The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Laboratory Animals
2.2. RNA Extraction, Library Construction, and High-Throughput Transcriptome Sequencing
2.3. In Silico Profiling of Liver Transcriptome Affected by BPA
2.3.1. Processing and Mapping of Raw Reads
2.3.2. Detection of Differentially Expressed Genes and Long Non-Coding RNAs and Interaction Analyses
2.3.3. Alternative Splicing Events and Differential Analysis
2.3.4. Functional Annotations of DEGs, DELs, and DASs
2.4. Real-Time PCR
3. Results
3.1. Liver Transcriptomic Statistics and the Abundance of Expression Profiles
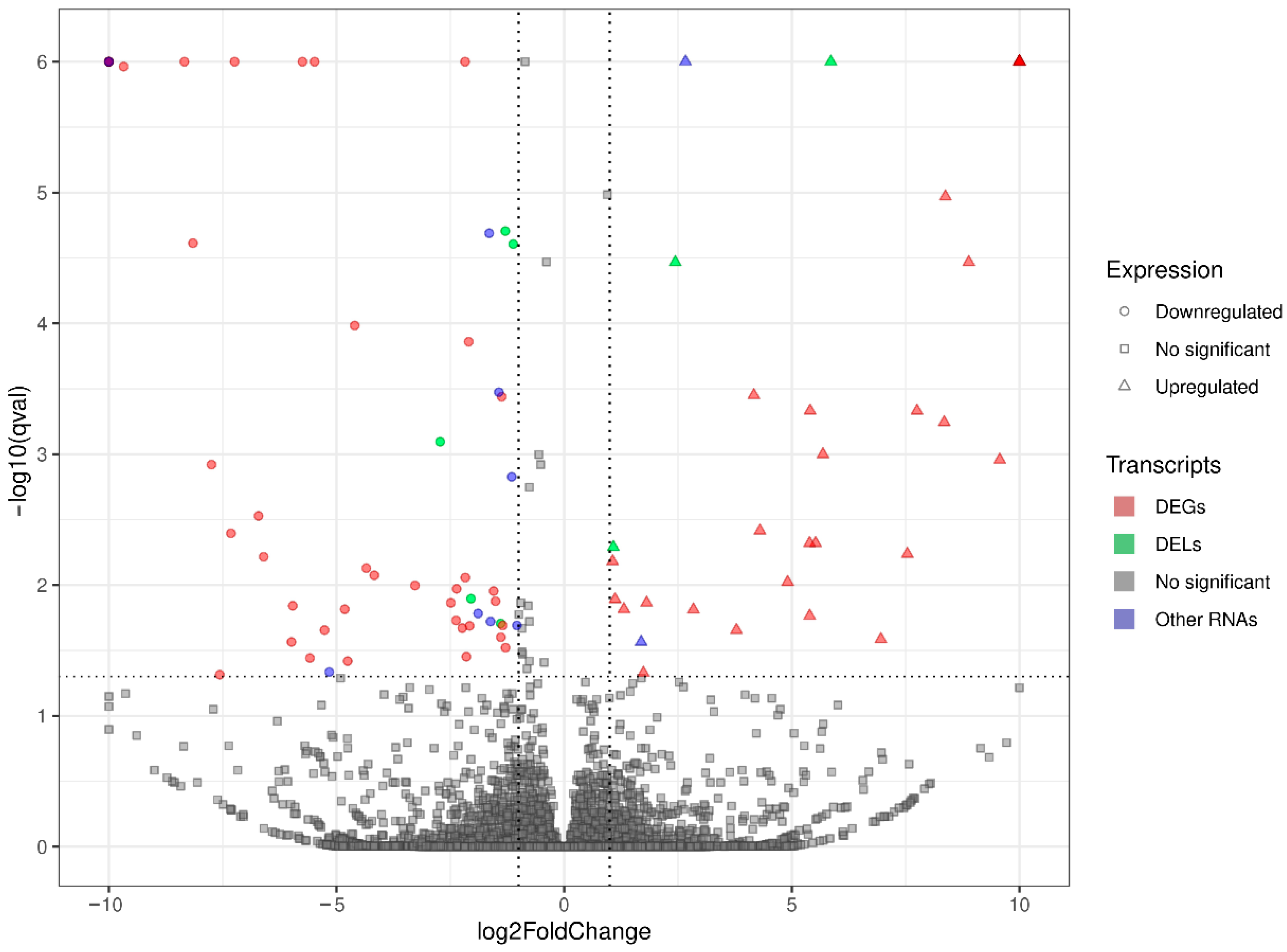
3.2. Transcriptomic Differences in the Liver after Oral Exposure to BPA
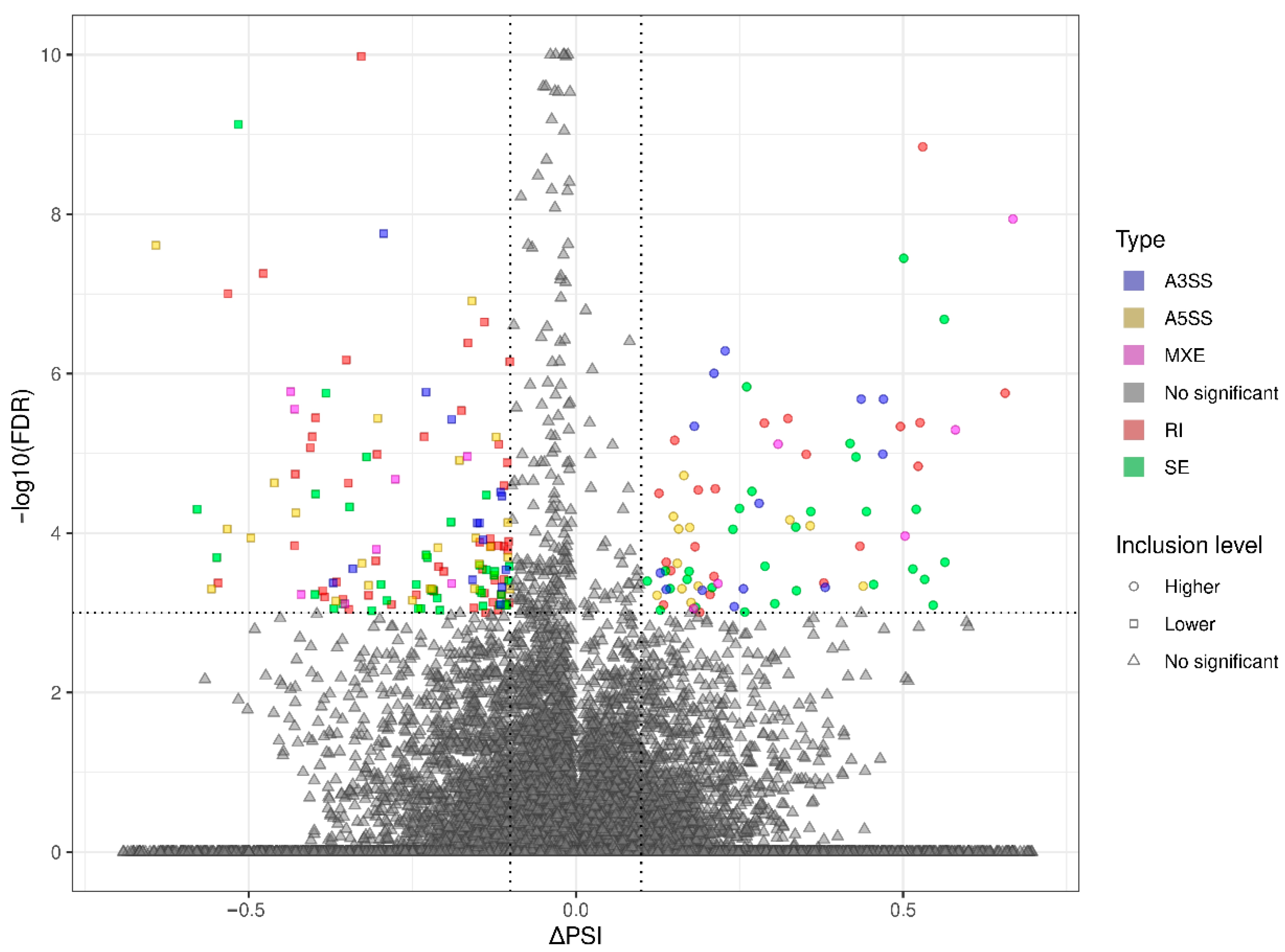
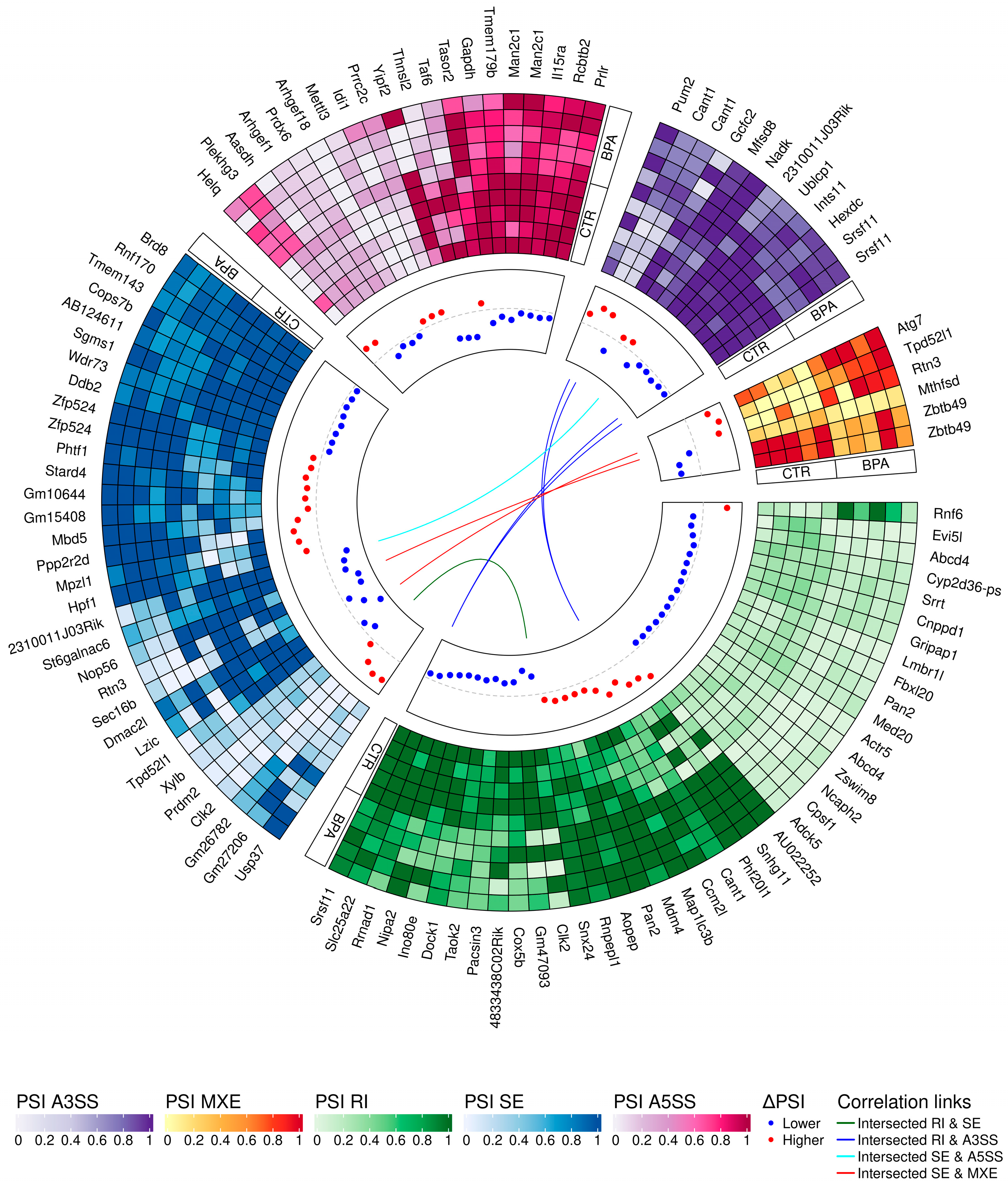
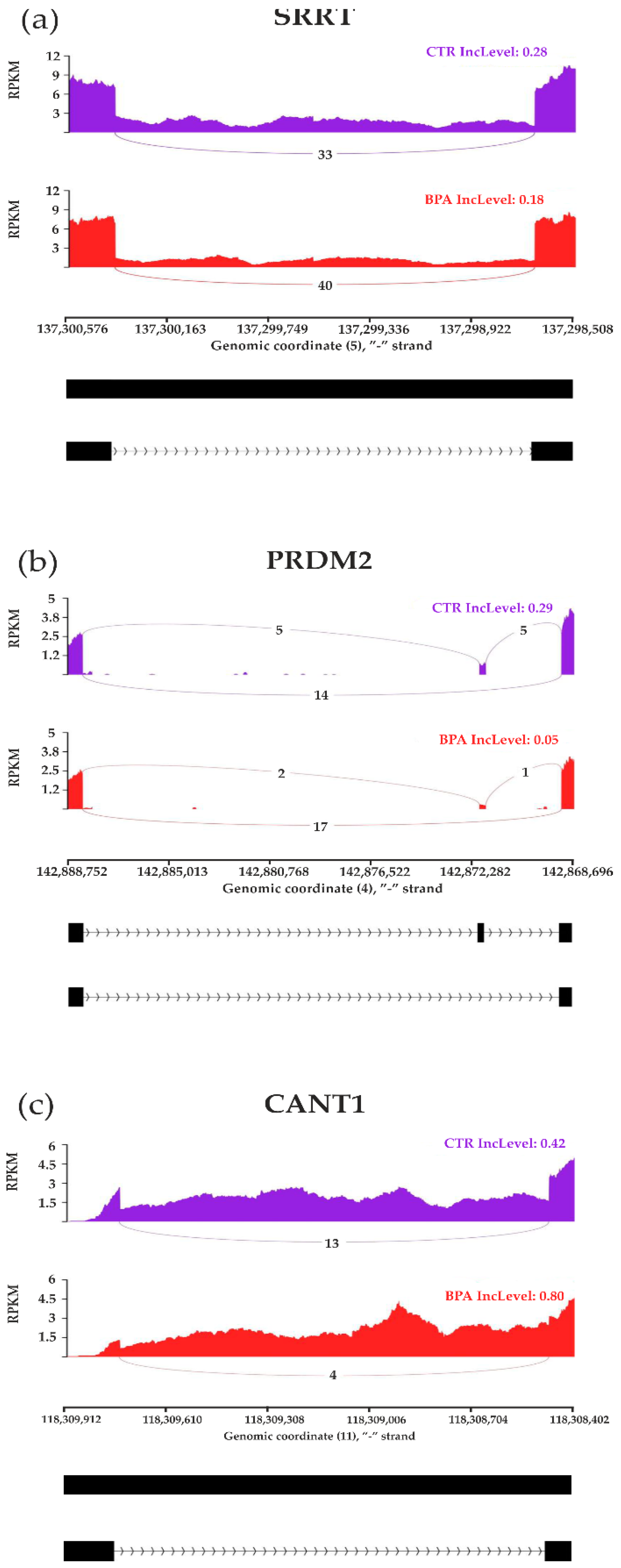
3.3. Transcriptomic Alternative Splicing Signatures of the Liver after Oral Exposure to BPA
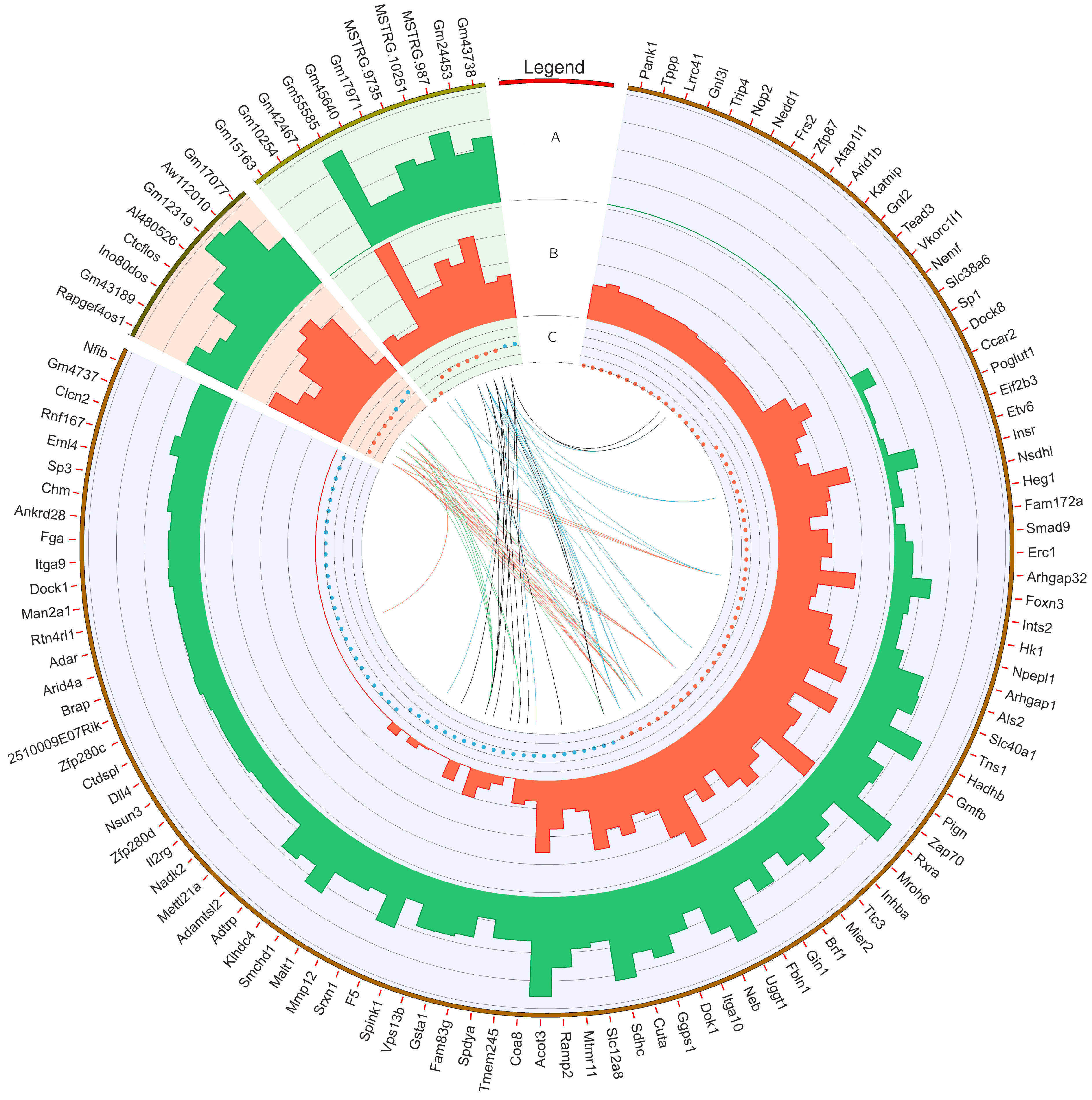
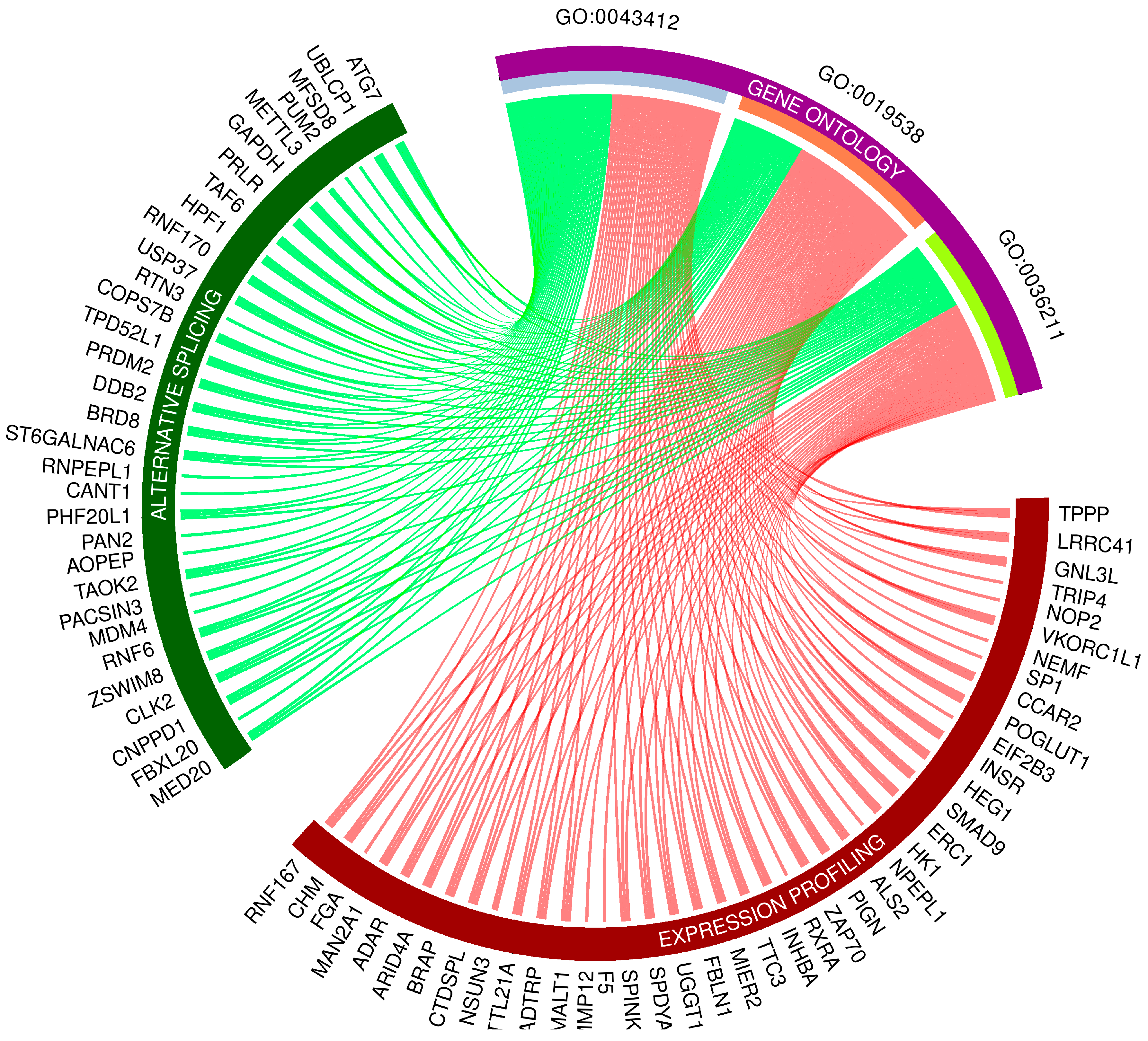
3.4. Gene Ontology Networks and Pathway Signaling Analysis of DEGs, DELs and DASs
3.5. Validation of the Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Michałowicz, J. Bisphenol A—Sources, Toxicity and Biotransformation. Environ. Toxicol. Pharmacol. 2014, 37, 738–758. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.S. Bisphenol A: An Endocrine Disruptor with Widespread Exposure and Multiple Effects. J. Steroid Biochem. Mol. Biol. 2011, 127, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhao, Z.; Ji, W. Bisphenol A Induces Apoptosis, Oxidative Stress and Inflammatory Response in Colon and Liver of Mice in a Mitochondria-Dependent Manner. Biomed. Pharmacother. 2019, 117, 109182. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Q.; Wong, C.K.C.; Zheng, J.S.; Bouwman, H.; Barra, R.; Wahlström, B.; Neretin, L.; Wong, M.H. Bisphenol A (BPA) in China: A Review of Sources, Environmental Levels, and Potential Human Health Impacts. Environ. Int. 2012, 42, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Liu, F.; Kannan, K. Bisphenol S, a New Bisphenol Analogue, in Paper Products and Currency Bills and Its Association with Bisphenol A Residues. Environ. Sci. Technol. 2012, 46, 6515–6522. [Google Scholar] [CrossRef]
- EU Commission Regulation (EU) 2016/2235 of 12 December 2016 Amending Annex XVII to Regulation (EC) No 1907/2006 of the European Parliament and of the Council Concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) as Regar. Off. J. Eur. Union 2016, 2001, 20–30.
- Hanaoka, T.; Kawamura, N.; Hara, K. Urinary Bisphenol A and Plasma Hormone Concentrations in Male Workers Exposed to Bisphenol A Diglycidyl Ether and Mixed Organic Solvents. Occup. Environ. Med. 2002, 59, 625–628. [Google Scholar] [CrossRef]
- Konieczna, A.R.A.; Rachoń, D. Health Risk of Exposure to Bisphenol A (BPA). Rocz. Państwowego Zakładu Hig. 2015, 66, 5–11. [Google Scholar]
- Noureddine El Moussawi, S.; Karam, R.; Cladière, M.; Chébib, H.; Ouaini, R.; Camel, V. Effect of Sterilisation and Storage Conditions on the Migration of Bisphenol A from Tinplate Cans of the Lebanese Market. Food Addit. Contam. Part A 2018, 35, 377–386. [Google Scholar] [CrossRef]
- Besaratinia, A. The State of Research and Weight of Evidence on the Epigenetic Effects of Bisphenol A. Int. J. Mol. Sci. 2023, 24, 7951. [Google Scholar] [CrossRef]
- Shmarakov, I.O.; Borschovetska, V.L.; Blaner, W.S. Hepatic Detoxification of Bisphenol A Is Retinoid-Dependent. Toxicol. Sci. 2017, 157, kfx022. [Google Scholar] [CrossRef] [PubMed]
- Hanioka, N.; Naito, T.; Narimatsu, S. Human UDP-Glucuronosyltransferase Isoforms Involved in Bisphenol A Glucuronidation. Chemosphere 2008, 74, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Andra, S.S.; Austin, C.; Yang, J.; Patel, D.; Arora, M.; Abstract, G. Recent Advances in Simultaneous Analysis of Bisphenol A and Its Conjugates in Human Matrices: Exposure Biomarker Perspectives HHS Public Access. Sci. Total Environ. 2016, 572, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Landolfi, A.; Troisi, J.; Savanelli, M.C.; Vitale, C.; Barone, P.; Amboni, M. Bisphenol A Glucuronidation in Patients with Parkinson’s Disease. Neurotoxicology 2017, 63, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, A.; Roy, D. In Vivo DNA Adduct Formation by Bisphenol A. Environ. Mol. Mutagen. 1995, 26, 60–66. [Google Scholar] [CrossRef]
- Kazemi, S.; Mousavi Kani, S.N.; Rezazadeh, L.; Pouramir, M.; Ghasemi-Kasman, M.; Moghadamnia, A.A. Low Dose Administration of Bisphenol A Induces Liver Toxicity in Adult Rats. Biochem. Biophys. Res. Commun. 2017, 494, 107–112. [Google Scholar] [CrossRef]
- Hassan, Z.K.; Elobeid, M.A.; Virk, P.; Omer, S.A.; ElAmin, M.; Daghestani, M.H.; AlOlayan, E.M. Bisphenol A Induces Hepatotoxicity through Oxidative Stress in Rat Model. Oxid. Med. Cell Longev. 2012, 2012, 194829. [Google Scholar] [CrossRef]
- Ma, L.; Hu, J.; Li, J.; Yang, Y.; Zhang, L.; Zou, L.; Gao, R.; Peng, C.; Wang, Y.; Luo, T.; et al. Bisphenol A Promotes Hyperuricemia via Activating Xanthine Oxidase. FASEB J. 2018, 32, 1007–1016. [Google Scholar] [CrossRef]
- Lee, S.; Min, H.; Ha, J.; Kim, B.; Choi, M.; Kim, S. Dysregulation of the MiRNA Biogenesis Components DICER1, DROSHA, DGCR8 and AGO2 in Clear Cell Renal Cell Carcinoma in Both a Korean Cohort and the Cancer Genome Atlas Kidney Clear Cell Carcinoma Cohort. Oncol. Lett. 2019, 18, 4337. [Google Scholar] [CrossRef]
- Dallio, M.; Diano, N.; Masarone, M.; Gravina, A.G.; Patanè, V.; Romeo, M.; Di Sarno, R.; Errico, S.; Nicolucci, C.; Abenavoli, L.; et al. Chemical Effect of Bisphenol A on Non-Alcoholic Fatty Liver Disease. Int. J. Environ. Res. Public Health 2019, 16, 3134. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, H.; Wu, J.; Yuan, L.; Wang, Y.; Du, X.; Wang, R.; Marwa, P.W.; Petlulu, P.; Chen, X.; et al. The Adverse Health Effects of Bisphenol A and Related Toxicity Mechanisms. Environ. Res. 2019, 176, 108575. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.-W.; Jeong, J.-Y.; Hwang, M.-S.; Jung, K.-K.; Lee, K.-H.; Lee, H.-M. Establishment of the Korean Tolerable Daily Intake of Bisphenol A Based on Risk Assessments by an Expert Committee. Toxicol. Res. 2010, 26, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Tyl, R.W. Three-Generation Reproductive Toxicity Study of Dietary Bisphenol A in CD Sprague-Dawley Rats. Toxicol. Sci. 2002, 68, 121–146. [Google Scholar] [CrossRef] [PubMed]
- Makowska, K.; Lepiarczyk, E.; Gonkowski, S. The Comparison of the Influence of Bisphenol A (BPA) and Its Analogue Bisphenol S (BPS) on the Enteric Nervous System of the Distal Colon in Mice. Nutrients 2022, 15, 200. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinform. 2010. Available online: www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 23 August 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Sequence Analysis STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.-X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. RMATS: Robust and Flexible Detection of Differential Alternative Splicing from Replicate RNA-Seq Data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Carbon, S.; Dietze, H.; Lewis, S.E.; Mungall, C.J.; Munoz-Torres, M.C.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Fey, P.; Thomas, P.D.; et al. Expansion of the Gene Ontology Knowledgebase and Resources: The Gene Ontology Consortium. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. G:Profiler—A Web Server for Functional Interpretation of Gene Lists. Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Prakash, A.; Tiwari, R. Environmental Endocrine-Disrupting Chemical Exposure: Role in Non-Communicable Diseases. Front. Public. Health 2020, 8, 553850. [Google Scholar] [CrossRef]
- Abdulhameed, A.-S.A.R.; Lim, V.; Bahari, H.; Khoo, B.Y.; Abdullah, M.N.H.; Tan, J.J.; Yong, Y.K. Adverse Effects of Bisphenol A on the Liver and Its Underlying Mechanisms: Evidence from In Vivo and In Vitro Studies. Biomed. Res. Int. 2022, 2022, 8227314. [Google Scholar] [CrossRef]
- Zou, Y.; Zhong, L.; Hu, C.; Sheng, G. Association between the Alanine Aminotransferase/Aspartate Aminotransferase Ratio and New-Onset Non-Alcoholic Fatty Liver Disease in a Nonobese Chinese Population: A Population-Based Longitudinal Study. Lipids Health Dis. 2020, 19, 245. [Google Scholar] [CrossRef]
- Elswefy, S.E.-S.; Abdallah, F.R.; Atteia, H.H.; Wahba, A.S.; Hasan, R.A. Inflammation, Oxidative Stress and Apoptosis Cascade Implications in Bisphenol A-Induced Liver Fibrosis in Male Rats. Int. J. Exp. Pathol. 2016, 97, 369–379. [Google Scholar] [CrossRef]
- Bindhumol, V.; Chitra, K.C.; Mathur, P.P. Bisphenol A Induces Reactive Oxygen Species Generation in the Liver of Male Rats. Toxicology 2003, 188, 117–124. [Google Scholar] [CrossRef]
- Murata, M.; Kang, J.H. Bisphenol A (BPA) and Cell Signaling Pathways. Biotechnol. Adv. 2018, 36, 311–327. [Google Scholar] [CrossRef]
- Deng1, Q.; He1, B.; Pan1, Y.; Sun2, H.; Liu1, X.; Chen2, J.; Ying3, H.; Lin1, K.; Peng3, H.; Wang1, S. Polymorphisms of GSTA1 Contribute to Elevated Cancer Risk: Evidence from 15 Studies. J. Buon 2015, 20, 287–295. [Google Scholar]
- Van Runnard Heimel, P.J.; Kavelaars, A.; Heijnen, C.J.; Peters, W.H.M.; Huisjes, A.J.M.; Franx, A.; Bruinse, H.W. HELLP Syndrome Is Associated with an Increased Inflammatory Response, Which May Be Inhibited by Administration of Prednisolone. Hypertens. Pregnancy 2008, 27, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.K.; Chang, W.C.; Chen, S.E.; Chang, L.C. DOCK1 Regulates Growth and Motility through the RRP1B-Claudin-1 Pathway in Claudin-Low Breast Cancer Cells. Cancers 2019, 11, 1762. [Google Scholar] [CrossRef]
- Cui, W.; Xue, J. Circular RNA DOCK1 Downregulates MicroRNA-124 to Induce the Growth of Human Thyroid Cancer Cell Lines. BioFactors 2020, 46, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, X.; Guo, X.; Chen, J.; Li, C.; Chen, M.; Liu, L.; Zhang, X.; Zu, X. Circular RNA DOCK1 Promotes Bladder Carcinoma Progression via Modulating CircDOCK1/Hsa-miR-132-3p/Sox5 Signalling Pathway. Cell Prolif. 2019, 52, e12614. [Google Scholar] [CrossRef]
- Wang, L.; Wei, Y.; Yan, Y.; Wang, H.; Yang, J.; Zheng, Z.; Zha, J.; Bo, P.; Tang, Y.; Guo, X.; et al. CircDOCK1 Suppresses Cell Apoptosis via Inhibition of MiR-196a-5p by Targeting BIRC3 in OSCC. Oncol. Rep. 2018, 39, 951–966. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, J.; Wu, Y. Interference with CircRNA DOCK1 Inhibits Hepatocellular Carcinoma Cell Proliferation, Invasion and Migration by Regulating the MiR-654-5p/SMAD2 Axis. Mol. Med. Rep. 2021, 24, 1–8. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Q.-L.; Mao, L.-H.; Chen, S.-Y.; Yang, Z.-H.; Liu, X.; Gao, Y.-H.; Li, X.-Q.; Zhou, Z.-H.; He, S. Hepatocyte-Specific Knock-Out of Nfib Aggravates Hepatocellular Tumorigenesis via Enhancing Urea Cycle. Front. Mol. Biosci. 2022, 9, 1–14. [Google Scholar] [CrossRef]
- Inokawa, Y.; Sonohara, F.; Kanda, M.; Hayashi, M.; Nishikawa, Y.; Sugimoto, H.; Kodera, Y.; Nomoto, S. Correlation Between Poor Prognosis and Lower TPPP Gene Expression in Hepatocellular Carcinoma. Anticancer Res. 2016, 36, 4639–4646. [Google Scholar] [CrossRef]
- Yu, W.; Dai, Y. LncRNA LOXL1-AS1 Promotes Liver Cancer Cell Proliferation and Migration by Regulating the MiR-377-3p/NFIB Axis. Oncol. Lett. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, L.Y.; Cheng, S.J.; Zhang, A.M.; Jin, X.S.; Li, Y. P53-Induced MicroRNA-1246 Inhibits the Cell Growth of Human Hepatocellular Carcinoma Cells by Targeting NFIB. Oncol. Rep. 2015, 33, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Kuang, J.; Yang, Y.; Zhang, L.; Leng, B.; She, R.; Zou, L. A Prognostic Model Based on NSUN3 Was Established to Evaluate the Prognosis and Response to Immunotherapy in Liver Hepatocellular Carcinoma. Mediat. Inflamm. 2023, 2023, 6645476. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Tsao, S.C.; Yuan, S.S.F.; Tsai, H.P.; Chai, C.Y. Serine Protease Inhibitor Kazal Type 1 (SPINK1) Promotes Proliferation of Colorectal Cancer Through the Epidermal Growth Factor as a Prognostic Marker. Pathol. Oncol. Res. 2015, 21, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Xie, W.; Wang, S.; Li, Q.; Wei, X.; Chen, B.; Hua, Y.; Li, S.; Peng, B.; Shen, S. High SPINK1 Expression Predicts Poor Prognosis and Promotes Cell Proliferation and Metastasis of Hepatocellular Carcinoma. J. Investig. Surg. 2021, 34, 1011–1020. [Google Scholar] [CrossRef]
- Wang, F.; Liu, H.; Bai, Y.; Li, H.; Wang, Z.; Xu, X. Performance of SPINK1 and SPINK1-Based Diagnostic Model in Detection of Hepatocellular Carcinoma. J. Clin. Lab. Anal. 2021, 35, e24025. [Google Scholar] [CrossRef]
- Lin, T.C. Functional Roles of Spink1 in Cancers. Int. J. Mol. Sci. 2021, 22, 3814. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Lin, P.-H.; Chen, C.-C.; Weng, W.-H.; Yu, K.-J.; Liu, C.-Y.; Hsieh, C.-H.; Chang, T.-H.; Shao, I.-H.; Kan, H.-C.; et al. Gain of TPPP as a Predictor of Progression in Patients with Bladder Cancer. Exp. Ther. Med. 2021, 22, 1204. [Google Scholar] [CrossRef]
- Schofield, A.V.; Gamell, C.; Suryadinata, R.; Sarcevic, B.; Bernard, O. Tubulin Polymerization Promoting Protein 1 (Tppp1) Phosphorylation by Rho-Associated Coiled-Coil Kinase (Rock) and Cyclin-Dependent Kinase 1 (Cdk1) Inhibits Microtubule Dynamics to Increase Cell Proliferation*. J. Biol. Chem. 2013, 288, 7907–7917. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The Human Gene Integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef]
- Schulte-Hermann, R.; Bursch, W.; Löw-Baselli, A.; Wagner, A.; Grasl-Kraupp, B. Apoptosis in the Liver and Its Role in Hepatocarcinogenesis. Cell Biol. Toxicol. 1997, 13, 339–348. [Google Scholar] [CrossRef]
- Rodgarkia-Dara, C.; Vejda, S.; Erlach, N.; Losert, A.; Bursch, W.; Berger, W.; Schulte-Hermann, R.; Grusch, M. The Activin Axis in Liver Biology and Disease. Mutat. Res./Rev. Mutat. Res. 2006, 613, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Frost, K.; Seir, K.; Lackner, A.; Grusch, M.; Grasl-Kraupp, B.; Schulte-Hermann, R.; Rodgarkia-Dara, C. Inhibin/Activin Expression in Human and Rodent Liver: Subunits α and Βb as New Players in Human Hepatocellular Carcinoma. Br. J. Cancer 2011, 104, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Dewdney, B.; Hebbard, L. A Novel Function for HEG1 in Promoting Metastasis in Hepatocellular Carcinoma. Clin. Sci. 2019, 133, 2019–2022. [Google Scholar] [CrossRef]
- Federico, A.; Rienzo, M.; Abbondanza, C.; Costa, V.; Ciccodicola, A.; Casamassimi, A. Pan-Cancer Mutational and Transcriptional Analysis of the Integrator Complex. Int. J. Mol. Sci. 2017, 18, 936. [Google Scholar] [CrossRef]
- Lim, B.; Kim, C.; Kim, J.-H.; Kwon, W.S.; Lee, W.S.; Kim, J.M.; Park, J.Y.; Kim, H.S.; Park, K.H.; Kim, T.S.; et al. Genetic Alterations and Their Clinical Implications in Gastric Cancer Peritoneal Carcinomatosis Revealed by Whole-Exome Sequencing of Malignant Ascites. Oncotarget 2016, 7, 8055–8066. [Google Scholar] [CrossRef]
- Han, X.; Zhao, L.; Li, X. HELQ in Cancer and Reproduction Minireview. Neoplasma 2016, 63, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Kong, Y.; Zhang, Y.; Sun, M.; Liu, S.; Wu, Z.; Gao, L.; Liang, X.; Ma, C. SREBF2–STARD4 Axis Confers Sorafenib Resistance in Hepatocellular Carcinoma by Regulating Mitochondrial Cholesterol Homeostasis. Cancer Sci. 2023, 114, 477–489. [Google Scholar] [CrossRef]
- Chu, Y.D.; Lin, W.R.; Lin, Y.H.; Kuo, W.H.; Tseng, C.J.; Lim, S.N.; Huang, Y.L.; Huang, S.C.; Wu, T.J.; Lin, K.H.; et al. Cox5b-Mediated Bioenergetic Alteration Regulates Tumor Growth and Migration by Modulating Ampk-Uhmk1-Erk Cascade in Hepatoma. Cancers 2020, 12, 1646. [Google Scholar] [CrossRef]
- He, Q.; Huang, Y.; Cai, L.; Zhang, S.; Zhang, C. Expression and Prognostic Value of Ars2 in Hepatocellular Carcinoma. Int. J. Clin. Oncol. 2014, 19, 880–888. [Google Scholar] [CrossRef]
- Tanadi, C.; Bambang, A.; Wendi, I.P.; Sidharta, V.M.; Hananta, L.; Sumarpo, A. The Predictive Value of PRDM2 in Solid Tumor: A Systematic Review and Meta-Analysis. PeerJ 2020, 8, e8826. [Google Scholar] [CrossRef]
- Yao, Q.; Yu, Y.; Wang, Z.; Zhang, M.; Ma, J.; Wu, Y.; Zheng, Q.; Li, J. CANT1 Serves as a Potential Prognostic Factor for Lung Adenocarcinoma and Promotes Cell Proliferation and Invasion In Vitro. BMC Cancer 2022, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, X.; Li, Y.; Chen, J.; Lv, Y.; Dai, S. Prognosis and Personalized Treatment Prediction in TP53 -Mutant Hepatocellular Carcinoma: An in Silico Strategy towards Precision Oncology. Brief. Bioinform. 2021, 22, bbaa164. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Q.; Zeng, Z.; Wu, P.; Jiang, Y.; Luo, T.; Ji, X.; Zhang, Q.; Hao, Y.; Chen, L. Increased Expression of G9A Contributes to Carcinogenesis and Indicates Poor Prognosis in Hepatocellular Carcinoma. Oncol. Lett. 2018, 15, 9757–9765. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Karsli-Uzunbas, G.; Poillet-Perez, L.; Sawant, A.; Hu, Z.S.; Zhao, Y.; Moore, D.; Hu, W.; White, E. Autophagy Promotes Mammalian Survival by Suppressing Oxidative Stress and P53. Genes. Dev. 2020, 34, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Taucher, E.; Mykoliuk, I.; Fediuk, M.; Smolle-Juettner, F.-M. Autophagy, Oxidative Stress and Cancer Development. Cancers 2022, 14, 1637. [Google Scholar] [CrossRef]
- Cimmino, I.; Fiory, F.; Perruolo, G.; Miele, C.; Beguinot, F.; Formisano, P.; Oriente, F. Potential Mechanisms of Bisphenol A (BPA) Contributing to Human Disease. Int. J. Mol. Sci. 2020, 21, 5761. [Google Scholar] [CrossRef]
- Burra, P.; Becchetti, C.; Germani, G. NAFLD and Liver Transplantation: Disease Burden, Current Management and Future Challenges. JHEP Rep. 2020, 2, 100192. [Google Scholar] [CrossRef]
- Nicolucci, C.; Errico, S.; Federico, A.; Dallio, M.; Loguercio, C.; Diano, N. Human Exposure to Bisphenol A and Liver Health Status: Quantification of Urinary and Circulating Levels by LC–MS/MS. J. Pharm. Biomed. Anal. 2017, 140, 105–112. [Google Scholar] [CrossRef]
- Masarone, M.; Rosato, V.; Dallio, M.; Gravina, A.G.; Aglitti, A.; Loguercio, C.; Federico, A.; Persico, M. Role of Oxidative Stress in Pathophysiology of Nonalcoholic Fatty Liver Disease. Oxidative Med. Cell. Longev. 2018, 2018, 9547613. [Google Scholar] [CrossRef]
- Marušić, M.; Paić, M.; Knobloch, M.; Liberati Pršo, A.M. NAFLD, Insulin Resistance, and Diabetes Mellitus Type 2. Can. J. Gastroenterol. Hepatol. 2021, 2021, 6613827. [Google Scholar] [CrossRef]
- Tanase, D.M.; Gosav, E.M.; Costea, C.F.; Ciocoiu, M.; Lacatusu, C.M.; Maranduca, M.A.; Ouatu, A.; Floria, M. The Intricate Relationship between Type 2 Diabetes Mellitus (T2DM), Insulin Resistance (IR), and Nonalcoholic Fatty Liver Disease (NAFLD). J. Diabetes Res. 2020, 2020, 3920196. [Google Scholar] [CrossRef] [PubMed]
- Nobakht, H.; Mahmoudi, T.; Sabzikarian, M.; Tabaeian, S.P.; Rezamand, G.; Asadi, A.; Farahani, H.; Dabiri, R.; Mansour-Ghanaei, F.; Maleki, I.; et al. Insulin and Insulin Receptor Gene Polymorphisms and Susceptibility to Nonalcoholic Fatty Liver Disease. Arq. Gastroenterol. 2020, 57, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.D.; Kulkarni, R.N.; Postic, C.; Previs, S.F.; Shulman, G.I.; Magnuson, M.A.; Kahn, C.R. Loss of Insulin Signaling in Hepatocytes Leads to Severe Insulin Resistance and Progressive Hepatic Dysfunction. Mol. Cell 2000, 6, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ouamrane, L.; Larrieu, G.; Gauthier, B.; Pineau, T. RXR Activators Molecular Signalling: Involvement of a PPARα-Dependent Pathway in the Liver and Kidney, Evidence for an Alternative Pathway in the Heart. Br. J. Pharmacol. 2003, 138, 845–854. [Google Scholar] [CrossRef]
- Yong-Mei Zhang, H.; Chohnan, S.; Virga, K.G.; Stevens, R.D.; Ilkayeva, O.R.; Wenner, B.R.; Bain, J.R.; Newgard, C.B.; Lee, R.E.; Rock, C.O.; et al. Chemical Knockout of Pantothenate Kinase Reveals the Metabolic and Genetic Program Responsible for Hepatic Coenzyme A. Chem. Biol. 2007, 14, 291–302. [Google Scholar] [CrossRef]
- Zi, Y.; Gao, J.; Wang, C.; Guan, Y.; Li, L.; Ren, X.; Zhu, L.; Mu, Y.; Chen, S.; Zeng, Z.; et al. Pantothenate Kinase 1 Inhibits the Progression of Hepatocellular Carcinoma by Negatively Regulating Wnt/β-Catenin Signaling. Int. J. Biol. Sci. 2022, 18, 1539–1554. [Google Scholar] [CrossRef]
- Moylan, C.A.; Mavis, A.M.; Jima, D.; Maguire, R.; Bashir, M.; Hyun, J.; Cabezas, M.N.; Parish, A.; Niedzwiecki, D.; Diehl, A.M.; et al. Alterations in DNA Methylation Associate with Fatty Liver and Metabolic Abnormalities in a Multi-Ethnic Cohort of Pre-Teenage Children. Epigenetics 2022, 17, 1446–1461. [Google Scholar] [CrossRef]

| Sample | CTR1 | CTR2 | CTR3 | CTR4 | CTR5 | BPA1 | BPA2 | BPA3 | BPA4 | BPA5 |
|---|---|---|---|---|---|---|---|---|---|---|
| Raw reads | 63,701,054 | 62,907,658 | 63,490,610 | 61,783,238 | 65,647,032 | 61,466,386 | 63,656,704 | 60,608,086 | 64,195,852 | 65,720,614 |
| Processed reads | 53,607,300 | 52,885,042 | 54,905,736 | 52,410,248 | 56,702,958 | 51,304,764 | 55,754,414 | 52,413,416 | 54,694,930 | 56,629,844 |
| All mapped reads | 48,317,860 | 44,886,888 | 50,268,208 | 48,843,756 | 51,240,802 | 48,998,204 | 51,833,736 | 49,518,666 | 50,821,082 | 51,468,034 |
| Uniquely mapped | 44,535,610 | 41,312,746 | 46,211,692 | 44,860,506 | 47,096,330 | 45,390,806 | 47,870,430 | 46,095,184 | 46,905,746 | 47,462,692 |
| Multi-mapped reads | 3,767,304 | 3,554,566 | 4,035,672 | 3,960,676 | 4,120,960 | 3,593,654 | 3,943,166 | 3,406,710 | 3,898,412 | 3,992,284 |
| Mapped to too many loci | 14,946 | 19,576 | 20,844 | 22,574 | 23,512 | 13,744 | 20,140 | 16,772 | 16,924 | 13,058 |
| % of CDS mapped bases | 67.74% | 68.16% | 68.71% | 67.99% | 68.70% | 70.27% | 68.98% | 68.11% | 69.05% | 69.02% |
| % of UTR mapped bases | 27.01% | 26.96% | 26.37% | 26.92% | 26.59% | 26.44% | 27.35% | 28.11% | 27.42% | 26.86% |
| % of Intron mapped bases | 3.37% | 3.01% | 3.05% | 3.31% | 2.96% | 1.86% | 2.05% | 2.23% | 1.99% | 2.37% |
| % of Intergenic mapped bases | 1.88% | 1.87% | 1.86% | 1.79% | 1.74% | 1.43% | 1.63% | 1.55% | 1.54% | 1.74% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiszpolska, M.; Lepiarczyk, E.; Maździarz, M.A.; Paukszto, Ł.; Makowczenko, K.G.; Lipka, A.; Łopieńska-Biernat, E.; Makowska, K.; Gonkowski, S.; Correia-de-Sá, P.; et al. The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies. Cancers 2023, 15, 5014. https://doi.org/10.3390/cancers15205014
Wiszpolska M, Lepiarczyk E, Maździarz MA, Paukszto Ł, Makowczenko KG, Lipka A, Łopieńska-Biernat E, Makowska K, Gonkowski S, Correia-de-Sá P, et al. The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies. Cancers. 2023; 15(20):5014. https://doi.org/10.3390/cancers15205014
Chicago/Turabian StyleWiszpolska, Marta, Ewa Lepiarczyk, Mateusz A. Maździarz, Łukasz Paukszto, Karol G. Makowczenko, Aleksandra Lipka, Elżbieta Łopieńska-Biernat, Krystyna Makowska, Sławomir Gonkowski, Paulo Correia-de-Sá, and et al. 2023. "The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies" Cancers 15, no. 20: 5014. https://doi.org/10.3390/cancers15205014
APA StyleWiszpolska, M., Lepiarczyk, E., Maździarz, M. A., Paukszto, Ł., Makowczenko, K. G., Lipka, A., Łopieńska-Biernat, E., Makowska, K., Gonkowski, S., Correia-de-Sá, P., & Majewska, M. (2023). The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies. Cancers, 15(20), 5014. https://doi.org/10.3390/cancers15205014








