Ductal Adenocarcinoma: Key Drivers of Disease Progression? Biology 2020, 9, 80. [Google Scholar] [CrossRef]
- Jones, L.E.; Humphreys, M.J.; Campbell, F.; Neoptolemos, J.P.; Boyd, M.T. Comprehensive Analysis of Matrix Metalloproteinase and Tissue Inhibitor Expression in Pancreatic Cancer. Clin. Cancer Res. 2004, 10, 2832–2845. [Google Scholar] [CrossRef]
- Shoucair, S.; Chen, J.; Martinson, J.R.; Habib, J.R.; Kinny-Köster, B.; Pu, N.; van Oosten, A.F.; Javed, A.A.; Shin, E.J.; Ali, S.Z.; et al. Association of Matrix Metalloproteinase 7 Expression with Pathologic Response After Neoadjuvant Treatment in Patients with Resected Pancreatic Ductal Adenocarcinoma. JAMA Surg. 2022, 157, e221362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Zhu, Y.; Xie, K.L.; Peng, Y.-P.; Tao, J.-Q.; Tang, J.; Li, Z.; Xu, Z.-K.; Dai, C.-C.; Qian, Z.-Y.; et al. Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol. Cancer 2014, 13, 130. [Google Scholar] [CrossRef] [PubMed]
- Giampazolias, E.; Schulz, O.; Lim, K.H.J.; Rogers, N.C.; Chakravarty, P.; Srinivasan, N.; Gordon, O.; Cardoso, A.; Buck, M.D.; Poirier, E.Z.; et al. Secreted gelsolin inhibits DNGR-1-dependent cross-presentation and cancer immunity. Cell 2021, 184, 4016–4031.e22. [Google Scholar] [CrossRef] [PubMed]
- Xue, N.; Du, T.; Lai, F.; Jin, J.; Ji, M.; Chen, X. Secreted HSP90α-LRP1 Signaling Promotes Tumor Metastasis and Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2022, 23, 5532. [Google Scholar] [CrossRef]
- Chen, Y.; Kang, M.; Lu, W.; Guo, Q.; Zhang, B.; Xie, Q.; Wu, Y. DJ-1, a novel biomarker and a selected target gene for overcoming chemoresistance in pancreatic cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhai, Q.; Bharadwaj, U.; Li, F.; Fisher, W.E.; Chen, C.; Yao, Q. Cyclophilin A is overexpressed in human pancreatic cancer cells and stimulates cell proliferation through CD147. Cancer 2006, 106, 2284–2294. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Jung, H.J. Cyclophilin A/CD147 Interaction: A Promising Target for Anticancer Therapy. Int. J. Mol. Sci. 2022, 23, 69341. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Z.; Zhang, K.; Liu, X.; Cao, W.; Zhang, L.; Zhang, S.; Yan, B.; Wang, Y.; Xia, C. Erratum to: Clusterin confers gemcitabine resistance in pancreatic cancer. World J. Surg. Oncol. 2013, 11, 149. [Google Scholar] [CrossRef]
- Rosati, A.; Bersani, S.; Tavano, F.; Dalla Pozza, E.; De Marco, M.; Palmieri, M.; De Laurenzi, V.; Franco, R.; Scognamiglio, G.; Palaia, R.; et al. Expression of the Antiapoptotic Protein BAG3 Is a Feature of Pancreatic Adenocarcinoma and Its Overexpression Is Associated with Poorer Survival. Am. J. Pathol. 2012, 181, 1524–1529. [Google Scholar] [CrossRef]
- Falco, A.; Rosati, A.; Festa, M.; Basile, A.; De Marco, M.; d’Avenia, M.; Pascale, M.; Dal Piaz, F.; Tavano, F.; Di Mola, F.F.; et al. BAG3 Is a Novel Serum Biomarker for Pancreatic Adenocarcinomas. Am. J. Gastroenterol. 2013, 108, 1178–1180. [Google Scholar] [CrossRef]
- Rosati, A.; Basile, A.; D’Auria, R.; d’Avenia, M.; De Marco, M.; Falco, A.; Festa, M.; Guerriero, L.; Iorio, V.; Parente, R.; et al. BAG3 promotes pancreatic ductal adenocarcinoma growth by activating stromal macrophages. Nat. Commun. 2015, 6, 8695. [Google Scholar] [CrossRef] [PubMed]
- Dufrusine, B.; Damiani, V.; Capone, E.; Pieragostino, D.; Dainese, E.; De Marco, M.; Reppucci, F.; Turco, M.C.; Rosati, A.; Marzullo, L.; et al. BAG3 induces fibroblasts to release key cytokines involved in pancreatic cell migration. J. Cell. Biochem. 2021, 123, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R.; Mohammad, J.; Orr, M.; Reindl, K.M. Glutathione S-Transferase pi-1 Knockdown Reduces Pancreatic Ductal Adenocarcinoma Growth by Activating Oxidative Stress Response Pathways. Cancers 2020, 12, 1501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yao, H.F.; Li, H.; Su, T.; Jiang, S.-H.; Wang, H.; Zhang, Z.-G.; Dong, F.-Y.; yang, Q.; Yang, X.-M. Transglutaminases Are Oncogenic Biomarkers in Human Cancers and Therapeutic Targeting of TGM2 Blocks Chemoresistance and Macrophage Infiltration in Pancreatic Cancer; Research Square Platform LLC.: Durham, NC, USA, 2023. [Google Scholar] [CrossRef]
- Prieto-Fernández, L.; Menéndez, S.T.; Otero-Rosales, M.; Montoro-Jiménez, I.; Hermida-Prado, F.; García-Pedrero, J.M.; Álvarez-Teijeiro, S. Pathobiological functions and clinical implications of annexin dysregulation in human cancers. Front. Cell Dev. Biol. 2022, 10, 1009908. [Google Scholar] [CrossRef] [PubMed]
- Bouter, A.; Gounou, C.; Bérat, R.; Tan, S.; Gallois, B.; Granier, T.; d’Estaintot, B.L.; Pöschl, E.; Brachvogel, B.; Brisson, A. RAnnexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nat. Commun. 2011, 2, 270. [Google Scholar] [CrossRef]
- Bouvet, F.; Ros, M.; Bonedeau, E.; Tan, S.; Gallois, B.; Granier, T.; d’Estaintot, B.L.; Pöschl, E.; Brachvogel, B.; Brisson, A.R. Defective Membrane Repair Machinery Impairs Survival of Invasive Cancer Cells; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2020. [Google Scholar] [CrossRef]
- Dong, X.; Javle, M.; Hess, K.R.; Shroff, R.; Abbruzzese, J.L.; Li, D. Insulin-Like Growth Factor Axis Gene Polymorphisms and Clinical Outcomes in Pancreatic Cancer. Gastroenterology 2010, 139, 464–473.e3. [Google Scholar] [CrossRef]
- Martínez-Bosch, N.; Cristóbal, H.; Iglesias, M.; Gironella, M.; Barranco, L.; Visa, L.; Calafato, D.; Jiménez-Parrado, S.; Earl, J.; Carrato, A.; et al. Soluble AXL is a novel blood marker for early detection of pancreatic ductal adenocarcinoma and differential diagnosis from chronic pancreatitis. eBioMedicine 2022, 75, 103797. [Google Scholar] [CrossRef]
- Miller, M.A.; Sullivan, R.J.; Lauffenburger, D.A. Molecular Pathways: Receptor Ectodomain Shedding in Treatment, Resistance, and Monitoring of Cancer. Clin. Cancer Res. 2017, 23, 623–629. [Google Scholar] [CrossRef]
- Jiang, W.; Li, X.; Xiang, C.; Zhou, W. Neutrophils in pancreatic cancer: Potential therapeutic targets. Front. Oncol. 2022, 12, 1025805. [Google Scholar] [CrossRef]
- Jin, W.; Xu, H.X.; Zhang, S.R.; Li, H.; Wang, W.-Q.; Gao, H.-L.; Wu, C.-T.; Xu, J.-Z.; Qi, Z.-H.; Li, S.; et al. Tumor-Infiltrating NETs Predict Postsurgical Survival in Patients with Pancreatic Ductal Adenocarcinoma. Ann. Surg. Oncol. 2018, 26, 635–643. [Google Scholar] [CrossRef]
- Yi, N.; Zhao, X.; Ji, J.; Xu, M.; Jiao, Y.; Qian, T.; Zhu, S.; Jiang, F.; Chen, J.; Xiao, M. Serum galectin-3 as a biomarker for screening, early diagnosis, prognosis and therapeutic effect evaluation of pancreatic cancer. J. Cell. Mol. Med. 2020, 24, 11583–11591. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Shen, R.; Yu, L.; Zheng, X.; Cui, R.; Song, Y.; Wang, D. Roles of galectin-3 in the tumor microenvironment and tumor metabolism (Review). Oncol. Rep. 2020, 44, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Teijeira, A.; Garasa, S.; Ochoa, M.C.; Villalba, M.; Olivera, I.; Cirella, A.; Eguren-Santamaria, I.; Berraondo, P.; Schalper, K.A.; de Andrea, C.E.; et al. IL8, Neutrophils, and NETs in a Collusion against Cancer Immunity and Immunotherapy. Clin. Cancer Res. 2020, 27, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Menzies-Gow, A.; Ying, S.; Sabroe, I.; Stubbs, V.L.; Soler, D.; Williams, T.J.; Kay, A.B. Eotaxin (CCL11) and Eotaxin-2 (CCL24) Induce Recruitment of Eosinophils, Basophils, Neutrophils, and Macrophages as Well As Features of Early- and Late-Phase Allergic Reactions Following Cutaneous Injection in Human Atopic and Nonatopic Volunteers. J. Immunol. 2002, 169, 2712–2718. [Google Scholar] [CrossRef] [PubMed]
- Sabrkhany, S.; Kuijpers, M.J.E.; van Kuijk, S.M.J.; Sanders, L.; Pineda, S.; Olde Damink, S.W.M.; Dingemans, A.-M.C.; Griffioen, A.W.; oude Egbrink, M.G.A. combination of platelet features allows detection of early-stage cancer. Eur. J. Cancer 2017, 80, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Lecot, P.; Ardin, M.; Dussurgey, S.; Alcazer, V.; Moudombi, L.; Hubert, M.; Swalduz, A.; Hernandez-Vargas, H.; Viari, A.; Caux, C.; et al. Gene Signature of Circulating Platelet-Bound Neutrophils Is Associated with Poor Prognosis in Cancer Patients; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2021. [Google Scholar] [CrossRef]
- Takehara, M.; Sato, Y.; Kimura, T.; Noda, K.; Miyamoto, H.; Fujino, Y.; Miyoshi, J.; Nakamura, F.; Wada, H.; Bando, Y.; et al. Cancer-associated adipocytes promote pancreatic cancer progression through SAA1 expression. Cancer Sci. 2020, 111, 2883–2894. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Yin, L.; Yang, X.; Yang, Y.; Gu, Y.; Sun, Y.; Yang, M.; Wang, Y.; Zhang, Q.; Ji, H. Serum amyloid A 1 induces suppressive neutrophils through the Toll-like receptor 2–mediated signaling pathway to promote progression of breast cancer. Cancer Sci. 2022, 113, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Hu, C.M.; Hsu, Y.S.; Lee, W.H. Interplays of glucose metabolism and KRAS mutation in pancreatic ductal adenocarcinoma. Cell Death Dis. 2022, 13, 817. [Google Scholar] [CrossRef]
- Yang, P.; Li, Z.; Wang, Y.; Zhang, L.; Wu, H.; Li, Z. Secreted pyruvate kinase M2 facilitates cell migration via PI3K/Akt and Wnt/β-catenin pathway in colon cancer cells. Biochem. Biophys. Res. Commun. 2015, 459, 327–332. [Google Scholar] [CrossRef]
- Lay, A.J.; Jiang, X.M.; Kisker, O.; Flynn, E.; Underwood, A.; Condron, R.; Hogg, P.J. Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature 2000, 408, 869–873. [Google Scholar] [CrossRef]
- Principe, M.; Borgoni, S.; Novelli, F. Commentary: “Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis”. J. Rare Dis. Res. Treat. 2017, 2, 18–21. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, J.; Yin, L.; Yang, J.; Zheng, Y.; Zhang, M.; Ni, B.; Wang, H. Upregulated GDF-15 expression facilitates pancreatic ductal adenocarcinoma progression through orphan receptor GFRAL. Aging 2020, 12, 22564–22581. [Google Scholar] [CrossRef] [PubMed]
- Albrengues, J.; Bourget, I.; Pons, C.; Butet, V.; Hofman, P.; Tartare-Deckert, S.; Feral, C.C.; Meneguzzi, G.; Gaggioli, C. LIF Mediates Proinvasive Activation of Stromal Fibroblasts in Cancer. Cell Rep. 2014, 7, 1664–1678. [Google Scholar] [CrossRef] [PubMed]
- Xelwa, N.; Candy, G.P.; Devar, J.; Omoshoro-Jones, J.; Smith, M.; Nweke, E.E. Targeting Growth Factor Signaling Pathways in Pancreatic Cancer: Towards Inhibiting Chemoresistance. Front. Oncol. 2021, 11, 683788. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, Y.W.; van den Hengel, L.G.; Myers, H.R.; Ayachi, O.; Jordanova, E.; Ruf, W.; Spek, C.A.; Reitsma, P.H.; Bogdanov, V.Y.; Versteeg, H.H. Alternatively spliced tissue factor induces angiogenesis through integrin ligation. Proc. Natl. Acad. Sci. USA 2009, 106, 19497–19502. [Google Scholar] [CrossRef] [PubMed]
- Winer, A.; Adams, S.; Mignatti, P. Matrix Metalloproteinase Inhibitors in Cancer Therapy: Turning Past Failures into Future Successes. Mol. Cancer Ther. 2018, 17, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Praharaj, P.P.; Patra, S.; Panigrahi, D.P.; Patra, S.K.; Bhutia, S.K. Clusterin as modulator of carcinogenesis: A potential avenue for targeted cancer therapy. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1875, 188500. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, L.; Song, F.; Zou, X.; Liu, Y.; Zheng, X.; Qian, J.; Gu, C.; Huang, P.; Yang, Y. Research progress on non-protein-targeted drugs for cancer therapy. J. Exp. Clin. Cancer Res. 2023, 42, 62. [Google Scholar] [CrossRef]
- Cardin, D.B.; Thota, R.; Goff, L.W.; Berlin, J.D.; Jones, C.M.; Ayers, G.D.; Whisenant, J.G.; Chan, E. A Phase II Study of Ganetespib as Second-line or Third-line Therapy for Metastatic Pancreatic Cancer. Am. J. Clin. Oncol. 2018, 41, 772–776. [Google Scholar] [CrossRef]
- Lang, J.E.; Forero-Torres, A.; Yee, D.; Yau, C.; Wolf, D.; Park, J.; Parker, B.A.; Chien, A.J.; Wallace, A.M.; Murthy, R.; et al. Safety and efficacy of HSP90 inhibitor ganetespib for neoadjuvant treatment of stage II/III breast cancer. NPJ Breast Cancer 2022, 8, 128. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Qin, Y.; Cong, Q.; Shao, C.; Du, Z.; Ni, X.; Li, P.; Ding, K. RN1, a novel galectin-3 inhibitor, inhibits pancreatic cancer cell growth in vitro and in vivo via blocking galectin-3 associated signaling pathways. Oncogene 2016, 36, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Kram, M. Galectin-3 inhibition as a potential therapeutic target in non-alcoholic steatohepatitis liver fibrosis. World J. Hepatol. 2023, 15, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Friedbichler, K.; Hofmann, M.H.; Kroez, M.; Ostermann, E.; Lamche, H.R.; Koessl, C.; Borges, E.; Pollak, M.N.; Adolf, G.; Adam, P.J. Data from Pharmacodynamic and Antineoplastic Activity of BI 836845, a Fully Human IGF Ligand-Neutralizing Antibody, and Mechanistic Rationale for Combination with Rapamycin; American Association for Cancer Research (AACR): Philadelphia, PA, USA, 2023. [Google Scholar] [CrossRef]
- Weyer-Czernilofsky, U.; Hofmann, M.H.; Friedbichler, K.; Baumgartinger, R.; Adam, P.J.; Solca, F.; Kraut, N.; Nguyen, H.M.; Corey, E.; Liu, G.; et al. Data from Antitumor Activity of the IGF-1/IGF-2–Neutralizing Antibody Xentuzumab (BI 836845) in Combination with Enzalutamide in Prostate Cancer Models; American Association for Cancer Research (AACR): Philadelphia, PA, USA, 2023. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Joaquim, A.; Jañez, N.M.; Morales, S.; Díaz-Redondo, T.; Blau, S.; Neven, P.; Lemieux, J.; García-Sáenz, J.Á.; et al. XENERA-1: A randomised double-blind Phase II trial of xentuzumab in combination with everolimus and exemestane versus everolimus and exemestane in patients with hormone receptor-positive/HER2-negative metastatic breast cancer and non-visceral disease. Breast Cancer Res. 2023, 25, 67. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.A.; Carbon, J.G.; Roland, C.L.; Toombs, J.E.; Nyquist-Andersen, M.; Kavlie, A.; Schlunegger, K.; Richardson, J.A.; Brekken, R.A. r84, a Novel Therapeutic Antibody against Mouse and Human VEGF with Potent Anti-Tumor Activity and Limited Toxicity Induction. PLoS ONE 2010, 5, e12031. [Google Scholar] [CrossRef] [PubMed]
- Kindler, H.L.; Niedzwiecki, D.; Hollis, D.; Sutherland, S.; Schrag, D.; Hurwitz, H.; Innocenti, F.; Mulcahy, M.F.; O’Reilly, E.; Wozniak, T.F.; et al. Gemcitabine Plus Bevacizumab Compared with Gemcitabine Plus Placebo in Patients with Advanced Pancreatic Cancer: Phase III Trial of the Cancer and Leukemia Group B (CALGB 80303). J. Clin. Oncol. 2010, 28, 3617–3622. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Vervenne, W.L.; Bennouna, J.; Humblet, Y.; Gill, S.; Van Laethem, J.L.; Verslype, C.; Scheithauer, W.; Shang, A.; Cosaert, J.; et al. Phase III Trial of Bevacizumab in Combination with Gemcitabine and Erlotinib in Patients with Metastatic Pancreatic Cancer. J. Clin. Oncol. 2009, 27, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.T.; Fer, N.; Galeas, J.; Collisson, E.A.; Kim, S.E.; Sharib, J.; McCormick, F. Blockade of leukemia inhibitory factor as a therapeutic approach to KRAS driven pancreatic cancer. Nat. Commun. 2019, 10, 3055. [Google Scholar] [CrossRef] [PubMed]
- Borazanci, E.; Schram, A.M.; Garralda, E.; Brana, I.; Vieito Villar, M.; Spreafico, A.; Oliva, M.; Lakhani, N.J.; Hoffman, K.; Hallett, R.M.; et al. Phase I, first-in-human study of MSC-1 (AZD0171), a humanized anti-leukemia inhibitory factor monoclonal antibody, for advanced solid tumors. ESMO Open 2022, 7, 100530. [Google Scholar] [CrossRef]
- Basile, A.; De Marco, M.; Festa, M.; Falco, A.; Iorio, V.; Guerriero, L.; Eletto, D.; Rea, D.; Arra, C.; Lamolinara, A.; et al. Development of an anti-BAG3 humanized antibody for treatment of pancreatic cancer. Mol. Oncol. 2019, 13, 1388–1399. [Google Scholar] [CrossRef]
- Iorio, V.; Rosati, A.; D’Auria, R.; De Marco, M.; Marzullo, L.; Basile, A.; Festa, M.; Pascale, M.; Remondelli, P.; Capunzo, M.; et al. Combined effect of anti-BAG3 and anti-PD-1 treatment on macrophage infiltrate, CD8+ Tcell number and tumour growth in pancreatic cancer. Gut 2017, 67, 314225. [Google Scholar] [CrossRef]
- De Marco, M.; Gauttier, V.; Pengam, S.; Mary, C.; Ranieri, B.; Basile, A.; Festa, M.; Falco, A.; Reppucci, F.; Cammarota, A.L.; et al. Concerted BAG3 and SIRPα blockade impairs pancreatic tumor growth. Cell Death Discov. 2022, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Iorio, V.; De Marco, M.; Basile, A.; Eletto, D.; Capunzo, M.; Remondelli, P.; Sala, G.; Marzullo, L.; Rosati, A.; De Laurenzi, V.; et al. CAF-Derived IL6 and GM-CSF Cooperate to Induce M2-like TAMs–Letter. Clin. Cancer Res. 2019, 25, 892–893. [Google Scholar] [CrossRef] [PubMed]
- Rosati, A.; Marzullo, L.; De Marco, M.; De Laurenzi, V.; D’Amico, M.F.; Turco, M.C. Toxicity in combined therapies for tumours treatments: A lesson from BAG3 in the TME? Front. Immunol. 2023, 14, 1241543. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Rozich, N.; Wang, J.; Wang, J.; Xu, Y.; Herbst, B.; Yu, R.; Muth, S.; Niu, N.; Li, K.; et al. Anti-IL-8 antibody activates myeloid cells and potentiates the anti-tumor activity of anti-PD-1 antibody in the humanized pancreatic cancer murine model. Cancer Lett. 2022, 539, 215722. [Google Scholar] [CrossRef]
- Bilusic, M.; Heery, C.R.; Collins, J.M.; Donahue, R.N.; Palena, C.; Madan, R.A.; Karzai, F.; Marté, J.L.; Strauss, J.; Gatti-Mays, M.E.; et al. Phase I trial of HuMax-IL8 (BMS-986253), an anti-IL-8 monoclonal antibody, in patients with metastatic or unresectable solid tumors. J. Immunotherapy Cancer 2019, 7, 240. [Google Scholar] [CrossRef]
- Lewis, C.S.; Karve, A.; Matiash, K.; Stone, T.; Li, J.; Wang, J.K.; Versteeg, H.H.; Aronow, B.J.; Ahmad, S.A.; Desai, P.B.; et al. A First-In-Class, Humanized Antibody Targeting Alternatively Spliced Tissue Factor: Preclinical Evaluation in an Orthotopic Model of Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2021, 11, 691685. [Google Scholar] [CrossRef]
- Ganguly, K.; Kimmelman, A.C. Reprogramming of tissue metabolism during cancer metastasis. Trends Cancer 2023, 9, 461–471. [Google Scholar] [CrossRef]
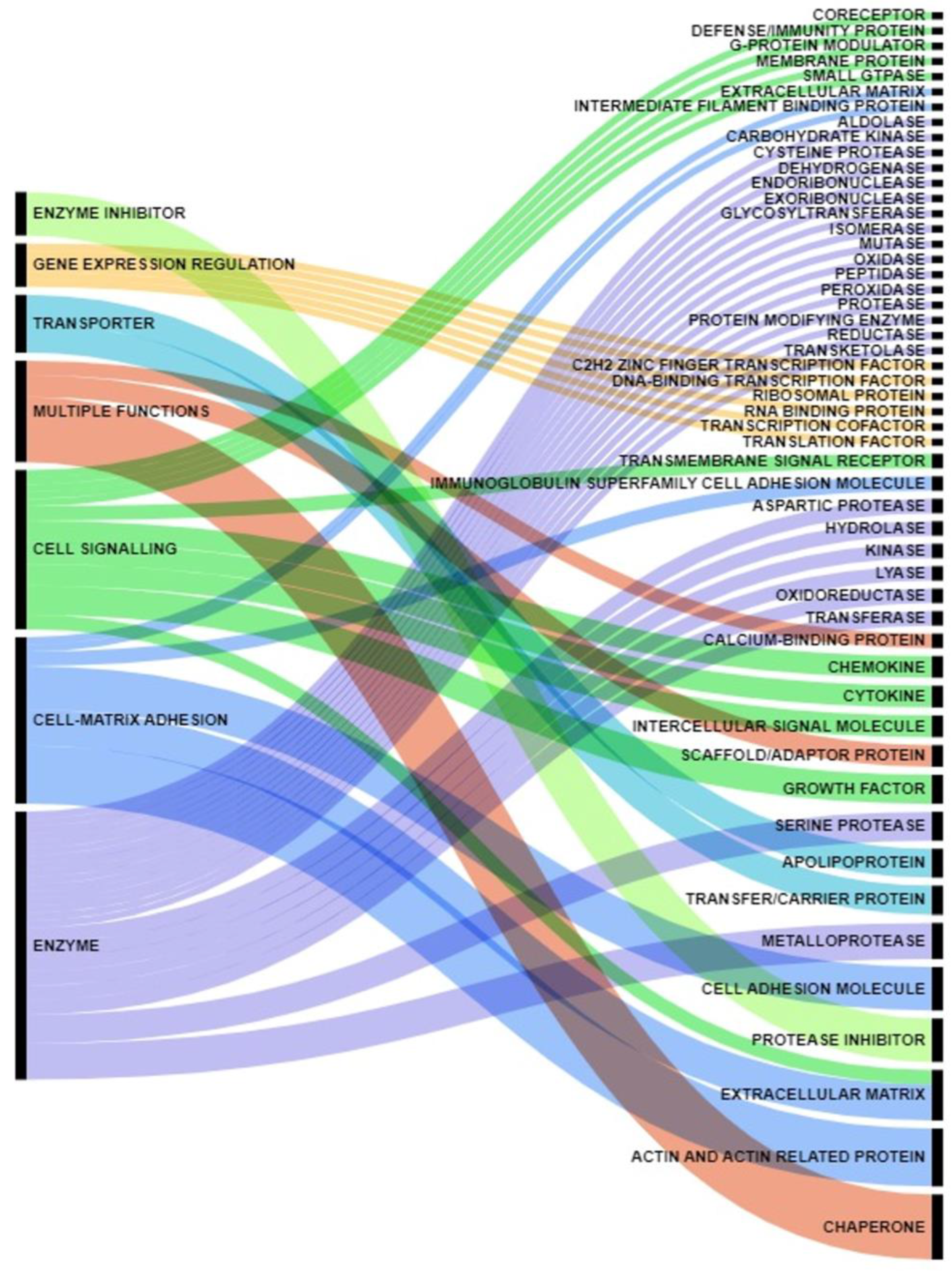
| Gene Names | References |
|---|---|
| Pancreatic cancer cells-secreted proteins | |
| LGALS1, ENO2, SERPINA1, NMI, PRDX4 | Chung J.C. et al. 2008 [34] |
| CP, LGALS3, MARCKS | Brandi J. et al. 2016 [35] |
| GDF15, TGM2, LIF, MMP2 | Li X. et al. 2022 [36] |
| SAA1, RC3H1, CCT8, ZNF518B, EXOSC8, IGF2, NPC2, HSP90AA1, PPIA, ENO1, | Liu P. et al. 2019 [37] |
| DNAJB11, PPT1, CTSD, CDH3, PLAU, LFNG | Liu P. et al. 2016 [38] |
| GLRX3 | Jo J.H. et al. 2021 [39] |
| PLEC | Kelly K.A. et al. 2008 [40] |
| MMP12, MMP10, LAMA5, WHAG, CPN1, THPH2 | Liu P. et al. 2020 [41] |
| ALB, ENO1, FN1, TF, LGALS1, APOE, CTSD, TPI1, GSTP1, PARK7, PRSS1, MSN, PGK1, ANXA5, PIN1, PKM, EEF1A1, THBS1, GSN, LGALS3, TIMP1, CFL1, FLNA, LGALS3BP, CALR, CLIC1, TAGLN2, LDHA, NME1, TKT, SFN, ALDOA, ENO2, PGAM1, ARHGDIA, ACTB, P4HB, ACTA1, AHSG | de Oliveira G. et al. 2020 [47] |
| Pancreatic cancer patients’ sera proteins | |
| CXCL8, LCN2, MUC5AC | Levink J.M. et al. 2022 [43] |
| ULBP2, NAPA, TGFBI, RAB14, ULBP2, CP, RPL22, PURB, C1S, ANXA11, ERO1L | Chang Y.T. et al. 2011 [44] |
| ALCAM, ANG, AXL, BAG3, BSG, CCL24, CEACAM5, CEACAM1, CLU, COL18A1, EPCAM, HP, ICAM1, IGFBP2, IGFBP4, LCN2, LRG1, MMP2, MMP7, MMP9, MSLN, PARK7, PF4, PPBP, PRG4, SPARCL1, SPP1, TGFBI, THBS1, TIMP1, TNFRSF1A, VEGF | Firpo M.A. et al. 2023 [45] |
| THBS2, IGFBP2, IGF1, ENPP2, LRG1, TTR, APOE, ITIH3, APOA1, APOL1, TFF1, TFF2, TFF3, GDF15 | Kapszewicz M. et al. 2021 [46] |
| Term | p-Value | q-Value | Overlaps Genes |
|---|---|---|---|
| Extracellular Matrix Disassembly (GO:0022617) | 7.05 × 10−9 | 2.09 × 10−6 | [MMP12, PRSS1, GSN, MMP7, MMP2, MMP9, MMP10] |
| Regulation of Apoptotic Process (GO:0042981) | 2.42 × 10−11 | 3.59 × 10−8 | [HSP90AA1, GSTP1, ANXA5, PARK7, IGF1, CLU, MMP9, THBS1, ACTB, NME1, LGALS1, AXL, BAG3, CEACAM5, ARHGDIA, CFL1, ALB, PPT1, FLNA, CALR, PPIA, CTSD, TGM2] |
| Neutrophil Chemotaxis (GO:0030593) | 1.94 × 10−7 | 2.88 × 10−5 | [LGALS3, CCL24, CXCL8, SAA1, PPBP, PPIA, PF4] |
| Carbohydrate Catabolic Process (GO:0016052) | 4.19 × 10−9 | 1.56 × 10−6 | [LDHA, TPI1, PKM, PGAM1, PGK1, ENO1, ENO2] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cammarota, A.L.; Falco, A.; Basile, A.; Molino, C.; Chetta, M.; D’Angelo, G.; Marzullo, L.; De Marco, M.; Turco, M.C.; Rosati, A. Pancreatic Cancer-Secreted Proteins: Targeting Their Functions in Tumor Microenvironment. Cancers 2023, 15, 4825. https://doi.org/10.3390/cancers15194825
Cammarota AL, Falco A, Basile A, Molino C, Chetta M, D’Angelo G, Marzullo L, De Marco M, Turco MC, Rosati A. Pancreatic Cancer-Secreted Proteins: Targeting Their Functions in Tumor Microenvironment. Cancers. 2023; 15(19):4825. https://doi.org/10.3390/cancers15194825
Chicago/Turabian StyleCammarota, Anna Lisa, Antonia Falco, Anna Basile, Carlo Molino, Massimiliano Chetta, Gianni D’Angelo, Liberato Marzullo, Margot De Marco, Maria Caterina Turco, and Alessandra Rosati. 2023. "Pancreatic Cancer-Secreted Proteins: Targeting Their Functions in Tumor Microenvironment" Cancers 15, no. 19: 4825. https://doi.org/10.3390/cancers15194825
APA StyleCammarota, A. L., Falco, A., Basile, A., Molino, C., Chetta, M., D’Angelo, G., Marzullo, L., De Marco, M., Turco, M. C., & Rosati, A. (2023). Pancreatic Cancer-Secreted Proteins: Targeting Their Functions in Tumor Microenvironment. Cancers, 15(19), 4825. https://doi.org/10.3390/cancers15194825









