Development of a Single-Neurosphere Culture to Assess Radiation Toxicity and Pre-Clinical Cancer Combination Therapy Safety
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Processing and Neurosphere Cultures
2.2. Radiation Exposure
2.3. Apoptosis Quantification Using Flow Cytometry
2.4. Immunostaining
2.5. Immunolabeled Cells and Area Quantification
2.6. RNA Extraction and RT-qPCR Analysis
2.7. Neurospheres’ Volume Quantification
2.8. cDNA Library Preparation and Sequencing
2.9. RNA-Seq Data Processing and Analysis
2.10. Statistical Analysis
3. Results
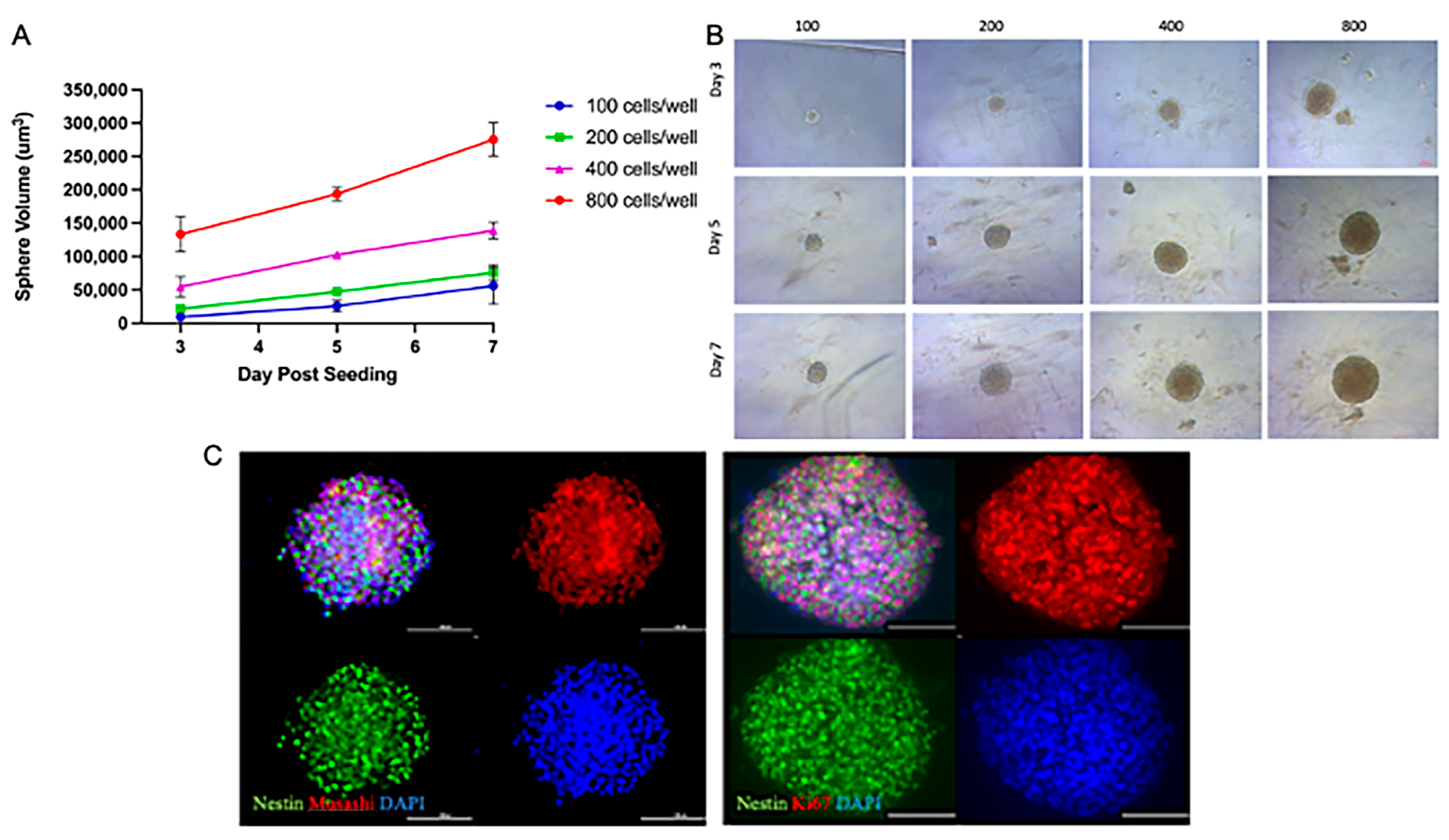
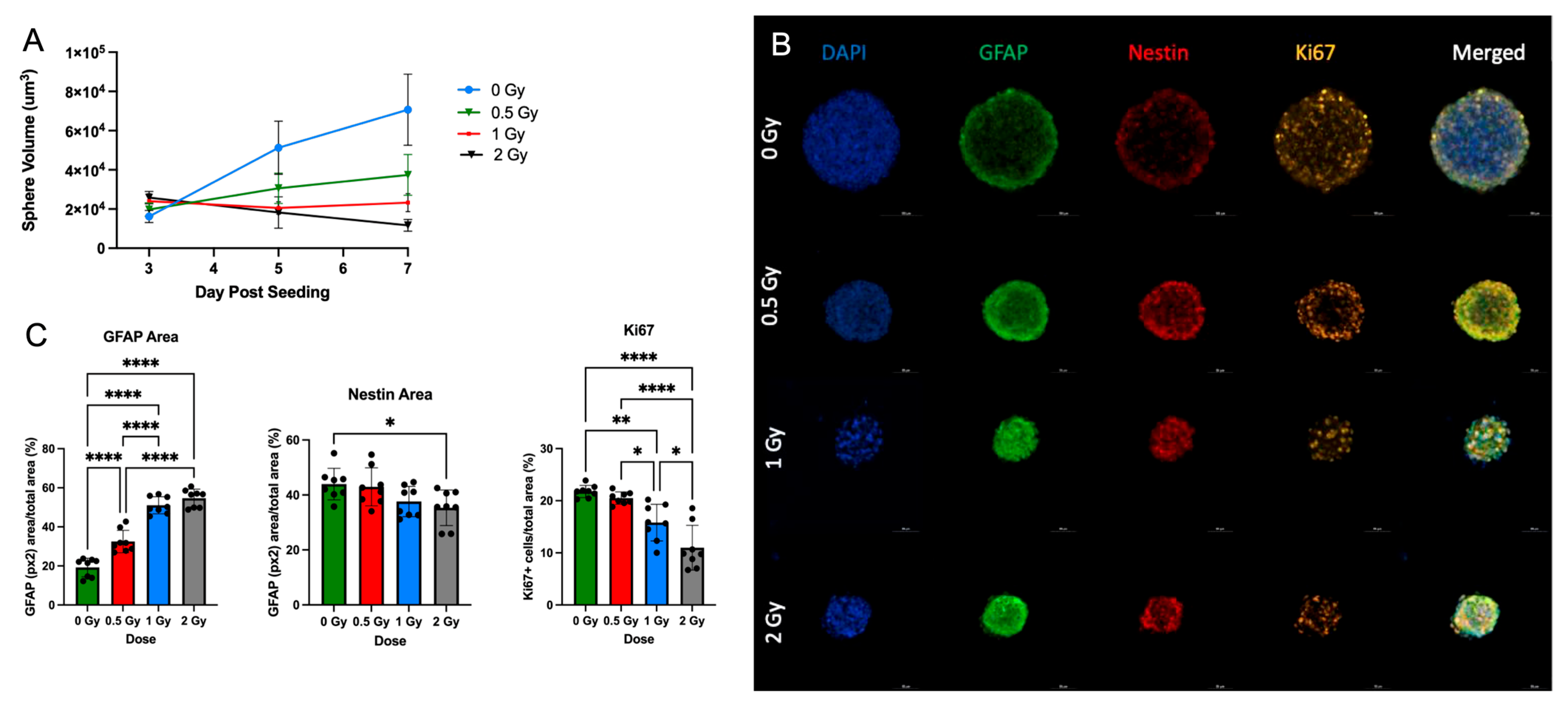
3.1. NSp Cultures Recapitulate NPC-Defining Characteristics
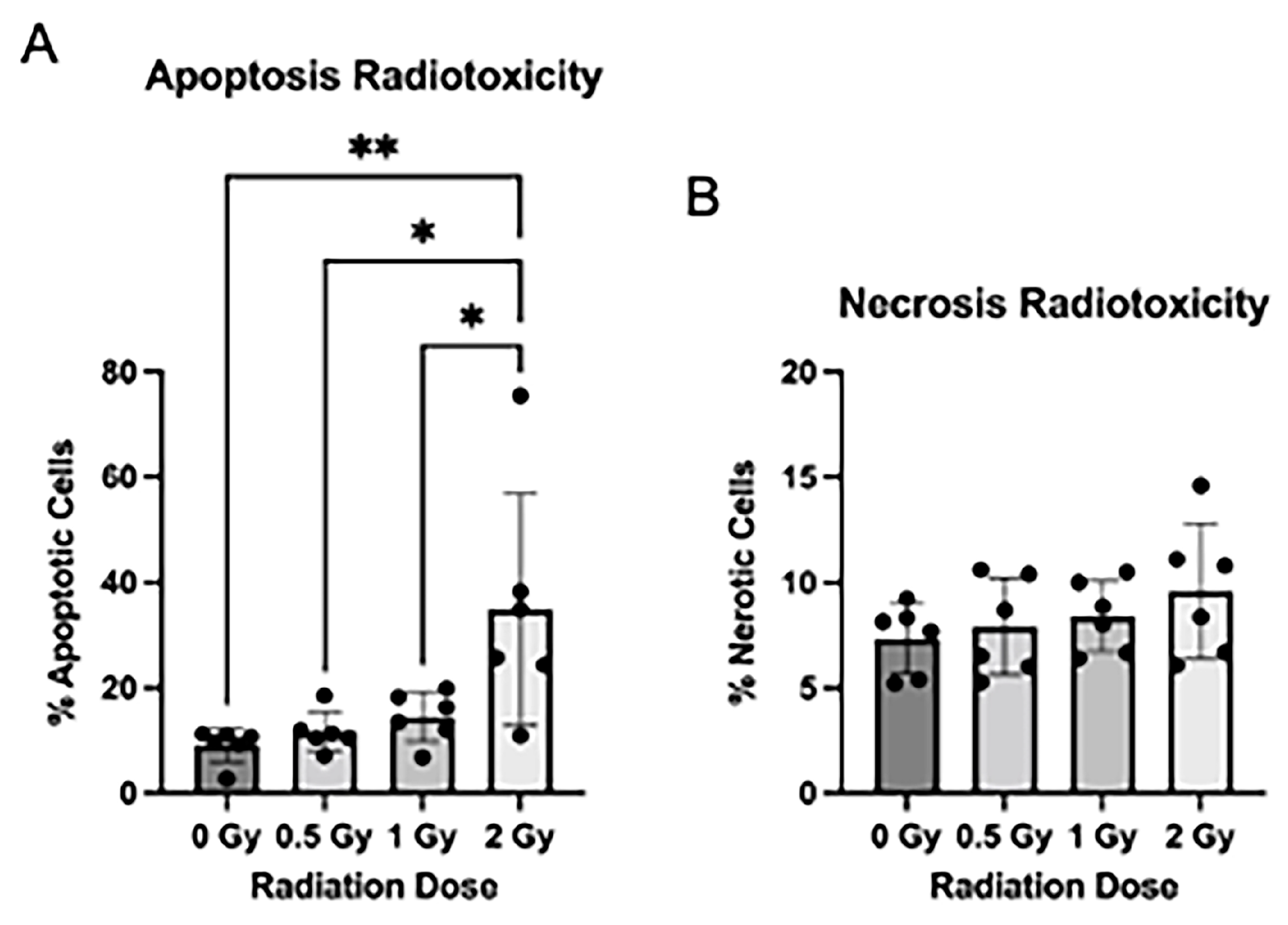
3.2. Radiation Induces Apoptosis after Radiation Exposure
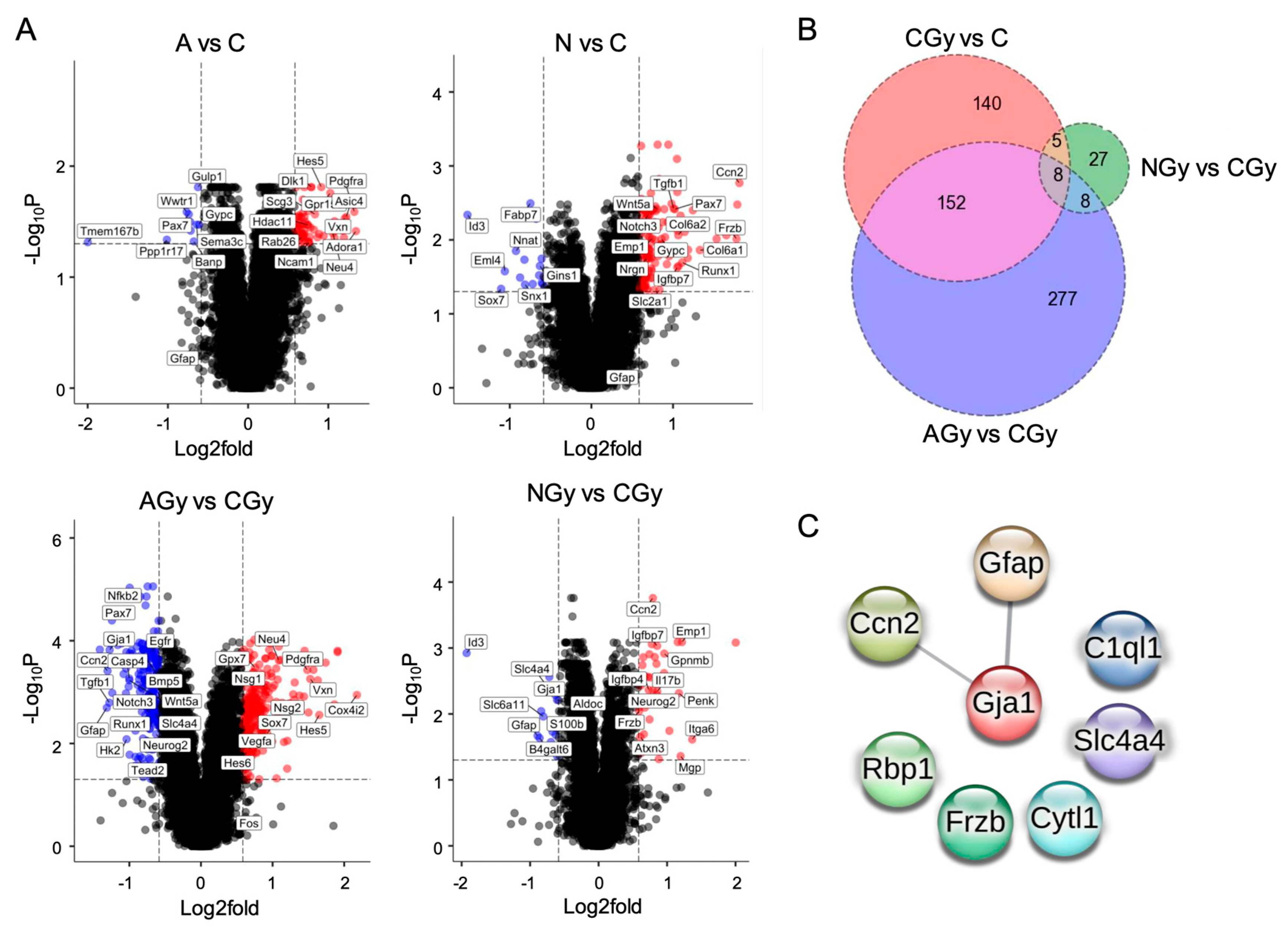
3.3. Radiation Treatment Increases NPC Differentiation in Neurospheres
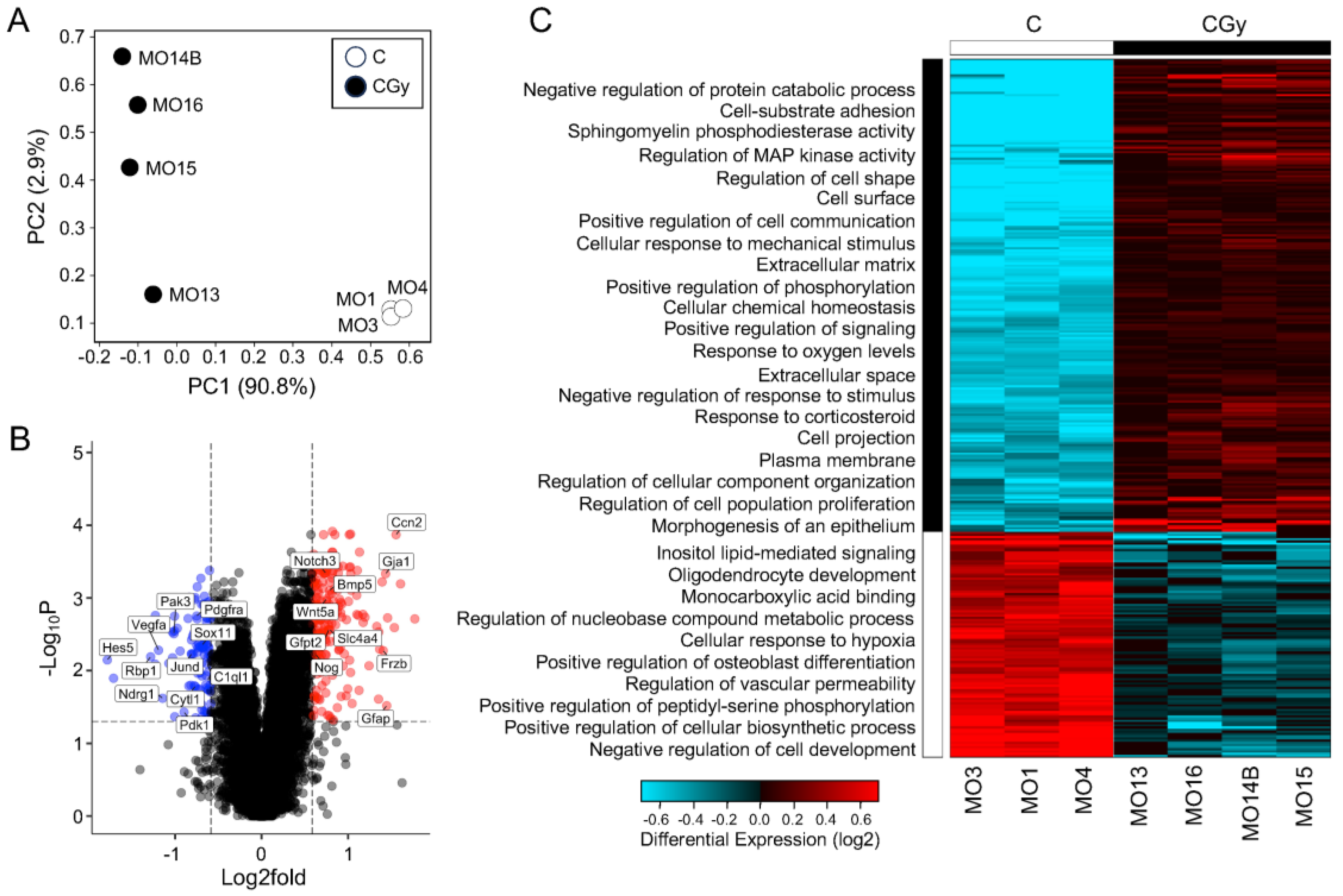
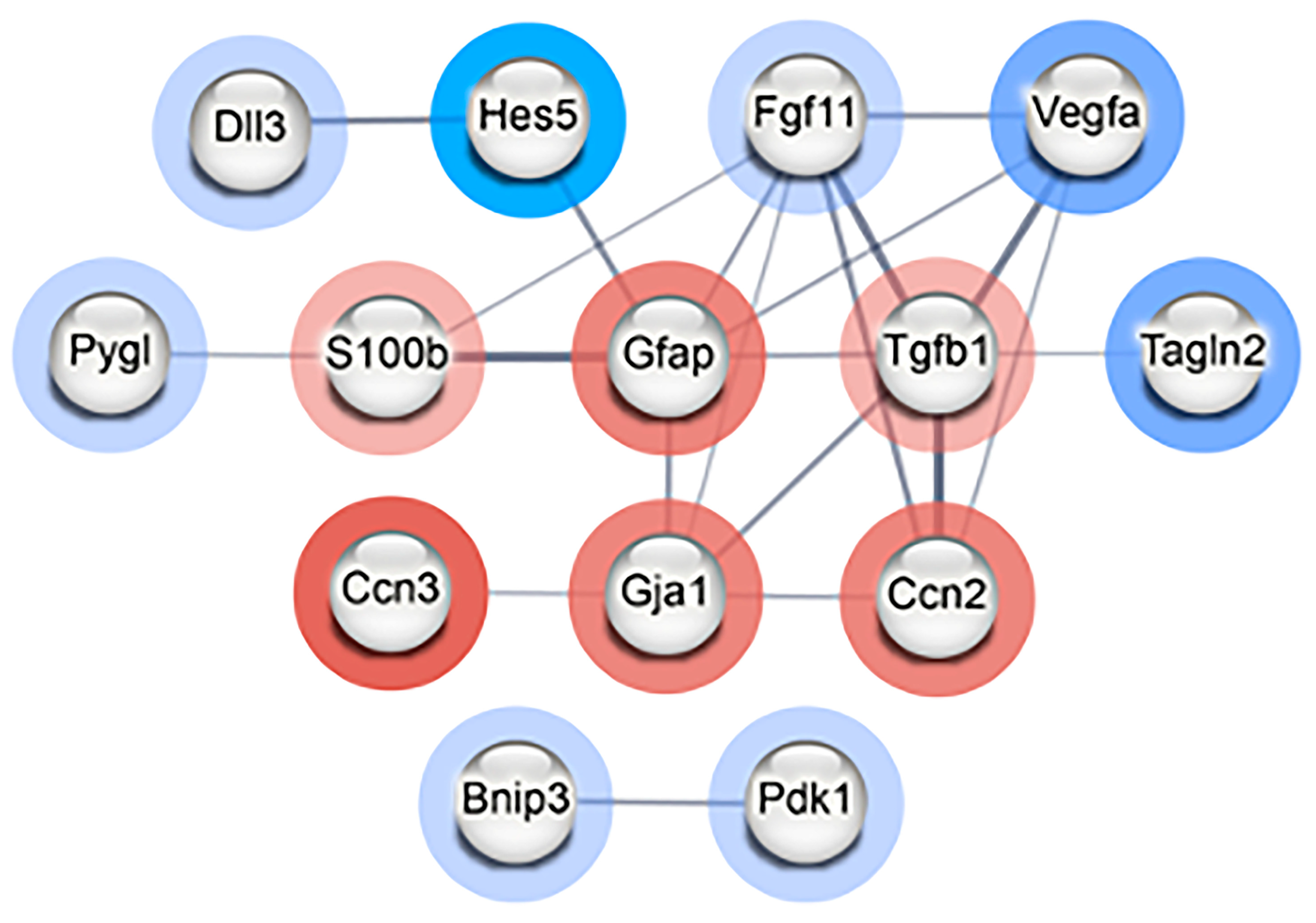
3.4. Neurogenesis Adaptation after Radiation of Single NSps
3.5. Inhibition of BMP Activity or Supplementation with Antioxidants Reduces Radiotoxicity in Neurospheres
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alessi, I.; Caroleo, A.M.; de Palma, L.; Mastronuzzi, A.; Pro, S.; Colafati, G.S.; Boni, A.; Della Vecchia, N.; Velardi, M.; Evangelisti, M.; et al. Short and Long-Term Toxicity in Pediatric Cancer Treatment: Central Nervous System Damage. Cancers 2022, 14, 1540. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Wernicke, A.G. Risk and survival outcomes of radiation-induced CNS tumors. J. Neurooncol. 2016, 129, 15–22. [Google Scholar] [CrossRef] [PubMed]
- McNerlin, C.; Guan, F.; Bronk, L.; Lei, K.; Grosshans, D.; Young, D.W.; Gaber, M.W.; Maletic-Savatic, M. Targeting hippocampal neurogenesis to protect astronauts’ cognition and mood from decline due to space radiation effects. Life Sci. Space Res. 2022, 35, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Belka, C.; Budach, W.; Kortmann, R.D.; Bamberg, M. Radiation induced CNS toxicity--molecular and cellular mechanisms. Br. J. Cancer. 2001, 85, 1233–1239. [Google Scholar] [CrossRef]
- Kim, J.H.; Brown, S.L.; Jenrow, K.A.; Ryu, S. Mechanisms of radiation-induced brain toxicity and implications for future clinical trials. J. Neurooncol. 2008, 87, 279–286. [Google Scholar] [CrossRef]
- Breunig, J.J.; Haydar, T.F.; Rakic, P. Neural stem cells: Historical perspective and future prospects. Neuron 2011, 70, 614–625. [Google Scholar] [CrossRef]
- Perry, A.; Schmidt, R.E. Cancer therapy-associated CNS neuropathology: An update and review of the literature. Acta Neuropathol. 2006, 111, 197–212. [Google Scholar] [CrossRef]
- Monje, M.L.; Mizumatsu, S.; Fike, J.R.; Palmer, T.D. Irradiation induces neural precursor-cell dysfunction. Nat. Med. 2002, 8, 955–962. [Google Scholar] [CrossRef]
- Pineda, J.R.; Daynac, M.; Chicheportiche, A.; Cebrian-Silla, A.; Felice, K.S.; Garcia-Verdugo, J.M.; Boussin, F.D.; Mouthon, M.A. Vascular-derived TGF-increases in the stem cell niche and perturbs neurogenesis during aging and following irradiation in the adult mouse brain. EMBO Mol. Med. 2013, 5, 548–562. [Google Scholar] [CrossRef]
- Schneider, L.; Pellegatta, S.; Favaro, R.; Pisati, F.; Roncaglia, P.; Testa, G.; Nicolis, S.K.; Finocchiaro, G.; Di Fagagna, F.D. DNA Damage in Mammalian Neural Stem Cells Leads to Astrocytic Differentiation Mediated by BMP2 Signaling through JAK-STAT. Stem Cell Rep. 2013, 1, 123–138. [Google Scholar] [CrossRef]
- Konirova, J.; Cupal, L.; Jarosova, S.; Michaelidesova, A.; Vachelova, J.; Davidkova, M.; Bartunek, P.; Zikova, M. Dierentiation Induction as a Response to Irradiation in Neural Stem Cells In Vitro. Cancers 2019, 11, 913. [Google Scholar] [CrossRef]
- Kobolak, J.; Teglasi, A.; Bellak, T.; Janstova, Z.; Molnar, K.; Zana, M.; Bock, I.; Laszlo, L.; Dunnyes, A. Human induced pluripotent stem cell-derived 3D-Neuropsheres are suitable for neurotoxicity screening. Cells 2020, 9, 1122. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Annika, G.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Jian, B.; Wang, D.; Chen, D.; Voss, J.; Chaudry, I.; Raju, R. Hypoxia-induced alteration of mitochondrial genes in cardiomyocytes: Role of Bnip3 and Pdk1. Shock 2010, 34, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, A.H.; Tracy, K.; Frankenberger, C.; Boland, M.L.; Sharifi, M.N.; Drake, L.E.; Sachleben, J.R.; Asara, J.M.; Locasale, J.W.; Karczmar, G.S.; et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015, 16, 1145–1163. [Google Scholar] [CrossRef]
- Bonaguidi, M.A.; Peng, C.Y.; McGuire, T.; Falciglia, G.; Gobeske, K.T.; Czeisler, C.; Kessler, J.A. Noggin expands neural stem cells in the adult hippocampus. J. Neurosci. 2008, 28, 9194–9204. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Chang, Y.; Saleem, A.; Wu, F.; Tian, L.; Zhang, S.; Li, Y.; Ma, S.; Dong, T.; Guo, T.; et al. Ascorbic acid can promote the generation and expansion of neuroepithelial-like stem cells derived from hiPS/ES cells under chemically defined conditions through promoting collagen synthesis. Stem. Cell Res. Ther. 2021, 12, 48. [Google Scholar] [CrossRef]
- Theparambil, S.M.; Hosford, P.S.; Ruminot, I.; Kopach, O.; Reynolds, J.R.; Sandoval, P.Y.; Rusakov, D.A.; Barros, L.F.; Gourine, A.V. Astrocytes regulate brain extracellular pH via a neuronal activity-dependent bicarbonate shuttle. Nat Commun. 2020, 11, 5073. [Google Scholar] [CrossRef]
- Mendes, F.A.; Coelho Aguiar, J.M.; Kahn, S.A.; Reis, A.H.; Dubois, L.G.; Romão, L.F.; Ferreira, L.S.; Chneiweiss, H.; Moura Neto, V.; Abreu, J.G. Connective-Tissue Growth Factor (CTGF/CCN2) Induces Astrogenesis and Fibronectin Expression of Embryonic Neural Cells In Vitro. PLoS ONE 2015, 10, e0133689. [Google Scholar] [CrossRef]
- Ris, M.D.; Packer, R.; Goldwein, J.; Jones-Wallace, D.; Boyett, J.M. Intellectual outcome after reduced-dose radiation therapy plus adjuvant chemotherapy for medulloblastoma: A Children’s Cancer Group study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2001, 19, 3470–3476. [Google Scholar] [CrossRef] [PubMed]
- Carbonara, R.; Di Rito, A.; Monti, A.; Rubini, G.; Sardaro, A. Proton versus Photon Radiotherapy for Pediatric Central Nervous System Malignancies: A Systematic Review and Meta-Analysis of Dosimetric Comparison Studies. J. Oncol. 2019, 2019, 5879723. [Google Scholar] [CrossRef] [PubMed]
- Greenberger, B.A.; Pulsifer, M.B.; Ebb, D.H.; MacDonald, S.M.; Jones, R.M.; Butler, W.E.; Huang, M.S.; Marcus, K.J.; Oberg, J.A.; Tarbell, N.J.; et al. Clinical outcomes and late endocrine, neurocognitive, and visual profiles of proton radiation for pediatric low-grade gliomas. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.T.; Sugimoto, C.; Regan, S.L.; Pitzer, E.M.; Fritz, A.L.; Sertorio, M.; Mascia, A.E.; Vatner, R.E.; Perentesis, J.P.; Vorhees, C.V. Cognitive and behavioral effects of whole brain conventional or high dose rate (FLASH) proton irradiation in a neonatal Sprague Dawley rat model. PLoS ONE 2022, 17, e0274007. [Google Scholar] [CrossRef]
- Montay-Gruel, P.; Acharya, M.M.; Petersson, K.; Alikhani, L.; Yakkala, C.; Allen, B.D.; Ollivier, J.; Petit, B.; Jorge, P.G.; Syage, A.R.; et al. Long-term neurocognitive benefits of FLASH radiotherapy driven by reduced reactive oxygen species. Proc. Natl. Acad Sci. USA 2019, 116, 10943–10951. [Google Scholar] [CrossRef]
- Montay-Gruel, P.; Markarian, M.; Allen, B.D.; Baddour, J.D.; Giedzinski, E.; Jorge, P.G.; Petit, B.; Bailat, C.; Vozenin, M.C.; Limoli, C.; et al. Ultra-High-Dose-Rate FLASH Irradiation Limits Reactive Gliosis in the Brain. Radiat. Res. 2020, 194, 636–645. [Google Scholar] [CrossRef]
- Chaklai, A.; Canaday, P.; O’neil, A.; Cucinotta, F.A.; Sloop, A.; Gladstone, D.; Pogue, B.; Zhang, R.; Sunnerberg, J.; Kheirollah, A.; et al. Effects of UHDR and Conventional Irradiation on Behavioral and Cognitive Performance and the Percentage of Ly6G+ CD45+ Cells in the Hippocampus. Int. J. Mol. Sci. 2023, 24, 12497. [Google Scholar] [CrossRef]
- Reynolds, B.A.; Weiss, S. Generation of Neurons and Astrocytes from Isolated Cells of the Adult Mammalian Central Nervous System. Science 1992, 255, 1707–1710. [Google Scholar] [CrossRef]
- Ulloa, F.; Briscoe, J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle 2007, 6, 2640–2649. [Google Scholar] [CrossRef]
- Molofsky, A.V.; Krenick, R.; Ullian, E.; Tsai, H.-H.; Deneen, B.; Richardson, W.D. Astrocytes and disease: A neurodevelopmental perspective. Genes Dev. 2012, 26, 891–907. [Google Scholar] [CrossRef]
- Dennis, D.J.; Han, S.; Schuurmans, C. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019, 1705, 48–65. [Google Scholar] [CrossRef]
- Okano-Uchida, T.; Naruse, M.; Ikezawa, T.; Shibasaki, K.; Ishizaki, Y. Cerebellar neural stem cells differentiate into two distinct types of astrocytes in response to CNTF and BMP2. Neurosci. Lett. 2013, 552, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Oria, M.; Pathak, B.; Li, Z.; Bakri, K.; Gouwens, K.; Varela, M.F.; Lampe, K.; Murphy, K.P.; Lin, C.-Y.; Peiro, J.L. Premature Neural Progenitor Cell Differentiation into Astrocytes in Retinoic Acid-Induced Spina Bifida Rat Model. Front. Mol. Neurosci. 2022, 17, 888351. [Google Scholar] [CrossRef]
- Furuta, Y.; Piston, D.W.; Hogan, B.L.M. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 1997, 124, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Mehler, M.F.; Mabie, P.C.; Zhang, D.; Kessler, J.A. Bone morphogenetic proteins in the nervous system. Trends Neurosci. 1997, 20, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Gross, R.E.; Mehler, M.F.; Mabie, P.C.; Zang, Z.; Santschi, L.; Kessler, J.A. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 1996, 17, 595–606. [Google Scholar] [CrossRef]
- Mabie, P.C.; Mehler, M.F.; Kessler, J.A. Multiple roles of bone morphogenetic protein signaling in the regulation of cortical cell number and phenotype. J. Neurosci. 1999, 19, 7077–7088. [Google Scholar] [CrossRef]
- Bonaguidi, M.A.; McGuire, T.; Hu, M.; Kan, L.; Samantha, J.; Kessler, J.A. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development 2005, 132, 5503–5514. [Google Scholar] [CrossRef]
- Miyagi, M.; Mikawa, S.; Sato, T.; Hasegawa, T.; Kobayashi, S.; Matsuyama, Y.; Sato, K. BMP2, BMP4, noggin, BMPRIA, BMPRIB, and BMPRII are differentially expressed in the adult rat spinal cord. Neuroscience 2012, 203, 12–26. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Yin, G.; Huang, J.R.; Xi, S.J.; Qian, F.; Lee, R.X.; Peng, X.C.; Tang, F.R. Ionizing Radiation-Induced Brain Cell Aging and the Potential Underlying Molecular Mechanisms. Cells 2021, 10, 3570. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Zhang, J.; He, X. Ionizing radiation-induced oxidative stress and its effects on stem cells. Stem Cell Res. Ther. 2013, 4, 26. [Google Scholar]
- Morshead, C.M.; Reynolds, B.A.; Craig, C.G.; McBurney, M.W.; Staines, W.A.; Morassutti, D.; Weiss, S. Radiation-induced cognitive decline is associated with decreased neurogenesis in the hippocampus. NeuroReport 1994, 5, 2565–2568. [Google Scholar]
- Yue, J.; Chen, J. Bone morphogenetic proteins and their receptors in neural development. Nat. Rev. Neurosci. 2008, 9, 898–909. [Google Scholar]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kwon, H.S.; Kim, J.Y.; Kim, Y.S.; Kim, J. Antioxidants and neuroprotection in neurodegenerative diseases. Antioxidants 2018, 7, 10. [Google Scholar]
- Jang, S.H.; Lee, H.J.; Shin, H.Y.; Lee, J.H.; Kim, H.J. N-acetylcysteine modulates neural stem/progenitor cell differentiation and neurogenesis. Neuroscience 2012, 201, 312–325. [Google Scholar]
- Zhou, Z.; Liu, Y.; Zhang, X.; Zhang, J. ROS scavenger tempol reduces oxidative stress and inhibits astrocyte activation in brain injury. Neuroscience 2015, 285, 221–232. [Google Scholar]
- Wei, X.; Xu, Y.; Xu, F.F.; Chaiswing, L.; Schnell, D.; Noel, T.; Wang, C.; Chen, J.; St Clair, D.K.; St Clair, W.H. RelB Expression Determines the Differential Effects of Ascorbic Acid in Normal and Cancer Cells. Cancer Res. 2017, 77, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, N.S.; Kuligina, E.V.; Dymova, M.A.; Savinovskaya, Y.I.; Zinchenko, N.D.; Ageenko, A.B.; Mishinov, S.V.; Dome, A.S.; Stepanov, G.A.; Richter, V.A.; et al. Transcriptome Changes in Glioma Cells Cultivated under Conditions of Neurosphere Formation. Cells 2022, 11, 3106. [Google Scholar] [CrossRef]


| C | CGy | N | NGy | A | Agy | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| C | ||||||||||||
| CGy | 209 | 97 | ||||||||||
| N | 138 | 15 | 132 | 53 | 150 | 52 | 322 | 247 | 329 | 209 | ||
| NGy | 244 | 147 | 35 | 13 | ||||||||
| A | 56 | 10 | 403 | 371 | 522 | 401 | 13 | 10 | ||||
| AGy | 65 | 33 | 224 | 222 | 282 | 241 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pathak, B.; Lange, T.E.; Lampe, K.; Hollander, E.; Oria, M.; Murphy, K.P.; Salomonis, N.; Sertorio, M.; Oria, M. Development of a Single-Neurosphere Culture to Assess Radiation Toxicity and Pre-Clinical Cancer Combination Therapy Safety. Cancers 2023, 15, 4916. https://doi.org/10.3390/cancers15204916
Pathak B, Lange TE, Lampe K, Hollander E, Oria M, Murphy KP, Salomonis N, Sertorio M, Oria M. Development of a Single-Neurosphere Culture to Assess Radiation Toxicity and Pre-Clinical Cancer Combination Therapy Safety. Cancers. 2023; 15(20):4916. https://doi.org/10.3390/cancers15204916
Chicago/Turabian StylePathak, Bedika, Taylor E. Lange, Kristin Lampe, Ella Hollander, Marina Oria, Kendall P. Murphy, Nathan Salomonis, Mathieu Sertorio, and Marc Oria. 2023. "Development of a Single-Neurosphere Culture to Assess Radiation Toxicity and Pre-Clinical Cancer Combination Therapy Safety" Cancers 15, no. 20: 4916. https://doi.org/10.3390/cancers15204916
APA StylePathak, B., Lange, T. E., Lampe, K., Hollander, E., Oria, M., Murphy, K. P., Salomonis, N., Sertorio, M., & Oria, M. (2023). Development of a Single-Neurosphere Culture to Assess Radiation Toxicity and Pre-Clinical Cancer Combination Therapy Safety. Cancers, 15(20), 4916. https://doi.org/10.3390/cancers15204916









