Simple Summary
Colorectal cancer (CRC) patients with peritoneal metastases (PM) have a poor prognosis. Currently, research has been ongoing to develop new treatment options for CRC patients with PM. DNA/RNA alterations identification in the primary tumor might help identify patients who are at high risk for developing PM postoperatively. These patients could benefit from preventive or early treatment. The aim of this systematic review is to create an overview of studies which analyzed genomic DNA and RNA expression alteration correlated to PM with the goal of identifying potentially predictive biomarkers. We included 32 studies investigating primary colorectal tumors of 18,906 patients. Only BRAF mutations were reported as significantly associated with PM in 10 of 17 studies. As no specific biomarkers in the primary tumors of CRC patients could have been identified, further research with comprehensive genomic profiling is still desirable.
Abstract
Background: As colorectal cancer (CRC) patients with peritoneal metastases (PM) have a poor prognosis, new treatment options are currently being investigated for CRC patients. Specific biomarkers in the primary tumor could serve as a prediction tool to estimate the risk of distant metastatic spread. This would help identify patients eligible for early treatment. Aim: To give an overview of previously studied DNA and RNA alterations in the primary tumor correlated to colorectal PM and investigate which gene mutations should be further studied. Methods: A systematic review of all published studies reporting genomic analyses on the primary tissue of CRC tumors in relation to PM was undertaken according to PRISMA guidelines. Results: Overall, 32 studies with 18,906 patients were included. BRAF mutations were analyzed in 17 articles, of which 10 found a significant association with PM. For all other reported genes, no association with PM was found. Two analyses with broader cancer panels did not reveal any new biomarkers. Conclusion: An association of specific biomarkers in the primary tumors of CRC patients with metastatic spread into peritoneum could not be proven. The role of BRAF mutations should be further investigated. In addition, studies searching for potential novel biomarkers are still required.
1. Introduction
Colorectal cancer (CRC) is the third most prevalent type of cancer worldwide and a common cause of morbidity and mortality, which is generally attributable to metastatic disease [1,2]. At initial diagnosis, almost one-fourth of patients with CRC present with synchronous metastases [2,3]. Liver metastases (LM) occur most frequently, followed by peritoneal metastases (PM) [2,4]. Colorectal PM are found in 5–15% of patients at primary diagnosis (synchronous PM) [2,4,5,6]. One can also develop PM after curative resection of the primary tumor (metachronous PM), usually within the first 3 years after the primary diagnosis [3]. Metachronous PM are reported in 4–12% of colon cancer patients and in 2–19% of rectal cancer patients [4,6]. However, the true incidence of PM might be underestimated. The preoperative diagnosis is mostly made by CT scan, but this has limited diagnostic accuracy for the assessment of the extent of PM [2,6,7].
CRC patients with PM have a poor prognosis. Currently, the only potentially life-prolonging treatment option involves surgical debulking of all visible metastases (cytoreductive surgery; CRS) followed by Hyperthermic Intraperitoneal Chemotherapy (HIPEC). Only a highly selected group of patients are eligible for this intervention. Patients with a poor physical condition and/or a too extensive metastatic disease are generally excluded and will undergo palliative systemic treatment or best supportive care only [2,8,9]. Without any treatment, the average life expectancy is 6 to 12 months after diagnosis [5,8,10].
Recently, research has been ongoing to develop new treatment options for locally advanced CRC patients [11]. Since these new treatment techniques could be invasive to a certain degree and be expensive, it would not be desirable to implement these routinely for all patients. A diagnostic tool able to identify patients who are at high risk of developing metachronous PM would allow targeted treatment in a preventive and/or curative setting [2]. According to previous research, a molecular profile of the primary tumor might help identify patients who are at high risk. It is hypothesized that specific biomarkers identified in the primary tumor can be incorporated in a prediction tool to estimate the risk of distant metastatic spread [12,13]. In patients with synchronous PM, genetic alterations could be interesting to determine prognosis or to predict response to therapy.
It is known that several pathogenic mutations occur during adenoma-to-carcinoma transformation in CRC. Important oncogenes are adenomatous polyposis coli (APC), tumor suppressor gene TP53, Kirsten rat sarcoma virus (KRAS), transforming growth factor beta (TGF-β), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) [14,15]. Recent data suggest mutations may also affect the metastatic dissemination of tumors [16]. Different omics techniques, such as genomics (e.g., next-generation sequencing (NGS), polymerase chain reaction (PCR), pyrosequencing (PS), Sanger sequencing (SS)) and transcriptomics (e.g., NGS), could be used to elucidate DNA markers and RNA transcripts, respectively. Furthermore, individual omics techniques can be integrated into multi-omics analyses, which capture the complexity of diseases on multiple levels. As sequencing technologies have become less expensive, tumor genotyping has become standard practice for metastatic CRC (mCRC) [14,16]. As a result, clinicians now often have information on the mutational status of several oncogenes, and investigating molecular changes in primary tumors concerning metastatic potential is becoming more common [16,17]. We hypothesize that specific biomarkers, based on DNA/RNA alterations identified in the primary tumor, might characterize colorectal PM patients. Once identified, these alterations can be incorporated into a prediction tool to estimate the risk of PM development, prognosis, and be helpful in choosing the appropriate treatment options [12,13].
In this paper, the authors aim to systematically review the available literature to: (1) create an overview of previously investigated DNA and RNA alterations in the primary tumor correlated to colorectal PM and (2) investigate which gene mutations are of potential biomarker value and should be further studied. This study focuses solely on CRC (stages I–IV) and does not include other types of neoplasms.
2. Methods
2.1. Study Protocol and Registration
This systematic review was conducted and reported according to the guidelines of the “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA) [18]. The study protocol was registered at PROSPERO (registration number CRD42021297366).
2.2. Search and Information Sources
A literature search was performed on the 6 January 2022 and repeated before submission on the 3rd of November 2022. PubMed, Embase, the Cochrane Library, and CINAHL Database were searched with the use of MeSH-, Emtree-, and free terms including “colorectal neoplasms”, “peritoneal neoplasms”, “mutations”, “genetic testing”, “genetic association studies”, “gene expression profiling” and “biomarkers, tumor” and additional search terms such as “colorectal”, “adenocarcinoma”, “carcinomatosis” and “predictive biomarker”. The full search strategy is displayed in Appendix A. A professional clinical librarian was involved to ensure an appropriate search strategy. Reference lists of all relevant publications were hand-searched for additional studies. This method of cross-referencing was continued until no further relevant publications were identified.
2.3. Selection Process
2.3.1. Inclusion and Exclusion Criteria
Articles containing original data concerning genomic analyses on patients with CRC and PM were considered eligible. The primary outcome measure was specific mutations on the DNA or RNA level in the primary colorectal tumor that might be associated with PM. Studies were excluded if the tumor samples were not from primary tumor tissue origin or if the researchers only investigated metastases other than peritoneal ones. The method of genomic analysis was not a criterion for exclusion. Secondary sources such as technical descriptions, letters to the editor, conference proceedings, and commentaries were not considered. Only articles in English, Dutch, French, Italian, or German were eligible.
2.3.2. Study Selection
All search results were imported into a free web tool designed for systematic reviewers (Rayyan) [19]. All duplicates were removed. The screening of studies for eligibility was performed by two reviewers (DH, JL) independently, using the predefined inclusion and exclusion criteria. First, articles were screened based on title and abstract. Disagreements between reviewers were resolved by initial discussion to create consensus. If the eligibility criteria were met after full-text screening by both reviewers, article inclusion followed. All references were stored in the Endnote Reference Management Tool.
2.3.3. Data Items and Collection Process
Two reviewers (DH, JL) independently extracted data from the text, tables, and figures in a standardized, predefined datasheet. Data extraction for each article included first author, year of publication, country, study design, study period, inclusion and exclusion criteria, aim of the study, number of patients and genes, general patient information, methods of genomic analysis, methods of tissue collection and sample information, and outcome of genetic analysis. Data acquired via the outlined search strategy are summarized in tables.
2.3.4. Study Risk of Bias Assessment
To assess the validity of the included studies, the bias risk was assessed independently by two reviewers (DH, JL). Since there is no standard bias assessment tool for the type of included studies, a suitable tool was designed based on the Risk of Bias using the Quality In Prognosis Studies (QUIPS) tool. All types of bias were evaluated and judged as low, moderate, or high risk.
3. Results
3.1. Study Selection
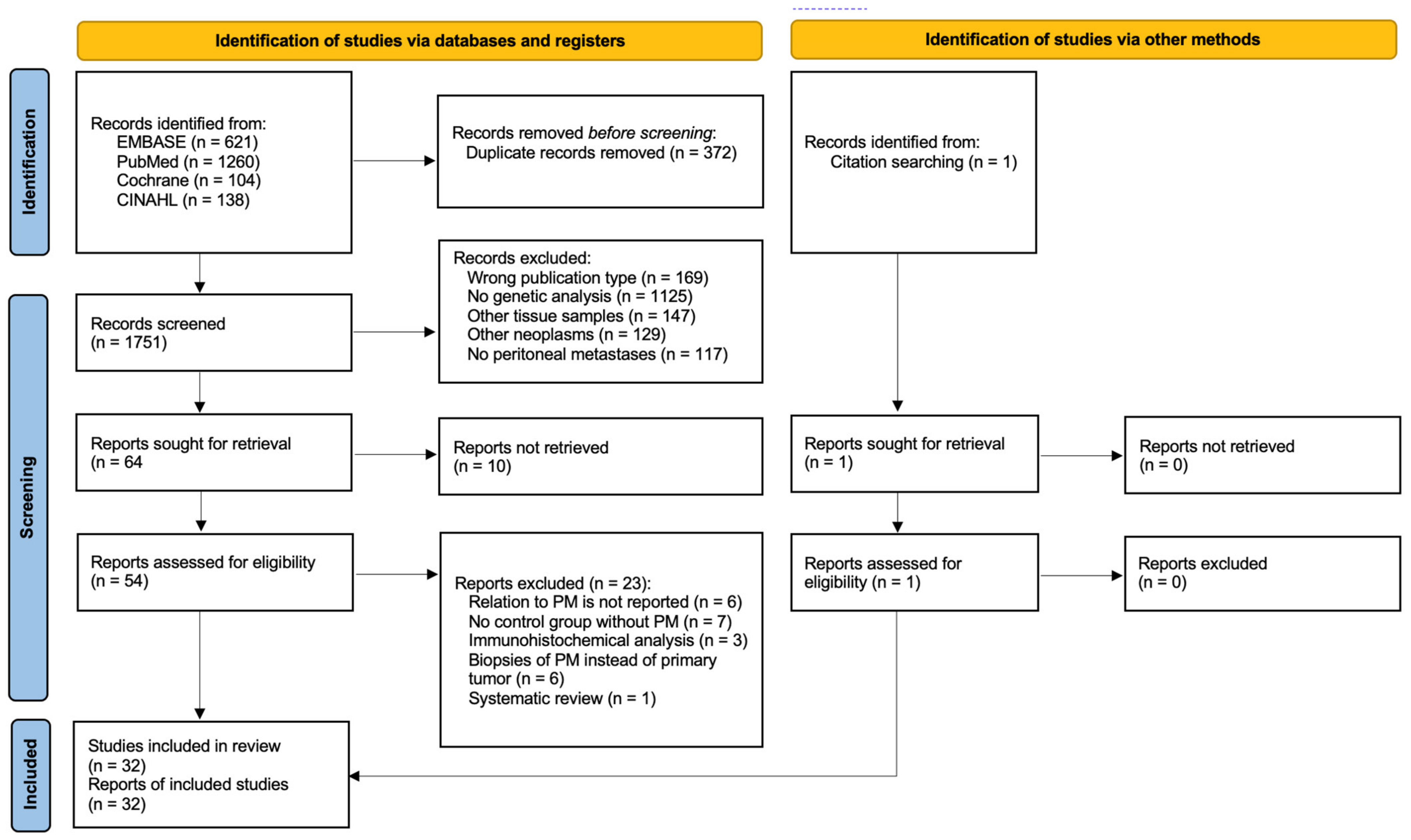
The electronic search yielded 1751 articles after removing duplicates. After abstract reading, 64 potentially eligible articles remained, based on the predefined inclusion and exclusion criteria. Full-text assessment from ten articles was not possible (e.g., language restrictions, congress submissions), whereafter 54 articles remained eligible. Reference checking resulted in one additional study, attaining 55 articles for full-text assessment. As 23 articles did not fulfill inclusion criteria, 32 studies were included for final analysis. No additional publications were identified after repeating the search before submission. The study selection process is summarized in Figure 1.
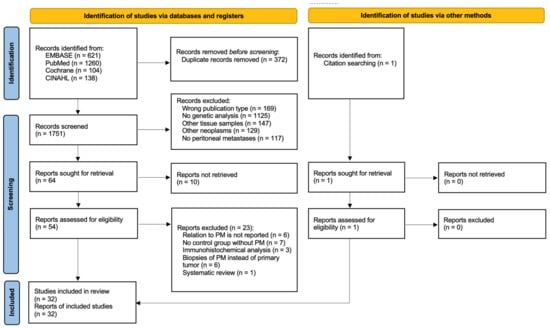
Figure 1.
PRISMA flowchart outlining study selection strategy [18].
3.2. Study Characteristics
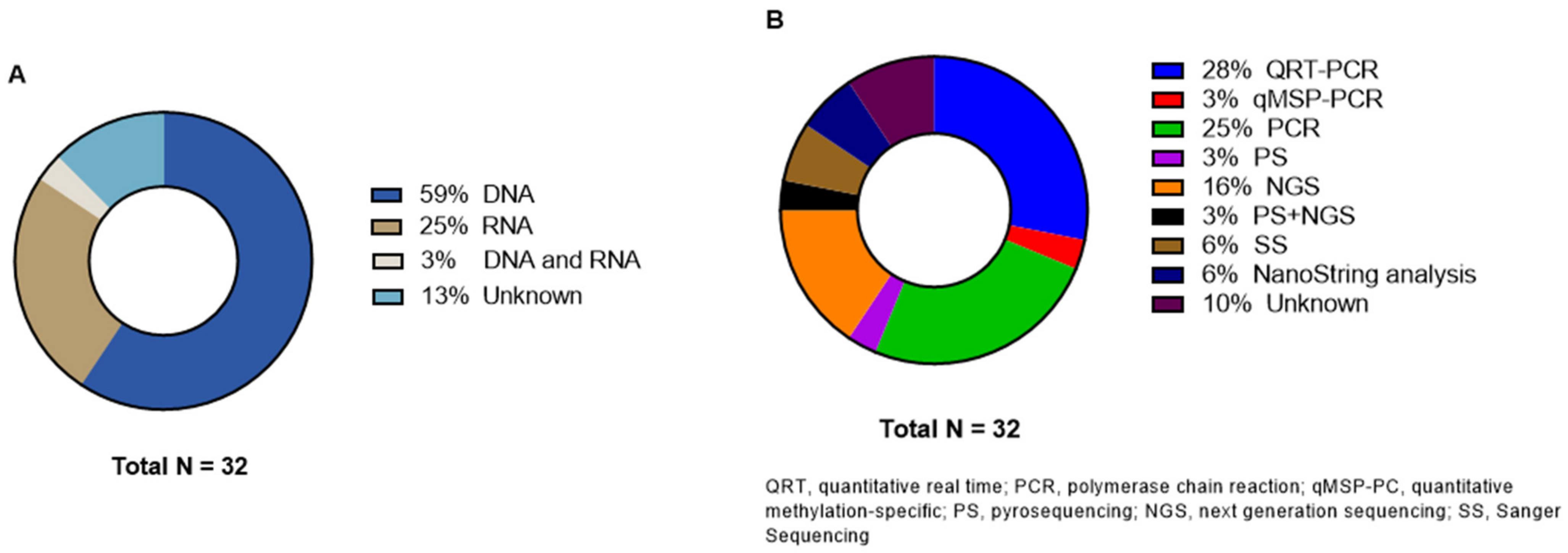
All 32 studies are observational cohort or case control studies published between 2008 and 2021. The number of subjects per study ranged from 15 to 5967, with a total of 18,906 patients. The main characteristics of the included studies are summarized in Table 1. In 21 studies, the tissue samples were retrieved retrospectively [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40]. Two studies collected tissue samples at time of surgical resection [41,42], and in nine studies, there was no need for tissue collection because the mutation status of genes of interest had already been analyzed as part of diagnostic reasons [43,44,45,46,47,48,49,50,51]. Most tissue samples used in the studies (n = 24) were formalin-fixed paraffin-embedded (FFPE), while the remaining eight studies used fresh frozen tumor samples [22,31,32,34,38,40,41,42]. All characteristics of patients and tissue samples are summarized in Table 2. Only two articles reported the time of PM occurrence, i.e., metachronous or synchronous metastases [23,44]. All other studies did not specify the time of onset of PM or only included synchronous metastases. Because of the heterogeneity among the included studies in terms of the study population, genetic analyses methods, level of genetic testing, and (number of) genes, pooling in a meta-analysis was not possible.

Table 1.
Characteristics of included studies.

Table 2.
Characteristics of patients and tissue samples in included studies.
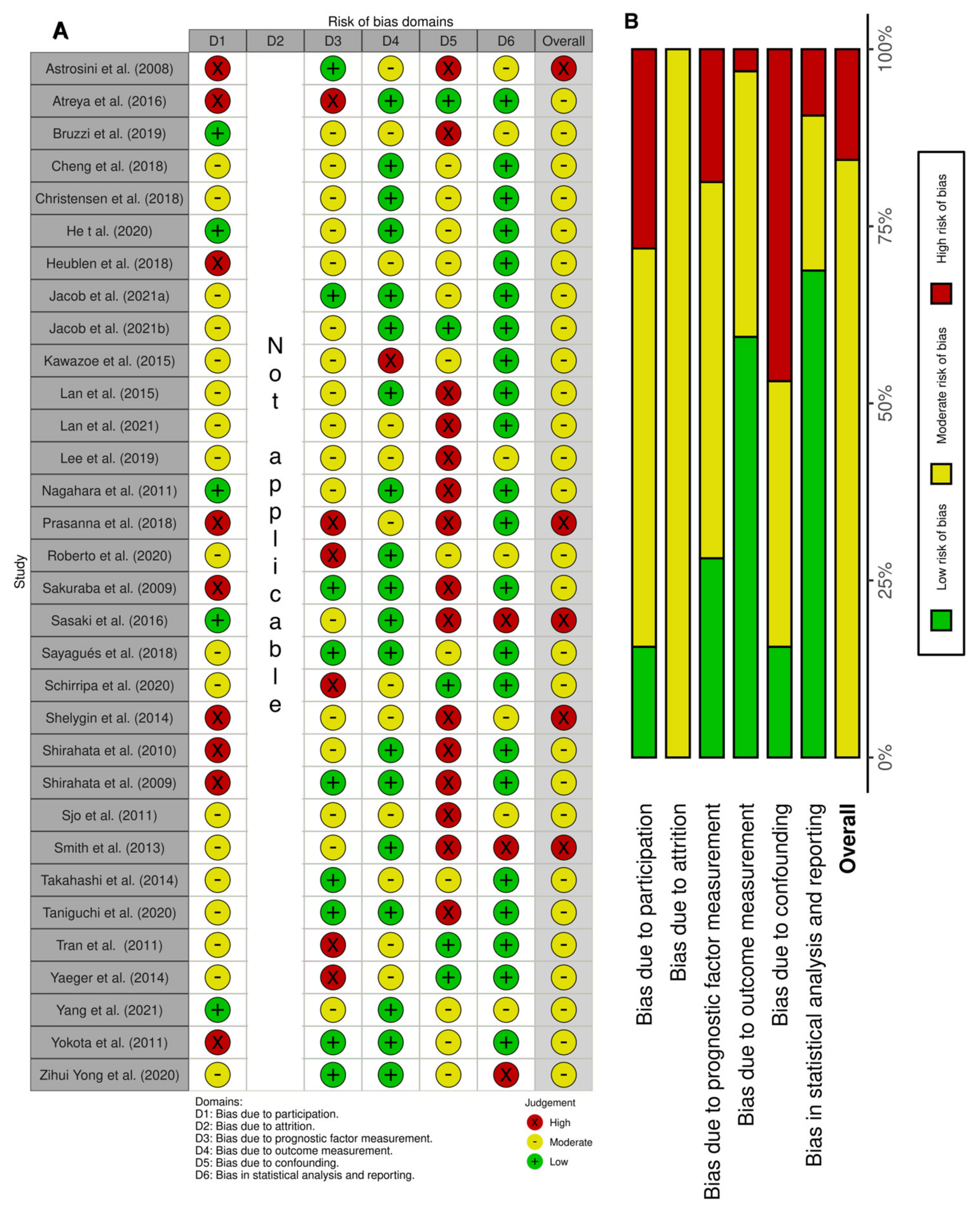
3.3. Risk of Bias in Studies
The relevant categories from the QUIPS tool were used to access the risk of bias; a score per domain per study is presented in Figure 2A. We reported a high risk of bias for five studies [20,33,35,37,45] and a moderate risk of bias for all other studies. The overall lowest risk of bias was found in the statistical and outcome measurement domains, while the highest was found in the confounding domain (Figure 2B).
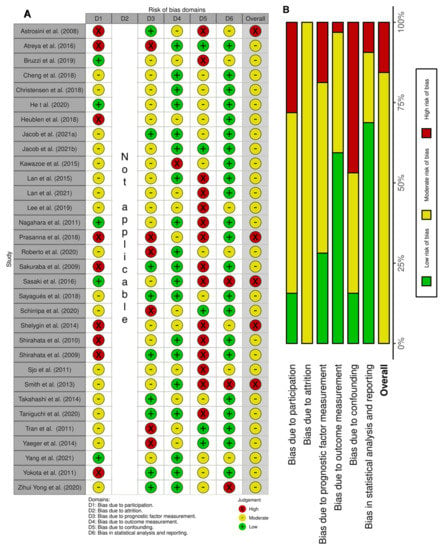
Figure 2.
Risk of bias based on the QUIPS tool. (A) Summary of the domain–level judgements for each study. (B) Risk–of–bias judgements within each bias domain.
3.4. Reported Genes
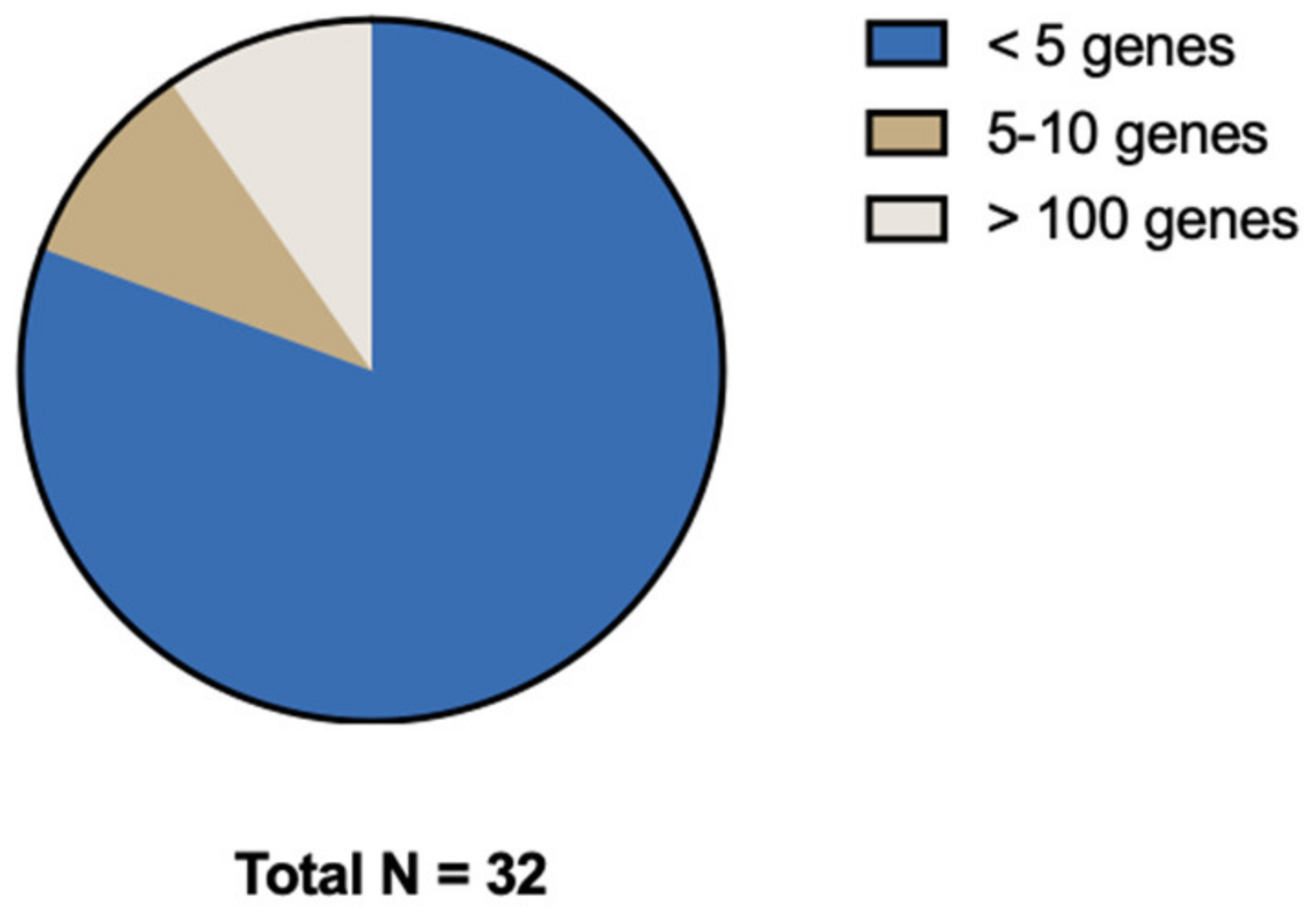
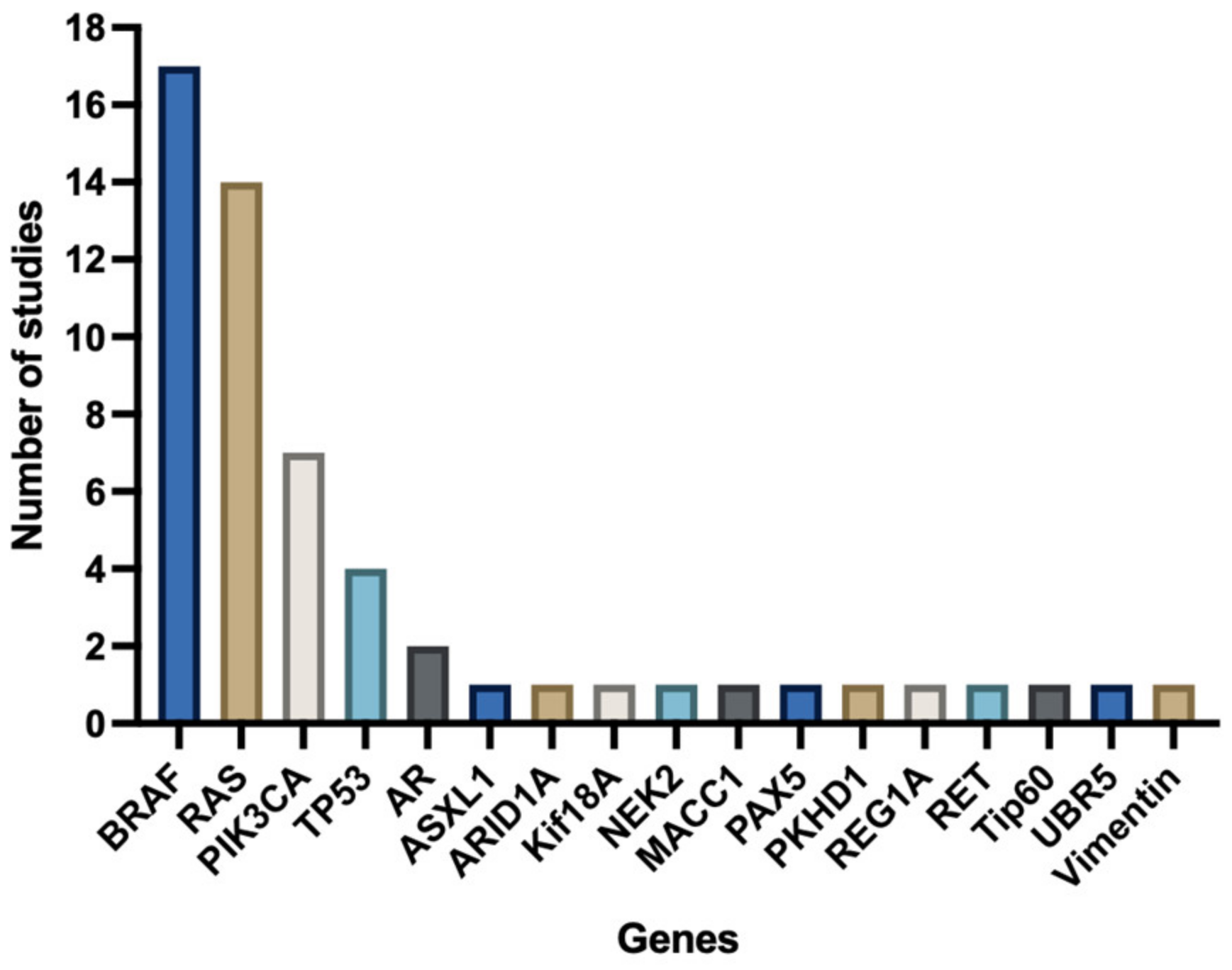
Most studies focused on a selected predefined group of genes (Figure 3). Genes that were predominantly studied were RAS (KRAS/NRAS), PIK3CA, TP53, and BRAF. The remaining 13 genes (e.g., androgen receptor (AR), ASXL Transcriptional Regulator 1 (ASXL1), AT-Rich Interaction Domain 1A (ARID1A), NIMA Related Kinase 2 (NEK2), MET Transcriptional Regulator MACC1 (MACC1), Paired Box 5 (PAX5), Ubiquitin Protein Ligase E3 Component N-Recognin 5 (UBR5), Vimentin, Ret Proto-Oncogene (RET), Histone acetyltransferase (Tip60), PKHD1 Ciliary IPT Domain Containing Fibrocystin/Polyductin (PKHD1), Regenerating Family Member 1 Alpha (REG1A), and Kinesin Family Member 18A (Kif18A)) were, except for ARID1A, all separately examined by individual studies (Figure 4). Three studies did a broader comprehensive genomic analysis on the tissue samples. Jacob et al. performed a PanCancer Progression Panel in 2 studies [25,26] including 770 genes, and Lee et al. used a Comprehensive Cancer Panel covering 409 genes [30]. All details about the reported genes are displayed in Table 3.
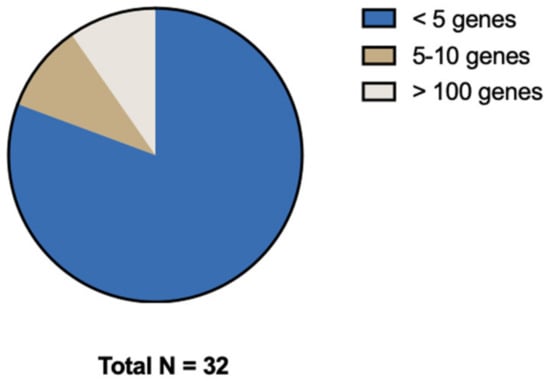
Figure 3.
Distribution of number of genes investigated.
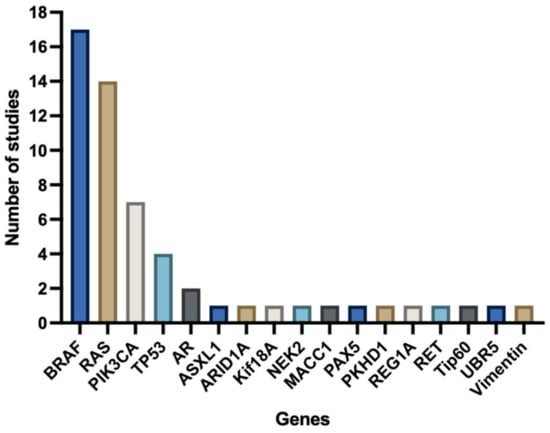
Figure 4.
Number of studies investigating specific genes.

Table 3.
Overview of genetic analysis and outcomes.
3.5. Genetic Analysis Methods
Primary tumor genetic analysis was performed on DNA level in 19 studies and on RNA level in seven studies (Figure 5). One study described the analysis on both levels [35]. Heublein et al. investigated MicroRNAs (miRNA) and the corresponding overexpression profiles [24]. Four articles did not specify if they performed testing on DNA or RNA level [43,46,47,52]; three of these articles did not even specify which method they used for genetic testing [43,46,52]. Nine articles reported the use of real-time polymerase chain reaction (RT-PCR) [20,21,24,31,32,35,38,42,48]. One study specified the PCR tool as quantitative methylation-specific PCR (qMSP) [41]. PS and NGS were used in one [37] and five [23,28,30,34,50] studies, respectively. Christensen et al. reported the use of both methods [44]. Two studies analyzed the samples with SS [47,49]. Jacob et al. described NanoString analysis in both their articles [25,26]. All details about the genetic analyses are displayed in Table 3.
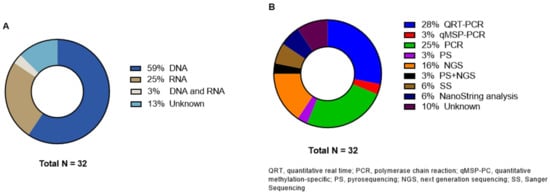
Figure 5.
Distribution of (A) genetic analysis level and (B) different molecular techniques.
3.6. DNA/RNA Alterations Outcomes and Association with PM
All details about the reported alterations are displayed in Table 3.
3.6.1. Mitogen-Activated Protein Kinase (MAPK) Pathway Outcomes
BRAF and RAS are both involved in the MAPK pathway and were most commonly reported. BRAF mutations were analyzed in 17 articles [21,22,23,27,29,33,34,35,37,39,40,43,44,45,46,48,49]. In ten studies, it was found on a statistically significant level that BRAF mutant tumors were more likely to develop PM and/or that patients with PM had more often BRAF mutated primary tumors compared to PM-free CRC patients [22,27,33,34,39,40,45,46,48,49]. Most studies conducted the BRAF mutation analysis on codon 600, exon 15 (n = 12). Taniguchi et al. reported that the frequencies of BRAF mutations, in combination with RAS wild-type (WT) tumors, were significantly higher in CRC patients with PM [39]. Smith et al. showed a statistically significant association when BRAF status in unresectable CRC patients with PM was compared to other metastatic sites. This result, however, did not remain significant after a post hoc Bonferroni correction [37]. The authors also mention that BRAF mutations were significantly more common in patients with peritoneal-only metastases compared to patients with liver-only metastases. This, however, did not withstand a correction for multiple testing [37]. Atreya et al. and Bruzzi et al. reported no statistically significant difference in metastatic sites and BRAF mutation, although PM were more commonly observed in patients whose tumors harbored a BRAF mutation [21,43]. He et al. investigated therapy-naïve synchronous mCRC patients and found no significant differences in mutation status [23]. Shelygin et al. found no association between PM and BRAF status when comparing patients, with and without PM, undergoing surgery for CRC [35]. Christensen et al. looked at the probability of developing PM while having a BRAF mutated tumor. The hazard ratio for developing PM and having a BRAF-mutated tumor was statistically not significant [44]. One article did not report any data about BRAF mutations and its relation to PM, although they intended to investigate this [28].
RAS pathway mutation analyses were reported in 14 studies. Seven studies focused on both KRAS and NRAS genes [21,27,28,29,34,37,44], and the other seven studies only described KRAS variants [23,33,35,40,45,47,51]. Lan et al. reported that the proportion of PM was significantly higher in stage I–IV CRC patients whose tumors carried a RAS pathway mutation, and KRAS-mutated tumors had a trend toward a higher proportion of PM, which was not significant [28]. Both Zihui Yong et al. and He et al. found a significant association between KRAS mutant tumors and PM [23,51]. He et al. also stated that therapy-naïve synchronous PM patients tend to carry a mutant KRAS codon 12 [23]. One article did not report any outcomes, although they aimed to do so [28]. All other studies did not find a significant association or trend between KRAS/NRAS mutant tumors and the development of PM [21,27,33,34,35,37,40,44,45,47].
To conclude, most articles (n = 10/17) state that BRAF mutant tumors are more likely to have PM and/or mutations in BRAF were more common in patients with PM compared to those without. Almost all articles (n = 10/14) state that RAS pathway mutated tumors are not likely to have PM and were not more common in patients with PM compared to without PM.
3.6.2. PIK3CA Outcomes
The potential association of PIK3CA mutations with PM was analyzed in seven studies. In five studies, the PIK3CA mutations were not significantly associated with PM [28,33,35,37,44]. Christensen et al. even found that PIK3CA mutations were associated with the absence of PM and a decreased hazard of developing PM (HR = 0.31; 95%CI = 0.11–0.86, p = 0.024) in mCRC patients who had received chemo- or immunotherapy treatments [44]. Two studies did not report any outcomes, although PIK3CA mutations were investigated [29,39].
3.6.3. TP53 Outcomes
TP53 mutations were analyzed in four studies. Two studies showed a significant association between PM and TP53 mutations. Lee et al. detected more TP53 mutations in patients with small obstructive CRC with PM compared to large non-obstructive tumors without PM [30]. Sjo et al. performed a multivariate analysis in stage IV CRC patients and showed that PM was significantly associated with TP53 mutations [36]. Lan et al. stated that stage IV CRC patients with PM had a higher frequency of TP53 mutations, although the authors did not perform statistical analysis on this association [29]. Sayagués et al. did not find a significant association between TP53 mutational status and PM in Caucasian patients diagnosed with CRC [34].
3.6.4. Other DNA Outcomes
AR, ASXL1, ARID1A, Kif18A, NEK2, MACC1, PAX5, PKHD1, REG1A, RET, Tip60, and UBR5 were mentioned as possible mutated genes associated with PM by several authors [20,29,30,31,32,38,41,50] but were, except for ARID1A, all investigated in only one study. NGS was performed by Yang et al. to detect RET mutations in mCRC without neoadjuvant treatment [50]. The presence of RET mutations was significantly associated with PM compared to WT tumors. Tip60 regulation analysis was performed with RT-PCR in patients undergoing surgery for CRC by Sakuraba et al. [32]. The authors found that a downregulation of Tip60 was significantly associated with PM. To conclude, all previous mentioned genes showed a significant association with PM, but all were studied by a single study only.
3.6.5. RNA Outcomes
Nagahara et al. report that Kif18A overexpression, measured by RT-PCR, in CRC patients without neoadjuvant treatment significantly correlates with PM [31]. The expression profile of NEK2 was analyzed by Takahashi et al. in patients with CRC who underwent surgical treatment [38], demonstrating that the high NEK2 expression group had significantly greater peritoneal dissemination compared to the low expression group. MACC1 expression was found to be significantly associated with PM by Shirahata et al. [42]. The expression of REG1A was explored in non-pretreated CRC patients by Astrosini et al. and showed a positive e correlation with the formation of PM [20]. In addition, Heublein et al. analyzed MicroRNAs (miRNAs) expression profiles and concluded that hsa-mri-31-5p seems to be overexpressed in patients with PM [24]. The authors reported a set of 31 miRNAs which were significantly upregulated in the PM group, while ten miRNAs were found to be repressed as compared to LM. Another set of two miRNAs was significantly upregulated in the PM group, while 25 were found to be repressed as compared to no metastases. Shirahata et al. discovered a trend toward preferentially developing PM in tumors with Vimentin methylation, although this was not significant [41].
3.6.6. Results of Broader Panel Analyses
Lee et al. performed a broader panel analysis of which the results (ARID1A, PKHD1, UBR5, PAX5, TP53, ASXL1 and AR) are already described in Section 3.6.4 [30]. Jacob et al. explored gene expression profiles with a broad cancer “panel” comparing four groups (without metastases, with LM, with PM, and with both LM and PM) [25]. They report that “18 genes had significantly different expression rates”, but they did not describe which genes. In another study, in which three groups were compared (without metastases, with LM, and with PM), the authors reported no significant down- or upregulation of distinct gene sets [26].
All details about the reported genes and corresponding conclusions are described in Supplementary Table S1. A conclusive summary for all genes is displayed in Figure 6.
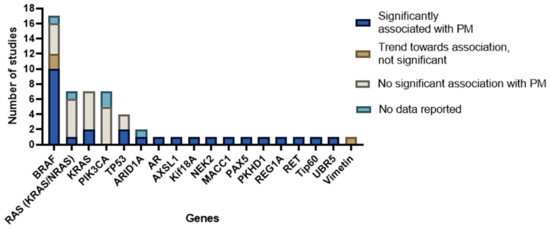
Figure 6.
Overview of genes investigated with conclusions formulated by the authors of included studies.
3.7. MSI Status
In addition to DNA and RNA alterations, microsatellite instability (MSI) status was reported in ten articles [21,28,34,35,37,43,45,46,48,50]. Tran et al. describe the impact of BRAF mutations in combination with MSI status on the pattern of metastatic spread and its prognosis [48]. The authors report that patients with MSI tumors show poorer survival in mCRC, and this is due to the association with BRAF mutations. Yang et al. state that MSI is associated with RET mutations [50].
4. Discussion
This systematic review provides an overview of the results of studies which analyzed genomic DNA and RNA expression alterations correlated to PM with the goal of identifying alterations that could potentially serve as a predictive biomarker in patients with CRC. Of the 17 studies investigating BRAF mutations, ten studies reported a significant association with PM. Mutations in ARID1A, ASXL1, Kif18A, NEK2, MACC1, PAX5, PKHD1, REG1A, RET, Tip60 and UBR5 were also reported to be associated with PM [20,29,30,31,32,38,41,50], although these results were only described in maximum of one study. A recent analysis with a cancer panel of 770 genes from Jacob et al. did not show a significant down- or upregulation of distinct gene sets between CRC patients with PM and without distant metastases. Their sample size was, however, small (n = 18) [26].
4.1. BRAF Mutations
BRAF gene mutations occur in 5–15% of the mCRC cases; over 95% of these mutations consist of a substitution of valine to glutamic acid at codon 600 (V600E) [13,16,53]. BRAF is a serine/threonine protein kinase that plays an important role in the MAPK pathway. This pathway drives cell proliferation, differentiation, migration, survival, and angiogenesis, and therefore, changes in this pathway are associated with tumorigenesis [54]. BRAF mutations can be considered as an independent negative prognostic factor in early-stage microsatellite stable tumors and as a negative predictive factor for therapeutic approaches [54]. Due to its chemoresistance and resistance to BRAF inhibitor therapy, BRAF-mutated tumors are difficult to treat [54,55]. Therefore, trials are currently going on with dual or triple drug therapy to enhance blockade of the MAPK pathway. Nowadays, CRC patients without metastases are not screened for BRAF mutations, and further molecular examination is only conducted in metastatic disease [56]. As only 55% of the studies reported a significant association between BRAF mutations and PM, we cannot conclude yet that BRAF mutations are specific enough to identify patients with colorectal PM.
4.2. Other Mutations
First, RAS pathway mutations are the most commonly investigated mutations in mCRC. Different codons of both KRAS and NRAS genes were included, thereby creating a broader overview of this pathway. KRAS is the most commonly activated oncogene in CRC, with mutations occurring in exon 2 codon 12 and 13, exon 3 codon 59 and 61, and exon 4 codon 117 and 146 [16,57]. Approximately 30–50% of the CRC patients carry a somatic KRAS mutation [16]. KRAS mutations have been associated with lung metastases but not with PM [16]. NRAS is mutually exclusive with BRAF and KRAS and occurs in approximately 3% of CRC patients [16]. There has been no previously described association with PM, which is in line with the findings of this review. Second, PIK3CA (exon 9 and 20) gene mutations occur in 10–18% of CRC patients [53]. They commonly co-occur with KRAS or BRAF mutations. Approximately 70% of PIK3CA mutant patients have concurrent mutations [16,58], although they have never been described to be associated with PM. The results of our study demonstrate this as well. Third, TP53 gene mutations are one of the most frequently described mutations as they occur in 35–75% of the colorectal PM patients [13]. Previous research shows the contradictory result of TP53 mutations and their prognostic value in CRC patients [53]. In this review, some authors showed a significant association, while others did not reach the significance.
4.3. MSI Status
Of the included studies, only 10 articles reported on MSI status, all without extensive analysis. This is unfortunate, as MSI status is the only prognostic molecular marker used in deciding adjuvant therapy options [56]. MSI originates from the inactivation of mismatch repair genes by either MLH1 hypermethylation or mutation. This results in the accumulation of somatic mutations and subsequent genomic instability, which is associated with nonhereditary CRC [53]. It is well reported that MSI is a good prognostic factor for some treatments in early-stage CRC [59]. We believe it is important to always report MSI status in biomarker research to incorporate all relevant characteristics.
4.4. Clinical Relevancy
Clinically, the known risk factors for metachronous colorectal PM are an advanced tumor stage, right-sided tumor, infiltrative or ulcero-infiltrative tumors, history of perforation, and obstruction [3,8,60]. A randomized trial (COLOPEC-1) investigating the therapeutic effectiveness of adjuvant HIPEC to prevent PM development in high-risk CRC patients showed that this treatment strategy did not improve PM-free survival [11]. In contrast, a Spanish study by Arjona-Sánchez et al. concluded that adjuvant HIPEC therapy might be useful in patients with T4 tumors [61]. Identifying genetic alterations in high-risk metachronous PM patients may have additional benefit on improving survival by additional targeted therapies such as adjuvant HIPEC. In synchronous PM patients, the alterations provide added value to determine prognosis or to predict response to therapy. For example, RAS pathway activating mutations are negative predictive markers for the efficacy of anti-epidermal growth factor receptor (EGFR) therapies [62], while MSI tumors with BRAF and PIK3CA mutations show survival benefit [39]. For CRS and HIPEC scheduled patients, a BRAF mutation is a marker for poor prognosis, whereas KRAS tumors do not influence the outcomes [63]. The choice of cytostatic in HIPEC can be based on mutation status, or specific therapy can be developed in the case of targetable mutations.
Unfortunately, most of the studies did not clearly specify whether the authors were using tumors from synchronous or metachronous PM patients. It was therefore hard to distinguish and separate these two scenarios in the results. Future studies should clearly specify the time of metastases onset, the aim of the genetic analysis, and clinical implications.
4.5. Techniques
In the studies evaluated in this review, several different genetic research techniques were applied. Since most studies used targeted PCR techniques to detect specific gene mutations, the number of studies that used comprehensive genetic analyses was scarce. The development and use of NGS technologies have revolutionized the speed and throughput of DNA and RNA sequencing [64,65]. However, since the number of relevant cancer genes guiding targeted therapy in CRC is still limited and costs per sample are substantial, NGS sequencing is not yet commonly used in clinical decision making or limited to mutation hotspot target regions [66]. This has most likely influenced the research to unmap PM predictive biomarkers so far, and we believe that more comprehensive NGS analyses are needed for this purpose. When we critically look at the choice of techniques used in the included studies, we believe these were too restricted to identify DNA/RNA biomarkers in the primary tumor of CRC patients with synchronous or metachronous PM.
As mCRC is a highly complex genetic disease, an understanding of how all aspects interact is required to achieve the prediction and treatment of colorectal PM. Single target techniques, mostly used in the included articles in this paper, might be insufficient for this purpose. We believe that omics techniques (i.e., techniques that generate high-throughput data [67]) might be a promising method for new CRC biomarkers research instead of most of the methods used in this paper. The integration of multiple omics techniques, by combining genomic data with data from other modalities such as transcriptomics, epigenetics, and proteomics, to measure gene expression, gene activation, and protein levels, could be helpful to reveal this problem in further research. This integration might bring us much closer to the prediction, prevention and tailored treatment of PM in CRC [68].
4.6. Limitations
This is the first systematic literature review of DNA/RNA biomarkers in relation to colorectal PM to the author’s knowledge. This study has also some limitations. First, almost all included studies were retrospective with a different number of patients and different patients’ characteristics (T-stage, number of metastatic sites, treatments, etc.). Second, comparisons between the studies are limited due to heterogeneity, and a meta-analysis was therefore not possible to perform. The standardization of techniques and analysis and more insight in the individual analysis outcomes via FAIR data sharing would be helpful. Third, most studies focused on the most commonly analyzed CRC target genes, i.e., KRAS, NRAS, BRAF, PIK3CA, and TP53 with simple sequencing methods and PCR technology. Only three studies performed a broader gene panel NGS analysis. Fourth, most of the included studies did not report if CRC patients received neoadjuvant systemic treatments and if they did, which type. Such treatments could namely affect the outcomes of the genetic analysis. Fifth, most of the studies lacked the MSI of the CRCs. Sixth, all studies showed a moderate to high risk of bias with a high risk for the confounding domain.
4.7. Future Perspectives
We believe the use of comprehensive genomic profiling with for example broader cancer gene panels is essential to identify new potential cancer genes for PM prediction. In addition to using an optimal technique, we recommend applying these in a homogenous patient population (e.g., strict synchronous or metachronous PM patients, tumor characteristics, etc.).
5. Conclusions
Increasing amount of data suggest that the presence of biomarkers in the primary tumor might have an impact on metastatic patterns. However, unfortunately, based on the given evidence, we cannot consider the genes (e.g., BRAF) possibly associated with PM as reliable enough to function as an individual biomarker in a clinical setting yet. Further investigation as well as more exploratory research questions leading to identify novel biomarkers, rather than performing analyses on panels consisting mostly of already established biomarkers, are still necessary. Techniques on DNA and RNA level are required to determine an association between genomic, epigenomic and transcriptomic changes and colorectal PM. Furthermore, future studies should include homogenous populations so that firm conclusions can be drawn. In that way, we might be able to identify biomarkers that can be incorporated in a prediction tool to estimate the risk of distant metastatic spread or to create targeted treatment options.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15020549/s1, Table S1: Overview investigated genes with conclusions.
Author Contributions
D.J.I.H., A.G.W.E.W., J.L., G.E.W.A.W., L.M., E.-J.M.S., I.H.J.T.d.H., N.D.B. and A.P. conceived, drafted, and revised the work. Study selection and data extraction was performed by D.J.I.H. and J.L. D.J.I.H. was responsible for writing the manuscript and original draft preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
Gregor Franssen was involved as a professional clinical librarian to ensure an appropriate search strategy.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Search strategy.
Table A1.
Search strategy.
| Database | Search Syntax |
|---|---|
| Pubmed | (((“Colorectal Neoplasms”[Mesh]) OR (((((((((“Neoplasms”[Mesh]) OR (carcinoma*[tw])) OR (adenocarcinoma*[tw])) OR (neoplas*[tw])) OR (tumour*[tw])) OR (tumor*[tw])) OR (oncolog*[tw])) OR (malignan*[tw])) AND ((((colorectal[tiab]) OR (colon[tw])) OR (rectal[tw])) OR (rectum[tw])))) AND ((“Peritoneal Neoplasms”[Mesh]) OR ((((“Peritoneum”[Mesh]) OR (peritoneal[tiab])) OR (peritoneum[tiab])) AND (((((((cancer[tiab]) OR (carcinomatos*[tiab])) OR (metastas*[tiab])) OR (neoplas*[tiab])) OR (tumor*[tiab])) OR (tumour*[tiab])) OR (malignan*[tiab]))))) AND ((((((((“Mutation”[Mesh]) OR (“Genetic Testing”[Mesh])) OR (“Genetic Association Studies”[Mesh])) OR (“Gene Expression Profiling”[Mesh])) OR (“Biomarkers, Tumor”[Mesh])) OR (((((tumor*[tw]) OR (tumour*[tw])) OR (cancer[tw])) OR (predictive[tw])) AND ((biomarker*[tw]) OR (marker*[tw])))) OR (((((mutation*[tiab]) OR (next generation sequencing[tiab])) OR (Gene*[tiab])) OR (RNA[tw])) OR (DNA[tw])))) |
| Embase | (exp colorectal tumor/ or ((neoplasm/ or (carcinoma* or adenocarcinoma* or neoplas* or tumour* or tumor* or oncolog* or malignan*).ti,ab,kw.) ADJ3 (colorectal or colon or rectal or rectum).ti,ab,kw.)) and (peritoneum tumor/ or ((exp peritoneum/ or (peritoneal or peritoneum).ti,ab,kw.) ADJ3 (cancer or carcinomatos* or metastas* or neoplas* or tumor or tumour or malignan*).ti,ab,kw.)) and ((exp mutation/ or exp sequence analysis/ or exp genetic association study/ or exp gene expression profiling/ or exp tumor marker/) or ((((tumour or tumor or cancer or predictive) ADJ3 (biomarker* or marker*)) or (mutation* or next generation sequencing or Gene* or RNA or DNA)).ti,ab,kw.)) |
| Cochrane | ([mh “Colorectal Neoplasms”] or (([mh Neoplasms] or (carcinoma* or adenocarcinoma* or neoplas* or tumour* or tumor* or oncolog* or malignan*):ti,ab,kw) and (colorectal OR colon OR rectal OR rectum):ti,ab,kw)) and ([mh “Peritoneal Neoplasms”] or ((peritoneal or peritoneum):ti,ab,kw and (cancer OR carcinomatos* OR metastas* OR neoplas* OR tumor* OR tumour* OR malignan*):ti,ab,kw)) and (([mh Mutation] or [mh “Genetic Testing”] or [mh “Genetic Association Studies”] or [mh “Gene Expression Profiling”] or [mh “Biomarkers, Tumor”]) or (((tumor* OR tumour* OR cancer* OR predictive):ti,ab,kw and (biomarker* OR marker*):ti,ab,kw) or (mutation* or Gene* or RNA or DNA):ti,ab,kw)) |
| CINAHL | (MH Colorectal Neoplasms OR (MH neoplasms OR carcinoma OR adenocarcinoma* OR neoplas* OR tumour* OR tumor* OR oncolog* OR malignan*) AND (colorectal OR colon OR rectal OR rectum)) AND (MH Peritoneal Neoplasms OR ((peritoneum OR peritoneal) AND (cancer OR carcinomatos* OR metastas* OR neoplas* OR tumor* OR tumour* OR malignan*))) AND ((MH mutation OR MH genetic testing OR MH Genetic Association Studied OR MH Gene Expression Profiling OR MH biomarkers) OR ((tumor* OR tumour* OR cancer OR predictive) AND (biomarker* OR marker*)) OR (mutation OR next-generation sequencing OR Gene* OR DNA OR RNA)) |
References
- Kamiyama, H.; Noda, H.; Konishi, F.; Rikiyama, T. Molecular biomarkers for the detection of metastatic colorectal cancer cells. World J. Gastroenterol. 2014, 20, 8928–8938. [Google Scholar] [CrossRef] [PubMed]
- Simkens, G.A.; Wintjens, A.; Rovers, K.P.; Nienhuijs, S.W.; de Hingh, I.H. Effective Strategies to Predict Survival of Colorectal Peritoneal Metastases Patients Eligible for Cytoreductive Surgery and HIPEC. Cancer Manag. Res. 2021, 13, 5239–5249. [Google Scholar] [CrossRef] [PubMed]
- Van Gestel, Y.R.; Thomassen, I.; Lemmens, V.E.; Pruijt, J.F.; van Herk-Sukel, M.P.; Rutten, H.J.; Creemers, G.J.; de Hingh, I.H. Metachronous peritoneal carcinomatosis after curative treatment of colorectal cancer. Eur. J. Surg. Oncol. 2014, 40, 963–969. [Google Scholar] [CrossRef]
- Lurvink, R.J.; Bakkers, C.; Rijken, A.; van Erning, F.N.; Nienhuijs, S.W.; Burger, J.W.; Creemers, G.J.; Verhoef, C.; Lemmens, V.E.; De Hingh, I.H. Increase in the incidence of synchronous and metachronous peritoneal metastases in patients with colorectal cancer: A nationwide study. Eur. J. Surg. Oncol. 2021, 47, 1026–1033. [Google Scholar] [CrossRef]
- Koppe, M.J.; Boerman, O.C.; Oyen, W.J.; Bleichrodt, R.P. Peritoneal carcinomatosis of colorectal origin: Incidence and current treatment strategies. Ann. Surg. 2006, 243, 212–222. [Google Scholar] [CrossRef]
- Klaver, Y.L.; Lemmens, V.E.; Nienhuijs, S.W.; Luyer, M.D.; de Hingh, I.H. Peritoneal carcinomatosis of colorectal origin: Incidence, prognosis and treatment options. World J. Gastroenterol. 2012, 18, 5489–5494. [Google Scholar] [CrossRef] [PubMed]
- Kranenburg, O.; van der Speeten, K.; de Hingh, I. Peritoneal Metastases From Colorectal Cancer: Defining and Addressing the Challenges. Front. Oncol. 2021, 11, 650098. [Google Scholar] [CrossRef] [PubMed]
- Jayne, D.G.; Fook, S.; Loi, C.; Seow-Choen, F. Peritoneal carcinomatosis from colorectal cancer. Br. J. Surg. 2002, 89, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, C.; Lurvink, R.J.; Rijken, A.; Nienhuijs, S.W.; Kok, N.F.; Creemers, G.J.; Verhoef, C.; Lemmens, V.E.; van Erning, F.N.; De Hingh, I.H. Treatment Strategies and Prognosis of Patients With Synchronous or Metachronous Colorectal Peritoneal Metastases: A Population-Based Study. Ann. Surg. Oncol. 2021, 28, 9073–9083. [Google Scholar] [CrossRef]
- Maggiori, L.; Elias, D. Curative treatment of colorectal peritoneal carcinomatosis: Current status and future trends. Eur. J. Surg. Oncol. 2010, 36, 599–603. [Google Scholar] [CrossRef]
- Klaver, C.E.L.; Wisselink, D.D.; Punt, C.J.A.; Snaebjornsson, P.; Crezee, J.; Aalbers, A.G.J.; Brandt, A.; Bremers, A.J.A.; Burger, J.W.A.; Fabry, H.F.J.; et al. Adjuvant hyperthermic intraperitoneal chemotherapy in patients with locally advanced colon cancer (COLOPEC): A multicentre, open-label, randomised trial. Lancet Gastroenterol. Hepatol. 2019, 4, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.; Löhrs, L.; Albertsmeier, M.; Reu, S.; Guba, M.; Werner, J.; Kirchner, T.; Angele, M. Cancer Stem Cell Markers Are Associated With Distant Hematogenous Liver Metastases But Not With Peritoneal Carcinomatosis in Colorectal Cancer. Cancer Investig. 2015, 33, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Karunasena, E.; Sham, J.; McMahon, K.W.; Ahuja, N. Genomics of Peritoneal Malignancies. Surg. Oncol. Clin. 2018, 27, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Roth, L.; Russo, L.; Ulugoel, S.; Freire Dos Santos, R.; Breuer, E.; Gupta, A.; Lehmann, K. Peritoneal Metastasis: Current Status and Treatment Options. Cancers 2021, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef]
- Lipsyc, M.; Yaeger, R. Impact of somatic mutations on patterns of metastasis in colorectal cancer. J. Gastrointest. Oncol. 2015, 6, 645–649. [Google Scholar] [CrossRef]
- Venkatachalam, R.; Ligtenberg, M.J.; Hoogerbrugge, N.; Geurts van Kessel, A.; Kuiper, R.P. Predisposition to colorectal cancer: Exploiting copy number variation to identify novel predisposing genes and mechanisms. Cytogenet. Genome Res. 2008, 123, 188–194. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Syst. Rev. 2021, 10, 89. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Astrosini, C.; Roeefzaad, C.; Dai, Y.Y.; Dieckgraefe, B.K.; Jöns, T.; Kemmner, W. REG1A expression is a prognostic marker in colorectal cancer and associated with peritoneal carcinomatosis. Int. J. Cancer 2008, 123, 409–413. [Google Scholar] [CrossRef]
- Bruzzi, M.; Auclin, E.; Lo Dico, R.; Voron, T.; Karoui, M.; Espin, E.; Cianchi, F.; Weitz, J.; Buggenhout, A.; Malafosse, R.; et al. Influence of Molecular Status on Recurrence Site in Patients Treated for a Stage III Colon Cancer: A Post Hoc Analysis of the PETACC-8 Trial. Ann. Surg. Oncol. 2019, 26, 3561–3567. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Lin, J.K.; Chen, W.S.; Jiang, J.K.; Yang, S.H.; Chang, S.C. Clinical significance of the BRAFV600E mutation in Asian patients with colorectal cancer. Int. J. Color. Dis. 2018, 33, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Wang, Y.; Zhong, Y.; Pan, X.; Si, L.; Lu, J. KRAS Codon 12 Mutation is Associated with More Aggressive Invasiveness in Synchronous Metastatic Colorectal Cancer (mCRC): Retrospective Research. Onco Targets Ther. 2020, 13, 12601–12613. [Google Scholar] [CrossRef] [PubMed]
- Heublein, S.; Albertsmeier, M.; Pfeifer, D.; Loehrs, L.; Bazhin, A.V.; Kirchner, T.; Werner, J.; Neumann, J.; Angele, M.K. Association of differential miRNA expression with hepatic vs. peritoneal metastatic spread in colorectal cancer. BMC Cancer 2018, 18, 201. [Google Scholar] [CrossRef]
- Jacob, S.; Bösch, F.; Schoenberg, M.B.; Pretzsch, E.; Lampert, C.; Haoyu, R.; Renz, B.W.; Michl, M.; Kumbrink, J.; Kirchner, T.; et al. Expression of CIB1 correlates with colorectal liver metastases but not with peritoneal carcinomatosis. BMC Cancer 2021, 21, 1243. [Google Scholar] [CrossRef]
- Jacob, S.; Jurinovic, V.; Lampert, C.; Pretzsch, E.; Kumbrink, J.; Neumann, J.; Haoyu, R.; Renz, B.W.; Kirchner, T.; Guba, M.O.; et al. The association of immunosurveillance and distant metastases in colorectal cancer. J. Cancer Res. Clin. Oncol. 2021, 147, 3333–3341. [Google Scholar] [CrossRef]
- Kawazoe, A.; Shitara, K.; Fukuoka, S.; Kuboki, Y.; Bando, H.; Okamoto, W.; Kojima, T.; Fuse, N.; Yamanaka, T.; Doi, T.; et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015, 15, 258. [Google Scholar] [CrossRef]
- Lan, Y.T.; Jen-Kou, L.; Lin, C.H.; Yang, S.H.; Lin, C.C.; Wang, H.S.; Chen, W.S.; Lin, T.C.; Jiang, J.K.; Chang, S.C. Mutations in the RAS and PI3K pathways are associated with metastatic location in colorectal cancers. J. Surg. Oncol. 2015, 111, 905–910. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chang, S.C.; Lin, P.C.; Lin, C.H.; Liang, W.Y.; Chen, W.S.; Jiang, J.K.; Yang, S.H.; Lin, J.K. High concordance of mutation patterns in 10 common mutated genes between tumor tissue and cell-free DNA in metastatic colorectal cancer. Am. J. Cancer Res. 2021, 11, 2228–2237. [Google Scholar]
- Lee, J.H.; Ahn, B.K.; Baik, S.S.; Lee, K.H. Comprehensive Analysis of Somatic Mutations in Colorectal Cancer With Peritoneal Metastasis. Vivo 2019, 33, 447–452. [Google Scholar] [CrossRef]
- Nagahara, M.; Nishida, N.; Iwatsuki, M.; Ishimaru, S.; Mimori, K.; Tanaka, F.; Nakagawa, T.; Sato, T.; Sugihara, K.; Hoon, D.S.; et al. Kinesin 18A expression: Clinical relevance to colorectal cancer progression. Int. J. Cancer 2011, 129, 2543–2552. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, K.; Yasuda, T.; Sakata, M.; Kitamura, Y.H.; Shirahata, A.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; et al. Down-regulation of Tip60 gene as a potential marker for the malignancy of colorectal cancer. Anticancer Res. 2009, 29, 3953–3955. [Google Scholar] [PubMed]
- Sasaki, Y.; Hamaguchi, T.; Yamada, Y.; Takahashi, N.; Shoji, H.; Honma, Y.; Iwasa, S.; Okita, N.; Takashima, A.; Kato, K.; et al. Value of KRAS, BRAF, and PIK3CA Mutations and Survival Benefit from Systemic Chemotherapy in Colorectal Peritoneal Carcinomatosis. Asian Pac. J. Cancer Prev. 2016, 17, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Sayagués, J.M.; Del Carmen, S.; Del Mar Abad, M.; Corchete, L.A.; Bengoechea, O.; Anduaga, M.F.; Baldeón, M.J.; Cruz, J.J.; Alcazar, J.A.; Angoso, M.; et al. Combined assessment of the TNM stage and BRAF mutational status at diagnosis in sporadic colorectal cancer patients. Oncotarget 2018, 9, 24081–24096. [Google Scholar] [CrossRef] [PubMed]
- Shelygin, Y.A.; Pospekhova, N.I.; Shubin, V.P.; Kashnikov, V.N.; Frolov, S.A.; Sushkov, O.I.; Achkasov, S.I.; Tsukanov, A.S. Epithelial-mesenchymal transition and somatic alteration in colorectal cancer with and without peritoneal carcinomatosis. Biomed. Res. Int. 2014, 2014, 629496. [Google Scholar] [CrossRef] [PubMed]
- Sjo, O.H.; Berg, M.; Merok, M.A.; Kolberg, M.; Svindland, A.; Lothe, R.A.; Nesbakken, A. Peritoneal carcinomatosis of colon cancer origin: Highest incidence in women and in patients with right-sided tumors. J. Surg. Oncol. 2011, 104, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.G.; Fisher, D.; Claes, B.; Maughan, T.S.; Idziaszczyk, S.; Peuteman, G.; Harris, R.; James, M.D.; Meade, A.; Jasani, B.; et al. Somatic profiling of the epidermal growth factor receptor pathway in tumors from patients with advanced colorectal cancer treated with chemotherapy ± cetuximab. Clin. Cancer Res. 2013, 19, 4104–4113. [Google Scholar] [CrossRef]
- Takahashi, Y.; Iwaya, T.; Sawada, G.; Kurashige, J.; Matsumura, T.; Uchi, R.; Ueo, H.; Takano, Y.; Eguchi, H.; Sudo, T.; et al. Up-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancer. Ann. Surg. Oncol. 2014, 21, 205–212. [Google Scholar] [CrossRef]
- Taniguchi, H.; Uehara, K.; Nakayama, G.; Nakayama, H.; Aiba, T.; Hattori, N.; Kataoka, M.; Nakano, Y.; Kawase, Y.; Okochi, O.; et al. Tumor Location Is Associated With the Prevalence of Braf And PIK3CA Mutations in Patients with Wild-Type Ras Colorectal Cancer: A Prospective Multi-Center Cohort Study in Japan. Transl. Oncol. 2020, 13, 100786. [Google Scholar] [CrossRef]
- Yokota, T.; Ura, T.; Shibata, N.; Takahari, D.; Shitara, K.; Nomura, M.; Kondo, C.; Mizota, A.; Utsunomiya, S.; Muro, K.; et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br. J. Cancer 2011, 104, 856–862. [Google Scholar] [CrossRef]
- Shirahata, A.; Sakata, M.; Sakuraba, K.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; Sanada, Y.; et al. Vimentin methylation as a marker for advanced colorectal carcinoma. Anticancer Res. 2009, 29, 279–281. [Google Scholar] [PubMed]
- Shirahata, A.; Shinmura, K.; Kitamura, Y.; Sakuraba, K.; Yokomizo, K.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; et al. MACC1 as a marker for advanced colorectal carcinoma. Anticancer Res. 2010, 30, 2689–2692. [Google Scholar] [PubMed]
- Atreya, C.E.; Greene, C.; McWhirter, R.M.; Ikram, N.S.; Allen, I.E.; Van Loon, K.; Venook, A.P.; Yeh, B.M.; Behr, S.C. Differential Radiographic Appearance of BRAF V600E-Mutant Metastatic Colorectal Cancer in Patients Matched by Primary Tumor Location. J. Natl. Compr. Cancer Netw. 2016, 14, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.D.; Palshof, J.A.; Larsen, F.O.; Poulsen, T.S.; Høgdall, E.; Pfeiffer, P.; Jensen, B.V.; Yilmaz, M.K.; Nielsen, D. Associations between primary tumor RAS, BRAF and PIK3CA mutation status and metastatic site in patients with chemo-resistant metastatic colorectal cancer. Acta Oncol. 2018, 57, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, T.; Karapetis, C.S.; Roder, D.; Tie, J.; Padbury, R.; Price, T.; Wong, R.; Shapiro, J.; Nott, L.; Lee, M.; et al. The survival outcome of patients with metastatic colorectal cancer based on the site of metastases and the impact of molecular markers and site of primary cancer on metastatic pattern. Acta Oncol. 2018, 57, 1438–1444. [Google Scholar] [CrossRef]
- Roberto, M.; Marchetti, P.; Arrivi, G.; Di Pietro, F.R.; Cascinu, S.; Gelsomino, F.; Caputo, F.; Cerma, K.; Ghidini, M.; Ratti, M.; et al. The treatment paradigm of right-sided metastatic colon cancer: Harboring BRAF mutation makes the difference. Int. J. Color. Dis. 2020, 35, 1513–1527. [Google Scholar] [CrossRef]
- Schirripa, M.; Nappo, F.; Cremolini, C.; Salvatore, L.; Rossini, D.; Bensi, M.; Businello, G.; Pietrantonio, F.; Randon, G.; Fucà, G.; et al. KRAS G12C Metastatic Colorectal Cancer: Specific Features of a New Emerging Target Population. Clin. Color. Cancer 2020, 19, 219–225. [Google Scholar] [CrossRef]
- Tran, B.; Kopetz, S.; Tie, J.; Gibbs, P.; Jiang, Z.Q.; Lieu, C.H.; Agarwal, A.; Maru, D.M.; Sieber, O.; Desai, J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011, 117, 4623–4632. [Google Scholar] [CrossRef]
- Yaeger, R.; Cercek, A.; Chou, J.F.; Sylvester, B.E.; Kemeny, N.E.; Hechtman, J.F.; Ladanyi, M.; Rosen, N.; Weiser, M.R.; Capanu, M.; et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer 2014, 120, 2316–2324. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Hu, W.M.; Xia, L.P.; He, W.Z. Association between somatic RET mutations and clinical and genetic characteristics in patients with metastatic colorectal cancer. Cancer Med. 2021, 10, 8876–8882. [Google Scholar] [CrossRef]
- Zihui Yong, Z.; Ching, G.T.H.; Ching, M.T.C. Metastatic Profile of Colorectal Cancer: Interplay Between Primary Tumor Location and KRAS Status. J. Surg. Res. 2020, 246, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, T.; Wong, R.; Price, T.; Shapiro, J.; Tie, J.; Wong, H.L.; Nott, L.; Roder, D.; Lee, M.; Kosmider, S.; et al. Metastasectomy and BRAF mutation; an analysis of survival outcome in metastatic colorectal cancer. Curr. Probl. Cancer 2021, 45, 100637. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.H.; Joo, Y.E. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest. Res. 2020, 18, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Caputo, F.; Santini, C.; Bardasi, C.; Cerma, K.; Casadei-Gardini, A.; Spallanzani, A.; Andrikou, K.; Cascinu, S.; Gelsomino, F. BRAF-Mutated Colorectal Cancer: Clinical and Molecular Insights. Int. J. Mol. Sci. 2019, 20, 5369. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Shi, Q.; Smyrk, T.C.; Thibodeau, S.N.; Dienstmann, R.; Guinney, J.; Bot, B.M.; Tejpar, S.; Delorenzi, M.; Goldberg, R.M.; et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015, 148, 88–99. [Google Scholar] [CrossRef]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Janakiraman, M.; Vakiani, E.; Zeng, Z.; Pratilas, C.A.; Taylor, B.S.; Chitale, D.; Halilovic, E.; Wilson, M.; Huberman, K.; Ricarte Filho, J.C.; et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010, 70, 5901–5911. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Popat, S.; Hubner, R.; Houlston, R.S. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005, 23, 609–618. [Google Scholar] [CrossRef]
- Segelman, J.; Granath, F.; Holm, T.; Machado, M.; Mahteme, H.; Martling, A. Incidence, prevalence and risk factors for peritoneal carcinomatosis from colorectal cancer. Br. J. Surg. 2012, 99, 699–705. [Google Scholar] [CrossRef]
- Arjona-Sánchez, A.; Barrios, P.; Boldo-Roda, E.; Camps, B.; Carrasco-Campos, J.; Concepción Martín, V.; García-Fadrique, A.; Gutiérrez-Calvo, A.; Morales, R.; Ortega-Pérez, G.; et al. HIPECT4: Multicentre, randomized clinical trial to evaluate safety and efficacy of Hyperthermic intra-peritoneal chemotherapy (HIPEC) with Mitomycin C used during surgery for treatment of locally advanced colorectal carcinoma. BMC Cancer 2018, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Sorich, M.J.; Wiese, M.D.; Rowland, A.; Kichenadasse, G.; McKinnon, R.A.; Karapetis, C.S. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized, controlled trials. Ann. Oncol. 2015, 26, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Graf, W.; Cashin, P.H.; Ghanipour, L.; Enblad, M.; Botling, J.; Terman, A.; Birgisson, H. Prognostic Impact of BRAF and KRAS Mutation in Patients with Colorectal and Appendiceal Peritoneal Metastases Scheduled for CRS and HIPEC. Ann. Surg. Oncol. 2020, 27, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, S.; Berger, M.F.; Roychowdhury, S. Clinical tumor sequencing: Opportunities and challenges for precision cancer medicine. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e175–e182. [Google Scholar] [CrossRef] [PubMed]
- Alorda-Clara, M.; Torrens-Mas, M.; Morla-Barcelo, P.M.; Martinez-Bernabe, T.; Sastre-Serra, J.; Roca, P.; Pons, D.G.; Oliver, J.; Reyes, J. Use of Omics Technologies for the Detection of Colorectal Cancer Biomarkers. Cancers 2022, 14, 817. [Google Scholar] [CrossRef]
- Lenos, K.J.; Bach, S.; Ferreira Moreno, L.; ten Hoorn, S.; Sluiter, N.R.; Bootsma, S.; Vieira Braga, F.A.; Nijman, L.E.; van den Bosch, T.; Miedema, D.M.; et al. Molecular characterization of colorectal cancer related peritoneal metastatic disease. Nat. Commun. 2022, 13, 4443. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).