Type 2 Cystatins and Their Roles in the Regulation of Human Immune Response and Cancer Progression
Abstract
:Simple Summary
Abstract
1. Introduction
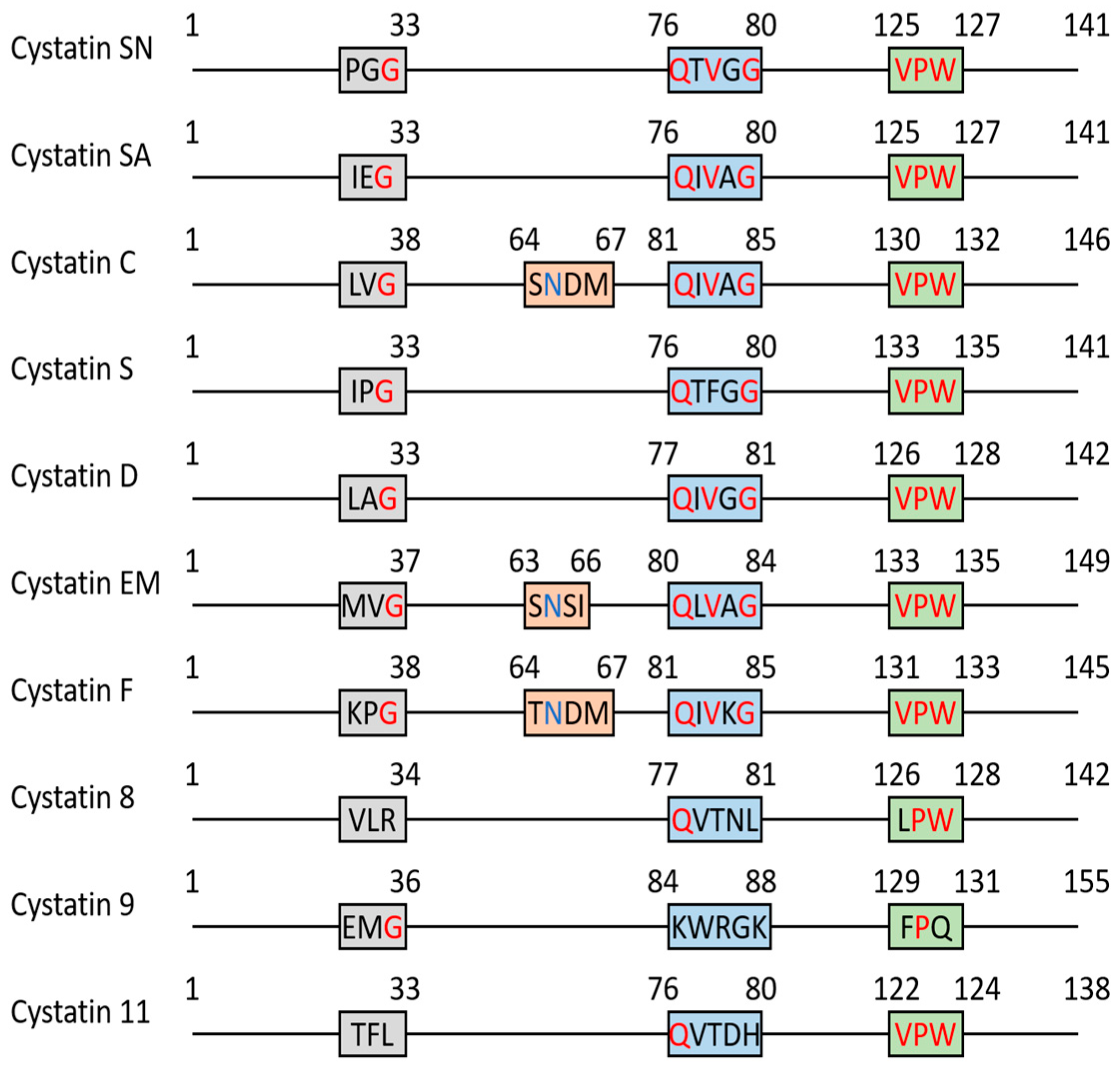
2. Cystatins as Protease Inhibitors
2.1. SD-Type Cystatins: Cystatin SN, SA, S, D (CST1, CST2, CST4, CST5)
2.2. Cystatin C (CST3)
2.3. Cystatin E/M (CST6)
2.4. Cystatin F (CST7)
2.5. Testatins: CST8, CST9, CST11
3. Cathepsin, Legumain, and Cystatins as Immune Modulators
3.1. Cystatin SN (CYS1)
3.2. Other SD-Type Cystatins (CST2, CST4, CST5)
3.3. Cystatin C (CYSC)
3.4. Cystatin E/M (CYS6)
3.5. Cystatin F (CYSF)
4. The Role of Cathepsins, Legumain, and Type 2 Cystatins in Cancer Development
4.1. Cystatin SN (CYS1)
4.2. Other SD-Type Cystatins (CST2, CST4, CST5)
4.3. Cystatin C (CYSC)
4.4. Cystatin E/M (CYS6)
4.5. Cystatin F (CYSF)
5. Type 2 Cystatin Studies on Mouse Model: A Brief Overview
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Novinec, M.; Lenarcic, B. Papain-like peptidases: Structure, function, and evolution. Biomol. Concepts 2013, 4, 287–308. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Fossum, K.; Whitaker, J.R. Ficin and papain inhibitor from chicken egg white. Arch. Biochem. Biophys. 1968, 125, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Abrahamson, M.; Alvarez-Fernandez, M.; Nathanson, C.M. Cystatins. Biochem. Soc. Symp. 2003, 70, 179–199. [Google Scholar] [CrossRef]
- Ochieng, J.; Chaudhuri, G. Cystatin superfamily. J. Health Care Poor Underserved 2010, 21, 51–70. [Google Scholar] [CrossRef]
- Kolodziejczyk, R.; Michalska, K.; Hernandez-Santoyo, A.; Wahlbom, M.; Grubb, A.; Jaskolski, M. Crystal structure of human cystatin C stabilized against amyloid formation. FEBS J 2010, 277, 1726–1737. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Nandy, S.K.; Seal, A. Structural Dynamics Investigation of Human Family 1 & 2 Cystatin-Cathepsin L1 Interaction: A Comparison of Binding Modes. PLoS ONE 2016, 11, e0164970. [Google Scholar] [CrossRef]
- Barrett, A.J.; Rawlings, N.D. Families and clans of cysteine peptidases. Perspect. Drug Discov. Des. 1996, 6, 1–11. [Google Scholar] [CrossRef]
- Turk, V.; Stoka, V.; Turk, D. Cystatins: Biochemical and structural properties, and medical relevance. Front. Biosci. J. Virtual Libr. 2008, 13, 5406–5420. [Google Scholar] [CrossRef]
- Lalmanach, G.; Kasabova-Arjomand, M.; Lecaille, F.; Saidi, A. Cystatin M/E (Cystatin 6): A Janus-Faced Cysteine Protease Inhibitor with Both Tumor-Suppressing and Tumor-Promoting Functions. Cancers 2021, 13, 1877. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Fernandez, M.; Barrett, A.J.; Gerhartz, B.; Dando, P.M.; Ni, J.; Abrahamson, M. Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J. Biol. Chem. 1999, 274, 19195–19203. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.P.; Thiesse, M.; Hicks, M.J. Expression of type 2 cystatin genes CST1-CST5 in adult human tissues and the developing submandibular gland. DNA Cell Biol. 2002, 21, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Zeeuwen, P.L.; van Vlijmen-Willems, I.M.; Egami, H.; Schalkwijk, J. Cystatin M / E expression in inflammatory and neoplastic skin disorders. Br. J. Dermatol. 2002, 147, 87–94. [Google Scholar] [CrossRef]
- Halfon, S.; Ford, J.; Foster, J.; Dowling, L.; Lucian, L.; Sterling, M.; Xu, Y.; Weiss, M.; Ikeda, M.; Liggett, D.; et al. Leukocystatin, a new Class II cystatin expressed selectively by hematopoietic cells. J. Biol. Chem. 1998, 273, 16400–16408. [Google Scholar] [CrossRef]
- Freije, J.P.; Abrahamson, M.; Olafsson, I.; Velasco, G.; Grubb, A.; Lopez-Otin, C. Structure and expression of the gene encoding cystatin D, a novel human cysteine proteinase inhibitor. J. Biol. Chem. 1991, 266, 20538–20543. [Google Scholar] [CrossRef]
- Alvarez-Fernandez, M.; Liang, Y.H.; Abrahamson, M.; Su, X.D. Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile. J. Biol. Chem. 2005, 280, 18221–18228. [Google Scholar] [CrossRef]
- Dickinson, D.P. Salivary (SD-type) cystatins: Over one billion years in the making—But to what purpose? Crit. Rev. Oral. Biol. Med. 2002, 13, 485–508. [Google Scholar] [CrossRef]
- de Sousa-Pereira, P.; Abrantes, J.; Pinheiro, A.; Colaco, B.; Vitorino, R.; Esteves, P.J. Evolution of C, D and S-type cystatins in mammals: An extensive gene duplication in primates. PLoS ONE 2014, 9, e109050. [Google Scholar] [CrossRef]
- Ganeshnarayan, K.; Velliyagounder, K.; Furgang, D.; Fine, D.H. Human salivary cystatin SA exhibits antimicrobial effect against Aggregatibacter actinomycetemcomitans. J. Periodontal. Res. 2012, 47, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Blankenvoorde, M.F.; van’t Hof, W.; Walgreen-Weterings, E.; van Steenbergen, T.J.; Brand, H.S.; Veerman, E.C.; Nieuw Amerongen, A.V. Cystatin and cystatin-derived peptides have antibacterial activity against the pathogen Porphyromonas gingivalis. Biol. Chem. 1998, 379, 1371–1375. [Google Scholar] [PubMed]
- Bosch, J.A.; de Geus, E.J.; Veerman, E.C.; Hoogstraten, J.; Nieuw Amerongen, A.V. Innate secretory immunity in response to laboratory stressors that evoke distinct patterns of cardiac autonomic activity. Psychosom. Med. 2003, 65, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Jasir, A.; Kasprzykowski, F.; Kasprzykowska, R.; Lindstrom, V.; Schalen, C.; Grubb, A. New antimicrobial cystatin C-based peptide active against gram-positive bacterial pathogens, including methicillin-resistant Staphylococcus aureus and multiresistant coagulase-negative staphylococci. APMIS 2003, 111, 1004–1010. [Google Scholar] [CrossRef]
- Blancas-Luciano, B.E.; Becker-Fauser, I.; Zamora-Chimal, J.; Delgado-Dominguez, J.; Ruiz-Remigio, A.; Leyva-Huerta, E.R.; Portilla-Robertson, J.; Fernandez-Presas, A.M. Antimicrobial and anti-inflammatory activity of Cystatin C on human gingival fibroblast incubated with Porphyromonas gingivalis. Peerj 2022, 10, e14232. [Google Scholar] [CrossRef] [PubMed]
- Szpak, M.; Trziszka, T.; Polanowski, A.; Gburek, J.; Golab, K.; Juszczynska, K.; Janik, P.; Malicki, A.; Szyplik, K. Evaluation of the antibacterial activity of cystatin against selected strains of Escherichia coli. Folia Biol. 2014, 62, 187–192. [Google Scholar] [CrossRef]
- Baron, A.; DeCarlo, A.; Featherstone, J. Functional aspects of the human salivary cystatins in the oral environment. Oral Dis. 1999, 5, 234–240. [Google Scholar] [CrossRef]
- Shintani, M.; Minaguchi, K.; Isemura, S.; Saitoh, E.; Sanada, K.; Semba, T. Genetic polymorphisms of the CST2 locus coding for cystatin SA. Hum. Genet. 1994, 94, 45–49. [Google Scholar] [CrossRef]
- Saitoh, E.; Minaguchi, K.; Ishibashi, O. Production and characterization of two variants of human cystatin SA encoded by two alleles at the CST2 locus of the type 2 cystatin gene family. Arch. Biochem. Biophys. 1998, 352, 199–206. [Google Scholar] [CrossRef]
- Isemura, S.; Saitoh, E.; Ito, S.; Isemura, M.; Sanada, K. Cystatin S: A cysteine proteinase inhibitor of human saliva. J. Biochem. 1984, 96, 1311–1314. [Google Scholar] [CrossRef]
- Isemura, S.; Saitoh, E.; Sanada, K. Isolation and amino acid sequence of SAP-1, an acidic protein of human whole saliva, and sequence homology with human gamma-trace. J. Biochem. 1984, 96, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Ramasubbu, N.; Reddy, M.S.; Bergey, E.J.; Haraszthy, G.G.; Soni, S.D.; Levine, M.J. Large-scale purification and characterization of the major phosphoproteins and mucins of human submandibular-sublingual saliva. Biochem. J. 1991, 280 Pt 2, 341–352. [Google Scholar] [CrossRef]
- Shaw, P.A.; Yu, W.A. Autonomic regulation of cystatin S gene expression in rat submandibular glands. Auton Neurosci. 2000, 83, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Laguna, M.; Holgado, M.; Hernandez, A.L.; Santamaria, B.; Lavin, A.; Soria, J.; Suarez, T.; Bardina, C.; Jara, M.; Sanza, F.J.; et al. Antigen-Antibody Affinity for Dry Eye Biomarkers by Label Free Biosensing. Comparison with the ELISA Technique. Sensors 2015, 15, 19819–19829. [Google Scholar] [CrossRef] [PubMed]
- Balbin, M.; Hall, A.; Grubb, A.; Mason, R.W.; Lopez-Otin, C.; Abrahamson, M. Structural and functional characterization of two allelic variants of human cystatin D sharing a characteristic inhibition spectrum against mammalian cysteine proteinases. J. Biol. Chem. 1994, 269, 23156–23162. [Google Scholar] [CrossRef]
- Dyhrfort, P.; Shen, Q.; Clausen, F.; Thulin, M.; Enblad, P.; Kamali-Moghaddam, M.; Lewen, A.; Hillered, L. Monitoring of Protein Biomarkers of Inflammation in Human Traumatic Brain Injury Using Microdialysis and Proximity Extension Assay Technology in Neurointensive Care. J. Neurotrauma 2019, 36, 2872–2885. [Google Scholar] [CrossRef]
- Saitoh, E.; Sabatini, L.M.; Eddy, R.L.; Shows, T.B.; Azen, E.A.; Isemura, S.; Sanada, K. The human cystatin C gene (CST3) is a member of the cystatin gene family which is localized on chromosome 20. Biochem. Biophys. Res. Commun. 1989, 162, 1324–1331. [Google Scholar] [CrossRef]
- Schnittger, S.; Rao, V.V.; Abrahamson, M.; Hansmann, I. Cystatin C (CST3), the candidate gene for hereditary cystatin C amyloid angiopathy (HCCAA), and other members of the cystatin gene family are clustered on chromosome 20p11.2. Genomics 1993, 16, 50–55. [Google Scholar] [CrossRef]
- Abrahamson, M.; Dalboge, H.; Olafsson, I.; Carlsen, S.; Grubb, A. Efficient production of native, biologically active human cystatin C by Escherichia coli. FEBS Lett. 1988, 236, 14–18. [Google Scholar] [CrossRef]
- Ferguson, T.W.; Komenda, P.; Tangri, N. Cystatin C as a biomarker for estimating glomerular filtration rate. Curr. Opin. Nephrol. Hypertens 2015, 24, 295–300. [Google Scholar] [CrossRef]
- Urbschat, A.; Obermuller, N.; Haferkamp, A. Biomarkers of kidney injury. Biomarkers 2011, 16 (Suppl. S1), S22–S30. [Google Scholar] [CrossRef] [PubMed]
- Dharnidharka, V.R.; Kwon, C.; Stevens, G. Serum cystatin C is superior to serum creatinine as a marker of kidney function: A meta-analysis. Am. J. Kidney Dis. 2002, 40, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Janowski, R.; Kozak, M.; Jankowska, E.; Grzonka, Z.; Grubb, A.; Abrahamson, M.; Jaskolski, M. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 2001, 8, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lindemann, P.; Vega-Ramos, J.; Zhang, J.G.; Villadangos, J.A. Developmental regulation of synthesis and dimerization of the amyloidogenic protease inhibitor cystatin C in the hematopoietic system. J. Biol. Chem. 2014, 289, 9730–9740. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhou, Y.; Halabisky, B.; Lo, I.; Cho, S.H.; Mueller-Steiner, S.; Devidze, N.; Wang, X.; Grubb, A.; Gan, L. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron 2008, 60, 247–257. [Google Scholar] [CrossRef]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef]
- Finckh, U.; von der Kammer, H.; Velden, J.; Michel, T.; Andresen, B.; Deng, A.; Zhang, J.; Muller-Thomsen, T.; Zuchowski, K.; Menzer, G.; et al. Genetic association of a cystatin C gene polymorphism with late-onset Alzheimer disease. Arch. Neurol. 2000, 57, 1579–1583. [Google Scholar] [CrossRef]
- Crawford, F.C.; Freeman, M.J.; Schinka, J.A.; Abdullah, L.I.; Gold, M.; Hartman, R.; Krivian, K.; Morris, M.D.; Richards, D.; Duara, R.; et al. A polymorphism in the cystatin C gene is a novel risk factor for late-onset Alzheimer’s disease. Neurology 2000, 55, 763–768. [Google Scholar] [CrossRef]
- Wahlbom, M.; Wang, X.; Lindstrom, V.; Carlemalm, E.; Jaskolski, M.; Grubb, A. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J. Biol. Chem. 2007, 282, 18318–18326. [Google Scholar] [CrossRef]
- Sotiropoulou, G.; Anisowicz, A.; Sager, R. Identification, cloning, and characterization of cystatin M, a novel cysteine proteinase inhibitor, down-regulated in breast cancer. J. Biol. Chem. 1997, 272, 903–910. [Google Scholar] [CrossRef]
- Ni, J.; Abrahamson, M.; Zhang, M.; Fernandez, M.A.; Grubb, A.; Su, J.; Yu, G.L.; Li, Y.; Parmelee, D.; Xing, L.; et al. Cystatin E is a novel human cysteine proteinase inhibitor with structural resemblance to family 2 cystatins. J. Biol. Chem. 1997, 272, 10853–10858. [Google Scholar] [CrossRef] [PubMed]
- Stenman, G.; Astrom, A.K.; Roijer, E.; Sotiropoulou, G.; Zhang, M.; Sager, R. Assignment of a novel cysteine proteinase inhibitor (CST6) to 11q13 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1997, 76, 45–46. [Google Scholar] [CrossRef] [PubMed]
- Lunde, N.N.; Haugen, M.H.; Bodin Larsen, K.B.; Damgaard, I.; Pettersen, S.J.; Kasem, R.; Rut, W.; Drag, M.; Poreba, M.; Johansen, H.T.; et al. Glycosylation is important for legumain localization and processing to active forms but not for cystatin E/M inhibitory functions. Biochimie 2017, 139, 27–37. [Google Scholar] [CrossRef]
- Dall, E.; Hollerweger, J.C.; Dahms, S.O.; Cui, H.; Haussermann, K.; Brandstetter, H. Structural and functional analysis of cystatin E reveals enzymologically relevant dimer and amyloid fibril states. J. Biol. Chem. 2018, 293, 13151–13165. [Google Scholar] [CrossRef] [PubMed]
- Tjabringa, G.S.; Vos, J.B.; Olthuis, D.; Ninaber, D.K.; Rabe, K.F.; Schalkwijk, J.; Hiemstra, P.S.; Zeeuwen, P.L. Host defense effector molecules in mucosal secretions. FEMS Immunol. Med. Microbiol. 2005, 45, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Zeeuwen, P.L.; van Vlijmen-Willems, I.M.; Cheng, T.; Rodijk-Olthuis, D.; Hitomi, K.; Hara-Nishimura, I.; John, S.; Smyth, N.; Reinheckel, T.; Hendriks, W.J.; et al. The cystatin M/E-cathepsin L balance is essential for tissue homeostasis in epidermis, hair follicles, and cornea. FASEB J. 2010, 24, 3744–3755. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Chen, J.; Xu, X.; Zhong, Y.; Xu, Y.; Lu, P.; Zhou, J.; Lin, Z.; Yang, B.; et al. Loss-of-function mutations in CST6 cause dry skin, desquamation and abnormal keratosis without hypotrichosis. Clin. Genet. 2023, 103, 301–309. [Google Scholar] [CrossRef]
- Eckl, K.M.; Gruber, R.; Brennan, L.; Marriott, A.; Plank, R.; Moosbrugger-Martinz, V.; Blunder, S.; Schossig, A.; Altmuller, J.; Thiele, H.; et al. Cystatin M/E Variant Causes Autosomal Dominant Keratosis Follicularis Spinulosa Decalvans by Dysregulating Cathepsins L and V. Front. Genet. 2021, 12, 689940. [Google Scholar] [CrossRef]
- Jansen, P.A.; van den Bogaard, E.H.; Kersten, F.F.; Oostendorp, C.; van Vlijmen-Willems, I.M.; Oji, V.; Traupe, H.; Hennies, H.C.; Schalkwijk, J.; Zeeuwen, P.L. Cystatin M/E knockdown by lentiviral delivery of shRNA impairs epidermal morphogenesis of human skin equivalents. Exp. Dermatol. 2012, 21, 889–891. [Google Scholar] [CrossRef]
- Zeeuwen, P.L.; van Vlijmen-Willems, I.M.; Hendriks, W.; Merkx, G.F.; Schalkwijk, J. A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum. Mol. Genet. 2002, 11, 2867–2875. [Google Scholar] [CrossRef]
- Morita, M.; Yoshiuchi, N.; Arakawa, H.; Nishimura, S. CMAP: A novel cystatin-like gene involved in liver metastasis. Cancer Res. 1999, 59, 151–158. [Google Scholar] [PubMed]
- Ni, J.; Fernandez, M.A.; Danielsson, L.; Chillakuru, R.A.; Zhang, J.; Grubb, A.; Su, J.; Gentz, R.; Abrahamson, M. Cystatin F is a glycosylated human low molecular weight cysteine proteinase inhibitor. J. Biol. Chem. 1998, 273, 24797–24804. [Google Scholar] [CrossRef] [PubMed]
- Langerholc, T.; Zavasnik-Bergant, V.; Turk, B.; Turk, V.; Abrahamson, M.; Kos, J. Inhibitory properties of cystatin F and its localization in U937 promonocyte cells. FEBS J. 2005, 272, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Maher, K.; Konjar, S.; Watts, C.; Turk, B.; Kopitar-Jerala, N. Cystatin F regulates proteinase activity in IL-2-activated natural killer cells. Protein Pept. Lett. 2014, 21, 957–965. [Google Scholar] [CrossRef]
- Dautovic, E.; Perisic Nanut, M.; Softic, A.; Kos, J. The transcription factor C/EBP alpha controls the role of cystatin F during the differentiation of monocytes to macrophages. Eur. J. Cell Biol. 2018, 97, 463–473. [Google Scholar] [CrossRef]
- Hamilton, G.; Colbert, J.D.; Schuettelkopf, A.W.; Watts, C. Cystatin F is a cathepsin C-directed protease inhibitor regulated by proteolysis. EMBO J. 2008, 27, 499–508. [Google Scholar] [CrossRef]
- Duff, M.O.; Olson, S.; Wei, X.; Garrett, S.C.; Osman, A.; Bolisetty, M.; Plocik, A.; Celniker, S.E.; Graveley, B.R. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 2015, 521, 376–379. [Google Scholar] [CrossRef]
- Colbert, J.D.; Plechanovova, A.; Watts, C. Glycosylation directs targeting and activation of cystatin f from intracellular and extracellular sources. Traffic 2009, 10, 425–437. [Google Scholar] [CrossRef]
- Parent, A.D.; Cornwall, G.A.; Liu, L.Y.; Smith, C.E.; Hermo, L. Alterations in the testis and epididymis associated with loss of function of the cystatin-related epididymal spermatogenic (CRES) protein. J. Androl. 2011, 32, 444–463. [Google Scholar] [CrossRef]
- Chau, K.M.; Cornwall, G.A. Reduced fertility in vitro in mice lacking the cystatin CRES (cystatin-related epididymal spermatogenic): Rescue by exposure of spermatozoa to dibutyryl cAMP and isobutylmethylxanthine. Biol. Reprod. 2011, 84, 140–152. [Google Scholar] [CrossRef]
- Belotti, Y.; Lim, E.H.; Lim, C.T. The Role of the Extracellular Matrix and Tumor-Infiltrating Immune Cells in the Prognostication of High-Grade Serous Ovarian Cancer. Cancers 2022, 14, 404. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.T. Neutrophil serine proteases: Specific regulators of inflammation. Nat. Rev. Immunol. 2006, 6, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.B.; Chen, X.; Zhang, Z.Y.; Wu, F.F.; Liu, X.H. Cathepsin C inhibitors as anti-inflammatory drug discovery: Challenges and opportunities. Eur. J. Med. Chem. 2021, 225, 113818. [Google Scholar] [CrossRef]
- Kos, J.; Nanut, M.P.; Prunk, M.; Sabotic, J.; Dautovic, E.; Jewett, A. Cystatin F as a regulator of immune cell cytotoxicity. Cancer Immunol. Immunother. 2018, 67, 1931–1938. [Google Scholar] [CrossRef] [PubMed]
- Senjor, E.; Kos, J.; Nanut, M.P. Cysteine Cathepsins as Therapeutic Targets in Immune Regulation and Immune Disorders. Biomedicines 2023, 11, 476. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.P.; Villadangos, J.A.; Dranoff, G.; Small, C.; Gu, L.; Haley, K.J.; Riese, R.; Ploegh, H.L.; Chapman, H.A. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity 1999, 10, 197–206. [Google Scholar] [CrossRef]
- Riese, R.J.; Wolf, P.R.; Bromme, D.; Natkin, L.R.; Villadangos, J.A.; Ploegh, H.L.; Chapman, H.A. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity 1996, 4, 357–366. [Google Scholar] [CrossRef]
- Driessen, C.; Bryant, R.A.; Lennon-Dumenil, A.M.; Villadangos, J.A.; Bryant, P.W.; Shi, G.P.; Chapman, H.A.; Ploegh, H.L. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J. Cell. Biol. 1999, 147, 775–790. [Google Scholar] [CrossRef]
- Nakagawa, T.Y.; Brissette, W.H.; Lira, P.D.; Griffiths, R.J.; Petrushova, N.; Stock, J.; McNeish, J.D.; Eastman, S.E.; Howard, E.D.; Clarke, S.R.; et al. Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity 1999, 10, 207–217. [Google Scholar] [CrossRef]
- House, I.G.; House, C.M.; Brennan, A.J.; Gilan, O.; Dawson, M.A.; Whisstock, J.C.; Law, R.H.; Trapani, J.A.; Voskoboinik, I. Regulation of perforin activation and pre-synaptic toxicity through C-terminal glycosylation. EMBO Rep. 2017, 18, 1775–1785. [Google Scholar] [CrossRef]
- Garnero, P.; Borel, O.; Byrjalsen, I.; Ferreras, M.; Drake, F.H.; McQueney, M.S.; Foged, N.T.; Delmas, P.D.; Delaisse, J.M. The collagenolytic activity of cathepsin K is unique among mammalian proteinases. J. Biol. Chem. 1998, 273, 32347–32352. [Google Scholar] [CrossRef]
- Solberg, R.; Lunde, N.N.; Forbord, K.M.; Okla, M.; Kassem, M.; Jafari, A. The Mammalian Cysteine Protease Legumain in Health and Disease. Int. J. Mol. Sci. 2022, 23, 15983. [Google Scholar] [CrossRef]
- Nocera, A.L.; Mueller, S.K.; Workman, A.D.; Wu, D.; McDonnell, K.; Sadow, P.M.; Amiji, M.M.; Bleier, B.S. Cystatin SN is a potent upstream initiator of epithelial-derived type 2 inflammation in chronic rhinosinusitis. J. Allergy Clin. Immunol. 2022, 150, 872–881. [Google Scholar] [CrossRef]
- Yan, B.; Lou, H.; Wang, Y.; Li, Y.; Meng, Y.; Qi, S.; Wang, M.; Xiao, L.; Wang, C.; Zhang, L. Epithelium-derived cystatin SN enhances eosinophil activation and infiltration through IL-5 in patients with chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 2019, 144, 455–469. [Google Scholar] [CrossRef]
- Higham, A.; Dungwa, J.; Pham, T.H.; McCrae, C.; Singh, D. Increased mast cell activation in eosinophilic chronic obstructive pulmonary disease. Clin. Transl. Immunol. 2022, 11, e1417. [Google Scholar] [CrossRef]
- Kato, Y.; Takabayashi, T.; Sakashita, M.; Imoto, Y.; Tokunaga, T.; Ninomiya, T.; Morikawa, T.; Yoshida, K.; Noguchi, E.; Fujieda, S. Expression and Functional Analysis of CST1 in Intractable Nasal Polyps. Am. J. Respir. Cell. Mol. Biol. 2018, 59, 448–457. [Google Scholar] [CrossRef]
- Wang, M.; Gong, L.; Luo, Y.; He, S.; Zhang, X.; Xie, X.; Li, X.; Feng, X. Transcriptomic analysis of asthma and allergic rhinitis reveals CST1 as a biomarker of unified airways. Front. Immunol. 2023, 14, 1048195. [Google Scholar] [CrossRef]
- Jakiela, B.; Soja, J.; Sladek, K.; Przybyszowski, M.; Plutecka, H.; Gielicz, A.; Licholai, S.; Aab, A.; Rebane, A.; Bochenek, G. Bronchial epithelial cell transcriptome shows endotype heterogeneity of asthma in patients with NSAID-exacerbated respiratory disease. J. Allergy Clin. Immunol. 2023, 151, 953–965. [Google Scholar] [CrossRef]
- Kack, U.; Einarsdottir, E.; van Hage, M.; Asarnoj, A.; James, A.; Nopp, A.; Krjutskov, K.; Katayama, S.; Kere, J.; Lilja, G.; et al. Nasal upregulation of CST1 in dog-sensitised children with severe allergic airway disease. ERJ Open Res. 2021, 7. [Google Scholar] [CrossRef]
- Wu, D.; Yan, B.; Wang, Y.; Wang, C.; Zhang, L. Prognostic and pharmacologic value of cystatin SN for chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 2021, 148, 450–460. [Google Scholar] [CrossRef]
- Liu, H.B.; Bai, J.; Wang, W.X. CST1 promotes the proliferation and migration of PDGF-BB-treated airway smooth muscle cells via the PI3K/AKT signaling pathway. Kaohsiung J. Med. Sci. 2023, 39, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Lee, I.T.; Le, W.; Tsunemi, Y.; Borchard, N.A.; Zarabanda, D.; Dholakia, S.S.; Gall, P.A.; Yang, A.; Kim, D.; et al. Inflammatory molecular endotypes of nasal polyps derived from White and Japanese populations. J. Allergy Clin. Immunol. 2022, 149, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Schraufstatter, I.U.; Zhao, M.; Khaldoyanidi, S.K.; Discipio, R.G. The chemokine CCL18 causes maturation of cultured monocytes to macrophages in the M2 spectrum. Immunology 2012, 135, 287–298. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Ren, H.; Dai, H.; Zhou, Y.; Ren, X.; Wang, Y.; Feng, S.; Deng, X.; Wu, J.; et al. Human endoderm stem cells reverse inflammation-related acute liver failure through cystatin SN-mediated inhibition of interferon signaling. Cell Res. 2023, 33, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Miyake, M.M.; Workman, A.D.; Nocera, A.L.; Wu, D.; Mueller, S.K.; Finn, K.; Amiji, M.M.; Bleier, B.S. Discriminant analysis followed by unsupervised cluster analysis including exosomal cystatins predict presence of chronic rhinosinusitis, phenotype, and disease severity. Int. Forum Allergy Rhinol. 2019, 9, 1069–1076. [Google Scholar] [CrossRef]
- Mueller, S.K.; Wendler, O.; Nocera, A.; Grundtner, P.; Schlegel, P.; Agaimy, A.; Iro, H.; Bleier, B.S. Escalation in mucus cystatin 2, pappalysin-A, and periostin levels over time predict need for recurrent surgery in chronic rhinosinusitis with nasal polyps. Int. Forum Allergy Rhinol. 2019, 9, 1212–1219. [Google Scholar] [CrossRef]
- Workman, A.D.; Nocera, A.L.; Mueller, S.K.; Otu, H.H.; Libermann, T.A.; Bleier, B.S. Translating transcription: Proteomics in chronic rhinosinusitis with nasal polyps reveals significant discordance with messenger RNA expression. Int. Forum Allergy Rhinol. 2019, 9, 776–786. [Google Scholar] [CrossRef]
- Boto de Los Bueis, A.; de la Fuente, M.; Montejano-Milner, R.; Del Hierro Zarzuelo, A.; Vecino, E.; Acera, A. A Pilot Study of a Panel of Ocular Inflammation Biomarkers in Patients with Primary Sjogren’s Syndrome. Curr. Issues Mol. Biol. 2023, 45, 2881–2894. [Google Scholar] [CrossRef]
- Soria, J.; Acera, A.; Merayo, L.J.; Duran, J.A.; Gonzalez, N.; Rodriguez, S.; Bistolas, N.; Schumacher, S.; Bier, F.F.; Peter, H.; et al. Tear proteome analysis in ocular surface diseases using label-free LC-MS/MS and multiplexed-microarray biomarker validation. Sci. Rep. 2017, 7, 17478. [Google Scholar] [CrossRef]
- Elam, M.B.; Majumdar, G.; Mozhui, K.; Gerling, I.C.; Vera, S.R.; Fish-Trotter, H.; Williams, R.W.; Childress, R.D.; Raghow, R. Patients experiencing statin-induced myalgia exhibit a unique program of skeletal muscle gene expression following statin re-challenge. PLoS ONE 2017, 12, e0181308. [Google Scholar] [CrossRef]
- Cowell, W.; Colicino, E.; Lee, A.G.; Bosquet Enlow, M.; Flom, J.D.; Berin, C.; Wright, R.O.; Wright, R.J. Data-driven discovery of mid-pregnancy immune markers associated with maternal lifetime stress: Results from an urban pre-birth cohort. Stress 2020, 23, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Panezai, J.; Ali, A.; Ghaffar, A.; Benchimol, D.; Altamash, M.; Klinge, B.; Engstrom, P.E.; Larsson, A. Upregulation of circulating inflammatory biomarkers under the influence of periodontal disease in rheumatoid arthritis patients. Cytokine 2020, 131, 155117. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R.; Grubb, A. Cystatin D, a natural salivary cysteine protease inhibitor, inhibits coronavirus replication at its physiologic concentration. Oral Microbiol. Immunol. 1998, 13, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Zavasnik-Bergant, T.; Repnik, U.; Schweiger, A.; Romih, R.; Jeras, M.; Turk, V.; Kos, J. Differentiation- and maturation-dependent content, localization, and secretion of cystatin C in human dendritic cells. J. Leukoc. Biol. 2005, 78, 122–134. [Google Scholar] [CrossRef]
- Pierre, P.; Mellman, I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell 1998, 93, 1135–1145. [Google Scholar] [CrossRef]
- Biasizzo, M.; Trstenjak-Prebanda, M.; Dolinar, K.; Pirkmajer, S.; Zavrsnik, J.; Turk, B.; Kopitar-Jerala, N. Cystatin C Deficiency Increases LPS-Induced Sepsis and NLRP3 Inflammasome Activation in Mice. Cells 2021, 10, 2071. [Google Scholar] [CrossRef]
- Hansen, T.; Petrow, P.K.; Gaumann, A.; Keyszer, G.; Brauer, R.; Kriegsmann, J. Synovial giant cells in rheumatoid arthritis: Expression of cystatin C, but not of cathepsin B. Exp. Toxicol. Pathol. 2000, 52, 312–316. [Google Scholar] [CrossRef]
- Longenecker, C.T.; Hileman, C.O.; Funderburg, N.T.; McComsey, G.A. Rosuvastatin preserves renal function and lowers cystatin C in HIV-infected subjects on antiretroviral therapy: The SATURN-HIV trial. Clin. Infect. Dis. 2014, 59, 1148–1156. [Google Scholar] [CrossRef]
- El-Sukkari, D.; Wilson, N.S.; Hakansson, K.; Steptoe, R.J.; Grubb, A.; Shortman, K.; Villadangos, J.A. The protease inhibitor cystatin C is differentially expressed among dendritic cell populations, but does not control antigen presentation. J. Immunol. 2003, 171, 5003–5011. [Google Scholar] [CrossRef]
- Frendeus, K.H.; Wallin, H.; Janciauskiene, S.; Abrahamson, M. Macrophage responses to interferon-gamma are dependent on cystatin C levels. Int. J. Biochem. Cell. Biol. 2009, 41, 2262–2269. [Google Scholar] [CrossRef]
- Xu, Y.; Schnorrer, P.; Proietto, A.; Kowalski, G.; Febbraio, M.A.; Acha-Orbea, H.; Dickins, R.A.; Villadangos, J.A. IL-10 controls cystatin C synthesis and blood concentration in response to inflammation through regulation of IFN regulatory factor 8 expression. J. Immunol. 2011, 186, 3666–3673. [Google Scholar] [CrossRef] [PubMed]
- Sokol, J.P.; Schiemann, W.P. Cystatin C antagonizes transforming growth factor beta signaling in normal and cancer cells. Mol. Cancer Res. 2004, 2, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Soh, H.; Venkatesan, N.; Veena, M.S.; Ravichandran, S.; Zinabadi, A.; Basak, S.K.; Parvatiyar, K.; Srivastava, M.; Liang, L.J.; Gjertson, D.W.; et al. Cystatin E/M Suppresses Tumor Cell Growth through Cytoplasmic Retention of NF-kappaB. Mol. Cell Biol. 2016, 36, 1776–1792. [Google Scholar] [CrossRef] [PubMed]
- Gai, D.; Chen, J.R.; Stewart, J.P.; Nookaew, I.; Habelhah, H.; Ashby, C.; Sun, F.; Cheng, Y.; Li, C.; Xu, H.; et al. CST6 suppresses osteolytic bone disease in multiple myeloma by blocking osteoclast differentiation. J. Clin. Investig. 2022, 132, e159527. [Google Scholar] [CrossRef] [PubMed]
- Prunk, M.; Nanut, M.P.; Sabotic, J.; Svajger, U.; Kos, J. Increased cystatin F levels correlate with decreased cytotoxicity of cytotoxic T cells. Radiol. Oncol. 2019, 53, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Magister, S.; Tseng, H.C.; Bui, V.T.; Kos, J.; Jewett, A. Regulation of split anergy in natural killer cells by inhibition of cathepsins C and H and cystatin F. Oncotarget 2015, 6, 22310–22327. [Google Scholar] [CrossRef]
- Magister, S.; Obermajer, N.; Mirkovic, B.; Svajger, U.; Renko, M.; Softic, A.; Romih, R.; Colbert, J.D.; Watts, C.; Kos, J. Regulation of cathepsins S and L by cystatin F during maturation of dendritic cells. Eur. J. Cell. Biol. 2012, 91, 391–401. [Google Scholar] [CrossRef]
- Jin, C.; Shao, Y.; Zhang, X.; Xiang, J.; Zhang, R.; Sun, Z.; Mei, S.; Zhou, J.; Zhang, J.; Shi, L. A Unique Type of Highly-Activated Microglia Evoking Brain Inflammation via Mif/Cd74 Signaling Axis in Aged Mice. Aging Dis. 2021, 12, 2125–2139. [Google Scholar] [CrossRef]
- Mangale, V.; Syage, A.R.; Ekiz, H.A.; Skinner, D.D.; Cheng, Y.; Stone, C.L.; Brown, R.M.; O’Connell, R.M.; Green, K.N.; Lane, T.E. Microglia influence host defense, disease, and repair following murine coronavirus infection of the central nervous system. Glia 2020, 68, 2345–2360. [Google Scholar] [CrossRef]
- Ma, K.; Chen, X.; Liu, W.; Chen, S.; Yang, C.; Yang, J. CTSB is a negative prognostic biomarker and therapeutic target associated with immune cells infiltration and immunosuppression in gliomas. Sci. Rep. 2022, 12, 4295. [Google Scholar] [CrossRef]
- Fonovic, M.; Turk, B. Cysteine cathepsins and extracellular matrix degradation. Biochim. Biophys. Acta. 2014, 1840, 2560–2570. [Google Scholar] [CrossRef]
- Mai, C.W.; Chung, F.F.; Leong, C.O. Targeting Legumain As a Novel Therapeutic Strategy in Cancers. Curr. Drug Targets 2017, 18, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Dong, C.; Chen, T.; Dong, A.; Ren, J.; Li, W.; Shu, G.; Yang, J.; Shen, W.; et al. CST1 inhibits ferroptosis and promotes gastric cancer metastasis by regulating GPX4 protein stability via OTUB1. Oncogene 2023, 42, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.N.; Li, Y.; Chen, B.; Du, Y.; Li, S.B.; Lu, S.X.; Zhao, Z.P.; Zhou, A.J.; Xue, N.; Xia, T.L.; et al. Elevated expression of CST1 promotes breast cancer progression and predicts a poor prognosis. J. Mol. Med. 2017, 95, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, Y.; Luo, R.Z.; Zhang, L.; Zhang, S.L.; Zeng, J.; Han, Y.J.; Wen, Z.S. Expression of Cystatin SN significantly correlates with recurrence, metastasis, and survival duration in surgically resected non-small cell lung cancer patients. Sci. Rep. 2015, 5, 8230. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, S.; Yin, X.; Tu, M.; Cai, L.; Zhang, Y.; Yu, L.; Zhang, S.; Pan, X.; Huang, Y. MiR-942-5p inhibits tumor migration and invasion through targeting CST1 in esophageal squamous cell carcinoma. PLoS ONE 2023, 18, e0277006. [Google Scholar] [CrossRef]
- Li, T.; Xiong, Q.; Zou, Z.; Lei, X.; Jiang, Q.; Liu, D. Prognostic significance of cystatin SN associated nomograms in patients with colorectal cancer. Oncotarget 2017, 8, 115153–115163. [Google Scholar] [CrossRef]
- Cui, Y.; Sun, D.; Song, R.; Zhang, S.; Liu, X.; Wang, Y.; Meng, F.; Lan, Y.; Han, J.; Pan, S.; et al. Upregulation of cystatin SN promotes hepatocellular carcinoma progression and predicts a poor prognosis. J. Cell Physiol. 2019, 234, 22623–22634. [Google Scholar] [CrossRef]
- Mullapudi, N.; Ye, B.; Suzuki, M.; Fazzari, M.; Han, W.; Shi, M.K.; Marquardt, G.; Lin, J.; Wang, T.; Keller, S.; et al. Genome Wide Methylome Alterations in Lung Cancer. PLoS ONE 2015, 10, e0143826. [Google Scholar] [CrossRef]
- Kim, J.T.; Lee, S.J.; Kang, M.A.; Park, J.E.; Kim, B.Y.; Yoon, D.Y.; Yang, Y.; Lee, C.H.; Yeom, Y.I.; Choe, Y.K.; et al. Cystatin SN neutralizes the inhibitory effect of cystatin C on cathepsin B activity. Cell Death Dis. 2013, 4, e974. [Google Scholar] [CrossRef]
- Zhang, W.P.; Wang, Y.; Tan, D.; Xing, C.G. Cystatin 2 leads to a worse prognosis in patients with gastric cancer. J. Biol. Regul. Homeost. Agents 2020, 34, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Zhang, Y.; Pan, Z.; Hu, X.; Yi, Y.; Zheng, X.; Wei, H.; Huang, P. Identification of novel key genes associated with the metastasis of prostate cancer based on bioinformatics prediction and validation. Cancer Cell. Int. 2021, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Zhou, Z.; Zhang, S. miRNA-6715-5p Inhibits Cellular Proliferation and Invasion in Colorectal Cancer by Directly Targeting CST4. J. Oncol. 2021, 2021, 7615712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Zhang, J.J.; Song, H.J.; Li, D.W. Overexpression of CST4 promotes gastric cancer aggressiveness by activating the ELFN2 signaling pathway. Am. J. Cancer Res. 2017, 7, 2290–2304. [Google Scholar] [PubMed]
- Yang, G.; Zhang, Y.; Lin, H.; Liu, J.; Huang, S.; Zhong, W.; Peng, C.; Du, L. CircRNA circ_0023984 promotes the progression of esophageal squamous cell carcinoma via regulating miR-134-5p/cystatin-s axis. Bioengineered 2022, 13, 10578–10593. [Google Scholar] [CrossRef]
- Wu, W.; Yong, W.W.; Chung, M.C. A simple biomarker scoring matrix for early gastric cancer detection. Proteomics 2016, 16, 2921–2930. [Google Scholar] [CrossRef]
- Berger, M.D.; Stintzing, S.; Heinemann, V.; Cao, S.; Yang, D.; Sunakawa, Y.; Matsusaka, S.; Ning, Y.; Okazaki, S.; Miyamoto, Y.; et al. A Polymorphism within the Vitamin D Transporter Gene Predicts Outcome in Metastatic Colorectal Cancer Patients Treated with FOLFIRI/Bevacizumab or FOLFIRI/Cetuximab. Clin. Cancer Res. 2018, 24, 784–793. [Google Scholar] [CrossRef]
- Munetsuna, E.; Kawanami, R.; Nishikawa, M.; Ikeda, S.; Nakabayashi, S.; Yasuda, K.; Ohta, M.; Kamakura, M.; Ikushiro, S.; Sakaki, T. Anti-proliferative activity of 25-hydroxyvitamin D3 in human prostate cells. Mol. Cell Endocrinol. 2014, 382, 960–970. [Google Scholar] [CrossRef]
- Alvarez-Diaz, S.; Valle, N.; Garcia, J.M.; Pena, C.; Freije, J.M.; Quesada, V.; Astudillo, A.; Bonilla, F.; Lopez-Otin, C.; Munoz, A. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J. Clin. Investig. 2009, 119, 2343–2358. [Google Scholar] [CrossRef]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef]
- Carlberg, C.; Munoz, A. An update on vitamin D signaling and cancer. Semin. Cancer Biol. 2022, 79, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Hunten, S.; Hermeking, H. p53 directly activates cystatin D/CST5 to mediate mesenchymal-epithelial transition: A possible link to tumor suppression by vitamin D3. Oncotarget 2015, 6, 15842–15856. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, J.; Kitamoto, S.; Yang, M.; Grubb, A.; Chapman, H.A.; Kalluri, R.; Shi, G.P. Cathepsin S controls angiogenesis and tumor growth via matrix-derived angiogenic factors. J. Biol. Chem. 2006, 281, 6020–6029. [Google Scholar] [CrossRef]
- Balaji, K.N.; Schaschke, N.; Machleidt, W.; Catalfamo, M.; Henkart, P.A. Surface cathepsin B protects cytotoxic lymphocytes from self-destruction after degranulation. J. Exp. Med. 2002, 196, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Indacochea, A.; Guerrero, S.; Urena, M.; Araujo, F.; Coll, O.; ME, L.L.; Gebauer, F. Cold-inducible RNA binding protein promotes breast cancer cell malignancy by regulating Cystatin C levels. RNA 2021, 27, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Mori, J.; Tanikawa, C.; Funauchi, Y.; Lo, P.H.; Nakamura, Y.; Matsuda, K. Cystatin C as a p53-inducible apoptotic mediator that regulates cathepsin L activity. Cancer Sci. 2016, 107, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Abrahamson, M.; Barrett, A.J.; Salvesen, G.; Grubb, A. Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J. Biol. Chem. 1986, 261, 11282–11289. [Google Scholar] [CrossRef]
- Nakabayashi, H.; Hara, M.; Shimuzu, K. Clinicopathologic significance of cystatin C expression in gliomas. Hum. Pathol. 2005, 36, 1008–1015. [Google Scholar] [CrossRef]
- Sokol, J.P.; Neil, J.R.; Schiemann, B.J.; Schiemann, W.P. The use of cystatin C to inhibit epithelial-mesenchymal transition and morphological transformation stimulated by transforming growth factor-beta. Breast Cancer Res. 2005, 7, R844–R853. [Google Scholar] [CrossRef]
- Saleh, Y.; Sebzda, T.; Warwas, M.; Kopec, W.; Ziolkowska, J.; Siewinski, M. Expression of cystatin C in clinical human colorectal cancer tissues. J. Exp. Ther. Oncol. 2005, 5, 49–53. [Google Scholar]
- Periyasamy, A.; Gopisetty, G.; Subramanium, M.J.; Velusamy, S.; Rajkumar, T. Identification and validation of differential plasma proteins levels in epithelial ovarian cancer. J. Proteom. 2020, 226, 103893. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Fan, Q.; Wang, L.; Zhou, Y.; Li, J.; Zhou, K. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget 2017, 8, 35750–35760. [Google Scholar] [CrossRef] [PubMed]
- Vancompernolle, K.; Van Herreweghe, F.; Pynaert, G.; Van de Craen, M.; De Vos, K.; Totty, N.; Sterling, A.; Fiers, W.; Vandenabeele, P.; Grooten, J. Atractyloside-induced release of cathepsin B, a protease with caspase-processing activity. FEBS Lett. 1998, 438, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Decock, J.; Obermajer, N.; Vozelj, S.; Hendrickx, W.; Paridaens, R.; Kos, J. Cathepsin B, cathepsin H, cathepsin X and cystatin C in sera of patients with early-stage and inflammatory breast cancer. Int. J. Biol. Markers 2008, 23, 161–168. [Google Scholar] [CrossRef]
- Lameire, N.; Vanholder, R.; Van Biesen, W.; Benoit, D. Acute kidney injury in critically ill cancer patients: An update. Crit. Care 2016, 20, 209. [Google Scholar] [CrossRef]
- Rosner, M.H.; Perazella, M.A. Acute Kidney Injury in Patients with Cancer. N. Engl. J. Med. 2017, 376, 1770–1781. [Google Scholar] [CrossRef]
- Jin, L.; Zhang, Y.; Li, H.; Yao, L.; Fu, D.; Yao, X.; Xu, L.X.; Hu, X.; Hu, G. Differential secretome analysis reveals CST6 as a suppressor of breast cancer bone metastasis. Cell. Res. 2012, 22, 1356–1373. [Google Scholar] [CrossRef]
- Pulukuri, S.M.; Gorantla, B.; Knost, J.A.; Rao, J.S. Frequent loss of cystatin E/M expression implicated in the progression of prostate cancer. Oncogene 2009, 28, 2829–2838. [Google Scholar] [CrossRef]
- Qiu, J.; Ai, L.; Ramachandran, C.; Yao, B.; Gopalakrishnan, S.; Fields, C.R.; Delmas, A.L.; Dyer, L.M.; Melnick, S.J.; Yachnis, A.T.; et al. Invasion suppressor cystatin E/M (CST6): High-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab Investig. 2008, 88, 910–925. [Google Scholar] [CrossRef]
- D’Costa, Z.C.; Higgins, C.; Ong, C.W.; Irwin, G.W.; Boyle, D.; McArt, D.G.; McCloskey, K.; Buckley, N.E.; Crawford, N.T.; Thiagarajan, L.; et al. TBX2 represses CST6 resulting in uncontrolled legumain activity to sustain breast cancer proliferation: A novel cancer-selective target pathway with therapeutic opportunities. Oncotarget 2014, 5, 1609–1620. [Google Scholar] [CrossRef]
- Wallin, H.; Apelqvist, J.; Andersson, F.; Ekstrom, U.; Abrahamson, M. Low-level internalization of cystatin E/M affects legumain activity and migration of melanoma cells. J. Biol. Chem. 2017, 292, 14413–14424. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, Y.; Lian, C.; Peng, F.; Xiao, Y.; He, Y.; Ma, C.; Wang, Y.; Zhang, P.; Deng, Y.; et al. CST6 protein and peptides inhibit breast cancer bone metastasis by suppressing CTSB activity and osteoclastogenesis. Theranostics 2021, 11, 9821–9832. [Google Scholar] [CrossRef] [PubMed]
- Fornetti, J.; Welm, A.L.; Stewart, S.A. Understanding the Bone in Cancer Metastasis. J. Bone Min. Res. 2018, 33, 2099–2113. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, Z.C.; Ni, C.J.; Jin, W.X.; Jin, Y.X.; Chen, Y.; Zhang, X.H.; Chen, E.D.; Cai, Y.F. Correlation of Cystatin E/M with Clinicopathological Features and Prognosis in Triple-Negative Breast Cancer. Ann. Clin. Lab. Sci. 2018, 48, 40–44. [Google Scholar] [PubMed]
- Zhou, X.; Wang, X.; Huang, K.; Liao, X.; Yang, C.; Yu, T.; Liu, J.; Han, C.; Zhu, G.; Su, H.; et al. Investigation of the clinical significance and prospective molecular mechanisms of cystatin genes in patients with hepatitis B virus-related hepatocellular carcinoma. Oncol. Rep. 2019, 42, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M.; Kashiwaya, K.; Eguchi, H.; Ohigashi, H.; Ishikawa, O.; Furihata, M.; Shinomura, Y.; Imai, K.; Nakamura, Y.; Nakagawa, H. Over-expression of cysteine proteinase inhibitor cystatin 6 promotes pancreatic cancer growth. Cancer Sci. 2008, 99, 1626–1632. [Google Scholar] [CrossRef]
- Khan, M.; Lin, J.; Wang, B.; Chen, C.; Huang, Z.; Tian, Y.; Yuan, Y.; Bu, J. A novel necroptosis-related gene index for predicting prognosis and a cold tumor immune microenvironment in stomach adenocarcinoma. Front. Immunol. 2022, 13, 968165. [Google Scholar] [CrossRef]
- Senjor, E.; Perisic Nanut, M.; Breznik, B.; Mitrovic, A.; Mlakar, J.; Rotter, A.; Porcnik, A.; Lah Turnsek, T.; Kos, J. Cystatin F acts as a mediator of immune suppression in glioblastoma. Cell. Oncol. 2021, 44, 1051–1063. [Google Scholar] [CrossRef]
- Utsunomiya, T.; Hara, Y.; Kataoka, A.; Morita, M.; Arakawa, H.; Mori, M.; Nishimura, S. Cystatin-like metastasis-associated protein mRNA expression in human colorectal cancer is associated with both liver metastasis and patient survival. Clin. Cancer Res. 2002, 8, 2591–2594. [Google Scholar]
- Chahal, M.S.; Ku, H.T.; Zhang, Z.; Legaspi, C.M.; Luo, A.; Hopkins, M.M.; Meier, K.E. Differential Expression of Ccn4 and Other Genes Between Metastatic and Non-metastatic EL4 Mouse Lymphoma Cells. Cancer Genom. Proteom. 2016, 13, 437–442. [Google Scholar] [CrossRef]
- Yang, C.; Yu, T.; Liu, Z.; Ye, X.; Liao, X.; Wang, X.; Han, C.; Zhu, G.; Qin, W.; Peng, T. Cystatin F as a key family 2 cystatin subunit and prognostic biomarker for early-stage pancreatic ductal adenocarcinoma. Oncol. Rep. 2019, 42, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Khalid, O.; Gabet, Y.; Shah, R.R.; Purcell, D.J.; Mav, D.; Kohn-Gabet, A.E.; Shi, Y.; Coetzee, G.A.; Frenkel, B. Runx2 transcriptome of prostate cancer cells: Insights into invasiveness and bone metastasis. Mol. Cancer 2010, 9, 258. [Google Scholar] [CrossRef]
- Qi, Q.; Chen, C.; Liu, C.; Zhang, B.; Ma, Y.; Zhang, H.; Huang, W.; Wang, C. Linc8087 predicts favorable prognosis and inhibits cell migration and invasion in NSCLC. Pathol. Res. Pract. 2021, 225, 153569. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Danjo, A.; Yamaza, T.; Kido, M.A.; Shimohira, D.; Tsukuba, T.; Kagiya, T.; Yamashita, Y.; Nishijima, K.; Masuko, S.; Goto, M.; et al. Cystatin C stimulates the differentiation of mouse osteoblastic cells and bone formation. Biochem. Biophys. Res. Commun. 2007, 360, 199–204. [Google Scholar] [CrossRef]
- Matthews, S.P.; McMillan, S.J.; Colbert, J.D.; Lawrence, R.A.; Watts, C. Cystatin F Ensures Eosinophil Survival by Regulating Granule Biogenesis. Immunity 2016, 44, 795–806. [Google Scholar] [CrossRef]


| Name of Gene | Encoded Protein | Gene Location | Site of Expression |
|---|---|---|---|
| CST1 | Cystatin SN | 20p11.21 | Parotid gland, lacrimal gland [13] |
| CST2 | Cystatin SA | 20p11.21 | Parotid gland [13] |
| CST3 | Cystatin C | 20p11.21 | Ubiquitous [14] |
| CST4 | Cystatin S | 20p11.21 | Parotid gland [13] |
| CST5 | Cystatin D | 20p11.21 | Parotid gland, lacrimal gland [13] |
| CST6 | Cystatin E/M | 11q13.1 | Epidermal tissue [15] |
| CST7 | Cystatin F | 20p11.21 | Hematopoietic cells [16] |
| CST8 | Cystatin 8 | 20p11.21 | Testis [14] |
| CST9 | Cystatin 9 | 20p11.21 | Testis [14] |
| CST11 | Cystatin 11 | 20p11.21 | Testis [14] |
| Name of Protein | Anti-Inflammatory Effect Pro-Tumor Effect | Pro-Inflammatory Effect Anti-Tumor Effect |
|---|---|---|
| CYS1 |
| |
| CYS2 |
|
|
| CYSC |
| |
| CYSS |
|
|
| CYSD |
| |
| CYS6 |
| |
| CYSF |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhan, F. Type 2 Cystatins and Their Roles in the Regulation of Human Immune Response and Cancer Progression. Cancers 2023, 15, 5363. https://doi.org/10.3390/cancers15225363
Zhang Z, Zhan F. Type 2 Cystatins and Their Roles in the Regulation of Human Immune Response and Cancer Progression. Cancers. 2023; 15(22):5363. https://doi.org/10.3390/cancers15225363
Chicago/Turabian StyleZhang, Zijun, and Fenghuang Zhan. 2023. "Type 2 Cystatins and Their Roles in the Regulation of Human Immune Response and Cancer Progression" Cancers 15, no. 22: 5363. https://doi.org/10.3390/cancers15225363




