Mutation Status and Glucose Availability Affect the Response to Mitochondria-Targeted Quercetin Derivative in Breast Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of Quercetin Derivatives
2.2. Preparation of Liposomes
2.3. Methodology of Autoxidation Measurements
2.4. Microcalorimetric Measurements
2.5. In Silico Analysis of Putative Cellular Targets of Quercetin Derivatives
2.6. Cell Lines and Culture Conditions
2.7. MitQ7 Uptake
2.8. Cell Cycle
2.9. Apoptosis
2.10. Intracellular pH
2.11. Imaging Cytometry
2.12. The Analysis of mitQ7-Induced Senescence
2.13. The Analysis of mitQ7-Mediated Senolytic Activity
2.14. The Analysis of Gene Mutation
2.15. Data Integration and Clustering Analysis (Heat Map)
2.16. Statistical Analysis
3. Results and Discussion
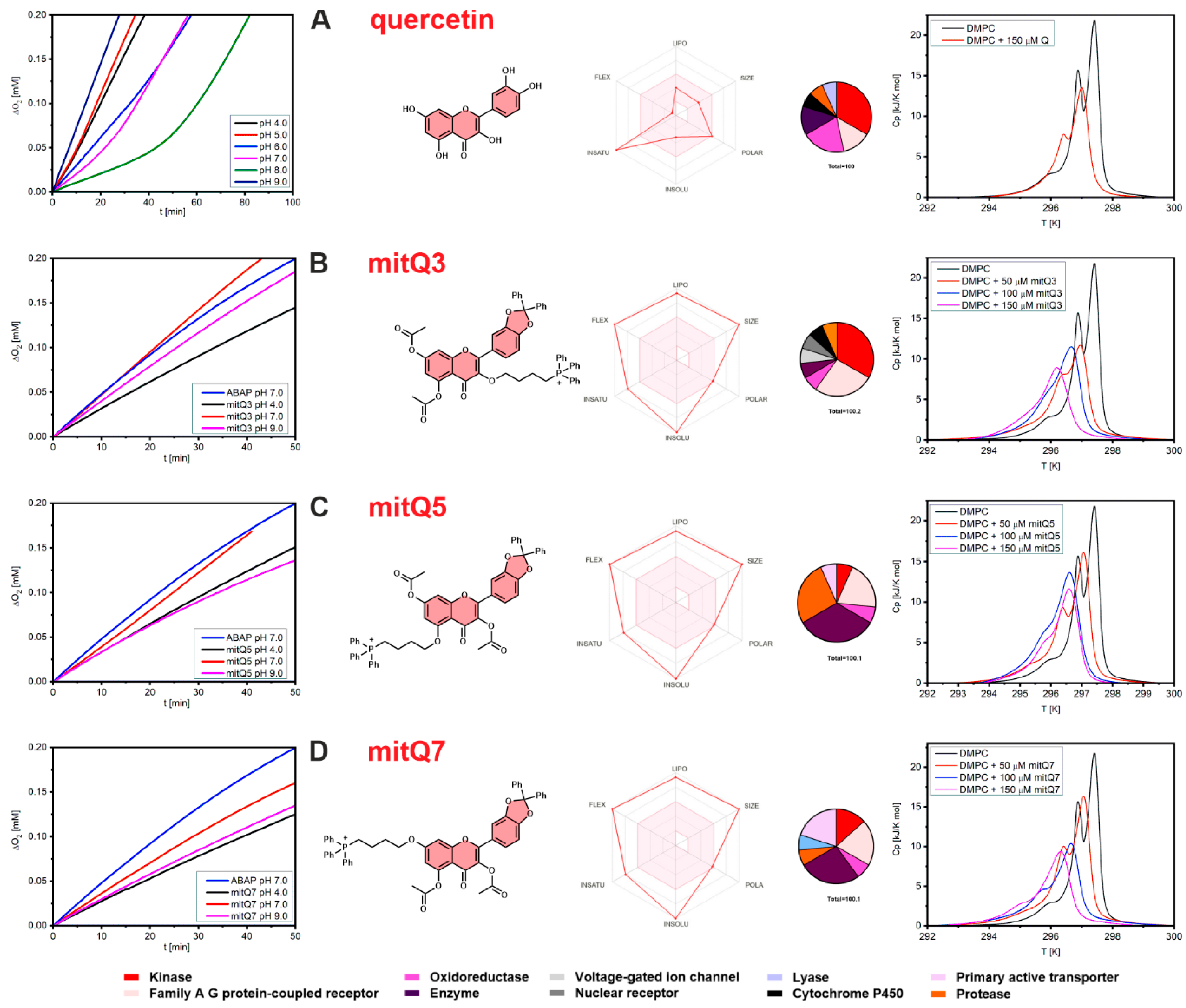
3.1. Synthesis of Quercetin Derivatives
3.2. Antioxidant Activity of Quercetin Derivatives and Their Interactions with DMPC Liposomes
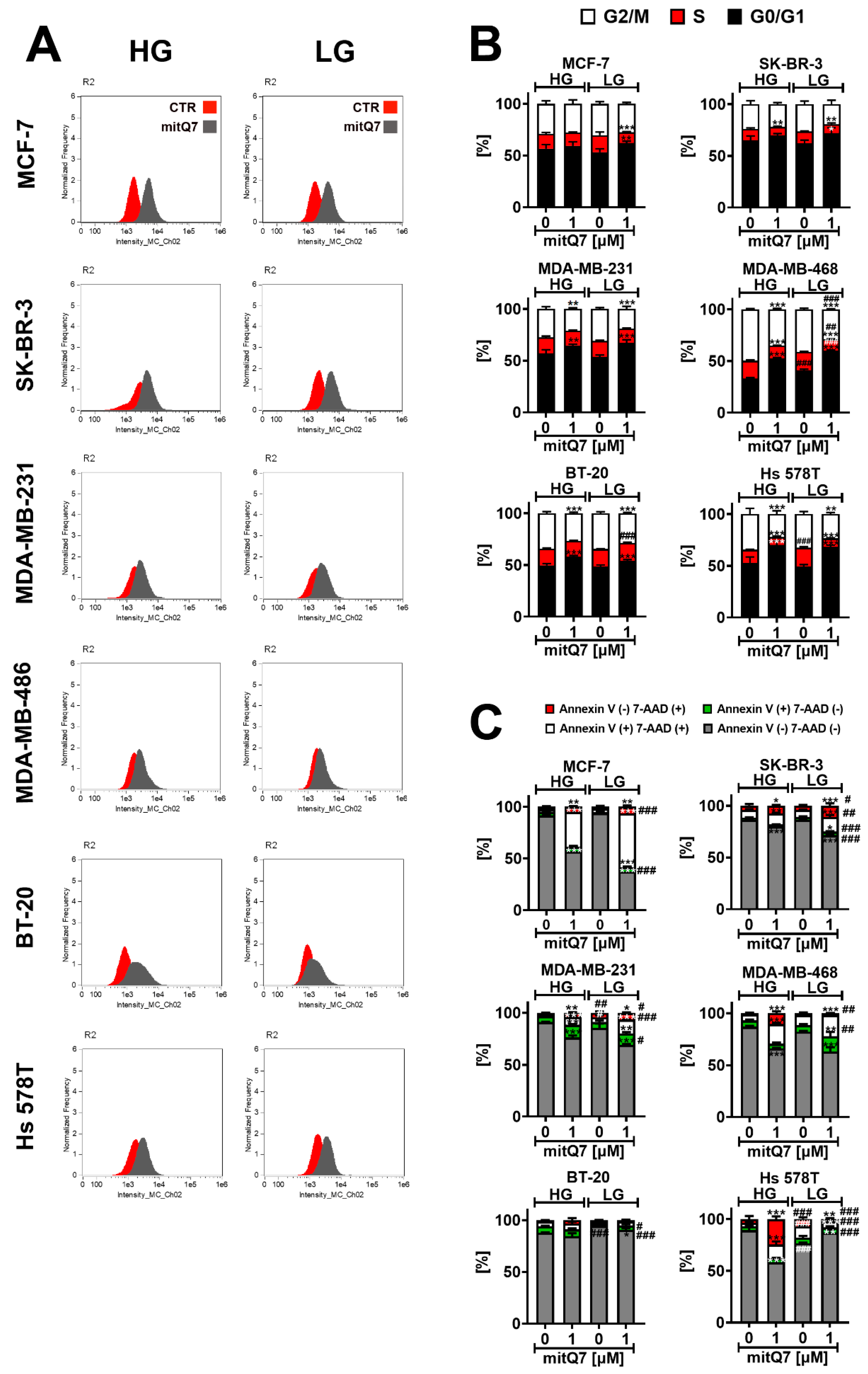
3.3. MitQ7-Mediated Cytotoxic and Cytostatic Effects in Breast Cancer Cells with Different Mutation and Receptor Status
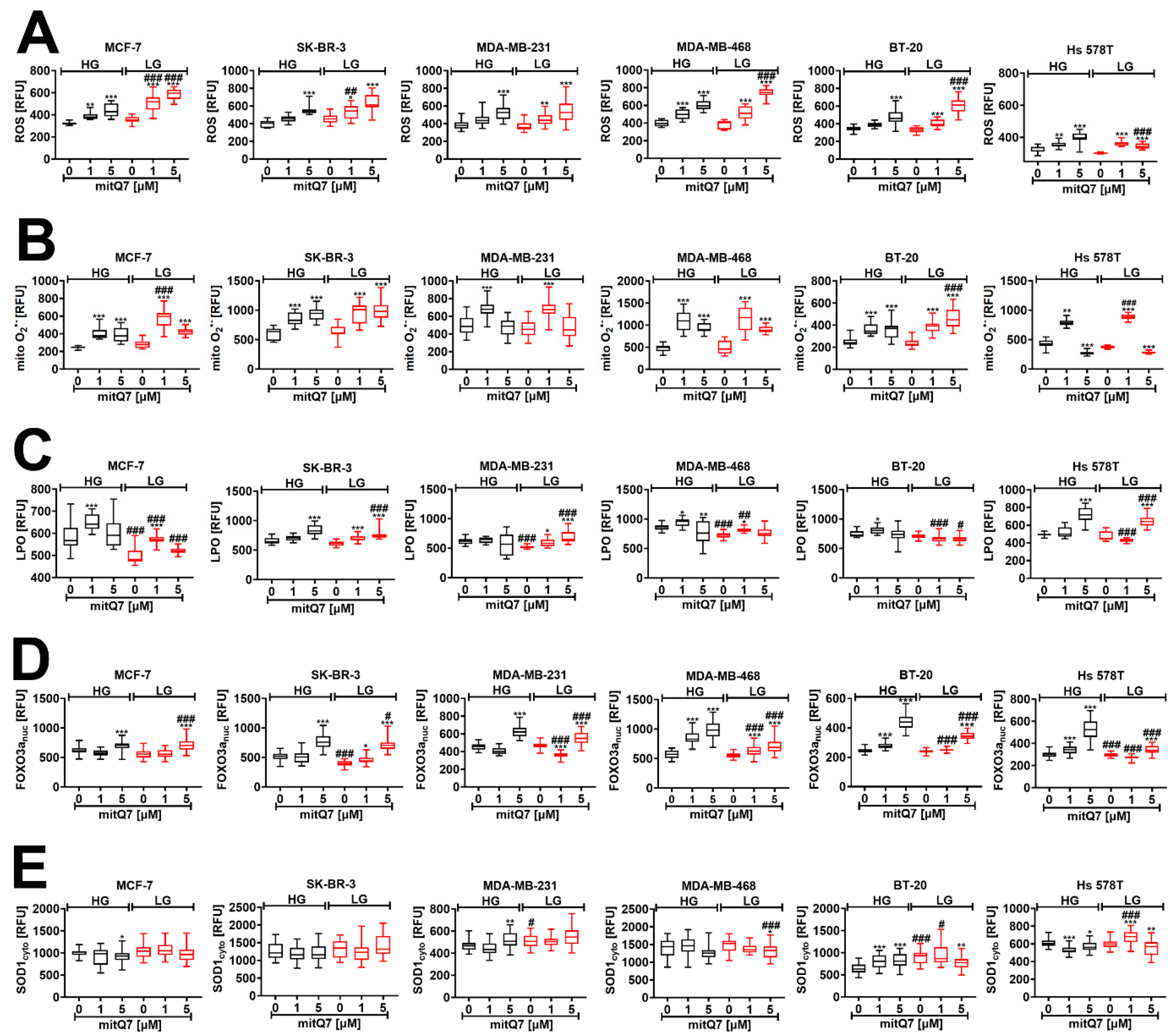
3.4. MitQ7 Treatment is Accompanied by Oxidative Stress in Breast Cancer Cells
3.5. MitQ7 Promoted Energetic Stress in Breast Cancer Cells
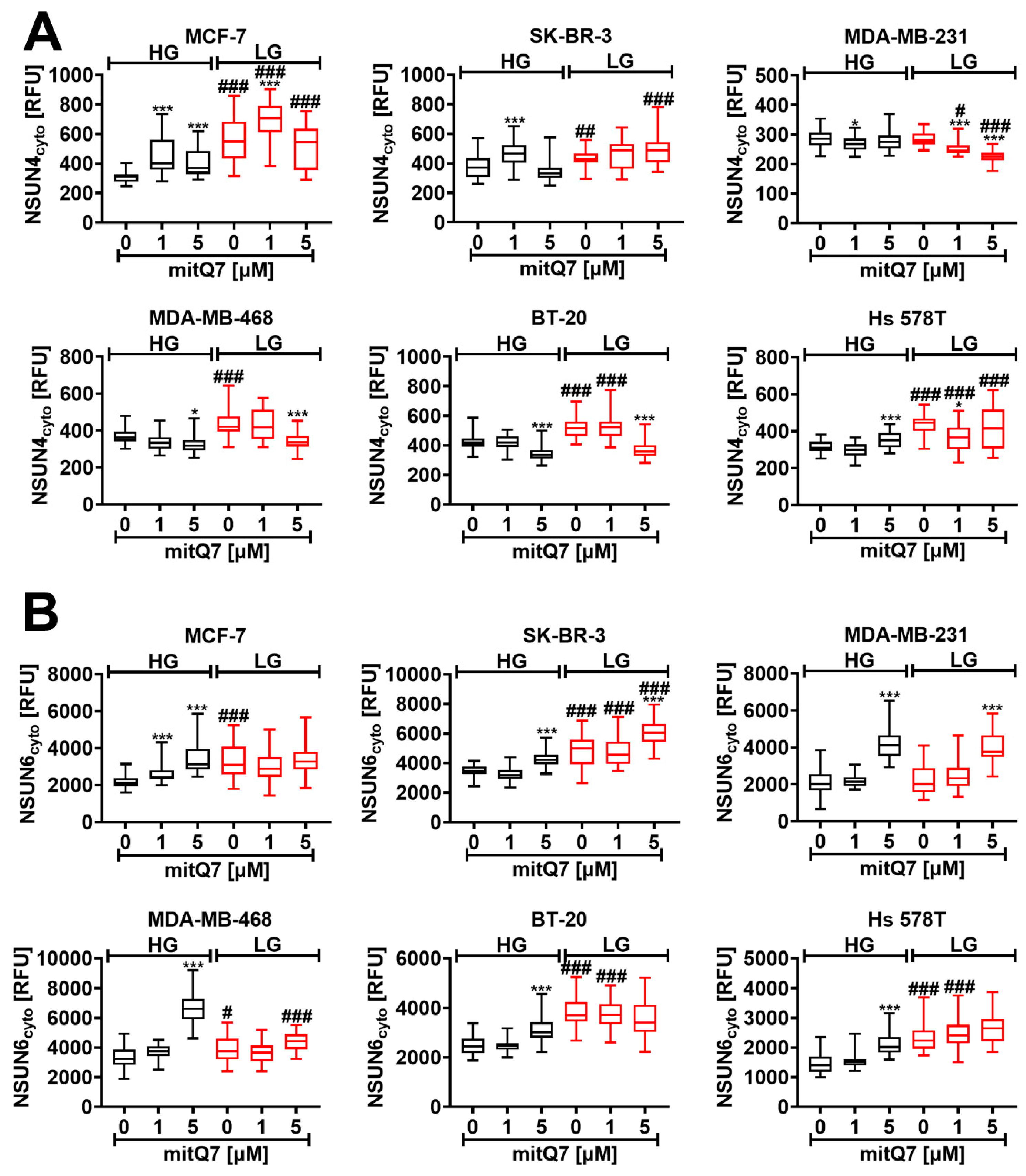
3.6. MitQ7-Mediated Elevation in NSUN4 and NSUN6 Levels
3.7. Limited Glucose Availability Is a Driver of Senescence in the Subpopulation of BT-20 and Hs 578T Breast Cancer Cell Survivals upon mitQ7 Stimulation
3.8. MitQ7-Mediated Senolytic Activity and Depolarization of Mitochondrial Transmembrane Potential in Drug-Induced Senescent Breast Cancer Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Denisenko, T.V.; Gorbunova, A.S.; Zhivotovsky, B. Mitochondrial Involvement in Migration, Invasion and Metastasis. Front. Cell Dev. Biol. 2019, 7, 355. [Google Scholar] [CrossRef] [PubMed]
- Sica, V.; Bravo-San Pedro, J.M.; Stoll, G.; Kroemer, G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int. J. Cancer 2020, 146, 10–17. [Google Scholar] [CrossRef]
- Tan, A.S.; Baty, J.W.; Dong, L.-F.; Bezawork-Geleta, A.; Endaya, B.; Goodwin, J.; Bajzikova, M.; Kovarova, J.; Peterka, M.; Yan, B.; et al. Mitochondrial Genome Acquisition Restores Respiratory Function and Tumorigenic Potential of Cancer Cells without Mitochondrial DNA. Cell Metab. 2015, 21, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Hubackova, S.; Magalhaes Novais, S.; Davidova, E.; Neuzil, J.; Rohlena, J. Mitochondria-driven elimination of cancer and senescent cells. Biol. Chem. 2019, 400, 141–148. [Google Scholar] [CrossRef]
- Reily, C.; Mitchell, T.; Chacko, B.K.; Benavides, G.A.; Murphy, M.P.; Darley-Usmar, V.M. Mitochondrially targeted compounds and their impact on cellular bioenergetics. Redox Biol. 2013, 1, 86–93. [Google Scholar] [CrossRef]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef]
- Boyle, K.A.; Van Wickle, J.; Hill, R.B.; Marchese, A.; Kalyanaraman, B.; Dwinell, M.B. Mitochondria-targeted drugs stimulate mitophagy and abrogate colon cancer cell proliferation. J. Biol. Chem. 2018, 293, 14891–14904. [Google Scholar] [CrossRef]
- Rohlenova, K.; Sachaphibulkij, K.; Stursa, J.; Bezawork-Geleta, A.; Blecha, J.; Endaya, B.; Werner, L.; Cerny, J.; Zobalova, R.; Goodwin, J.; et al. Selective Disruption of Respiratory Supercomplexes as a New Strategy to Suppress Her2high Breast Cancer. Antioxid. Redox Signal. 2017, 26, 84–103. [Google Scholar] [CrossRef]
- Halliwell, B. Are polyphenols antioxidants or pro-oxidants? What do we learn from cell culture and in vivo studies? Arch. Biochem. Biophys. 2008, 476, 107–112. [Google Scholar] [CrossRef]
- De Marchi, U.; Biasutto, L.; Garbisa, S.; Toninello, A.; Zoratti, M. Quercetin can act either as an inhibitor or an inducer of the mitochondrial permeability transition pore: A demonstration of the ambivalent redox character of polyphenols. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1787, 1425–1432. [Google Scholar] [CrossRef]
- Zheng, J.; Ramirez, V.D. Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals: Interaction of phytochemicals with ATP synthase. Br. J. Pharmacol. 2000, 130, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.-Y.; Wu, Y.-C.; Chung, J.-G.; Yang, J.-S.; Lu, H.-F.; Tsou, M.-F.; Wood, W.; Kuo, S.-J.; Chen, D.-R. Quercetin-induced apoptosis acts through mitochondrial- and caspase-3-dependent pathways in human breast cancer MDA-MB-231 cells. Hum. Exp. Toxicol. 2009, 28, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-C.; Yang, J.-S.; Lu, H.-F.; Ip, S.-W.; Lo, C.; Wu, C.-C.; Lin, J.-P.; Tang, N.-Y.; Chung, J.-G.; Chou, M.-J.; et al. Quercetin-mediated cell cycle arrest and apoptosis involving activation of a caspase cascade through the mitochondrial pathway in human breast cancer MCF-7 cells. Arch. Pharm. Res. 2010, 33, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Rivera Rivera, A.; Castillo-Pichardo, L.; Gerena, Y.; Dharmawardhane, S. Anti-Breast Cancer Potential of Quercetin via the Akt/AMPK/Mammalian Target of Rapamycin (mTOR) Signaling Cascade. PLoS ONE 2016, 11, e0157251. [Google Scholar] [CrossRef]
- Mattarei, A.; Biasutto, L.; Marotta, E.; De Marchi, U.; Sassi, N.; Garbisa, S.; Zoratti, M.; Paradisi, C. A Mitochondriotropic Derivative of Quercetin: A Strategy to Increase the Effectiveness of Polyphenols. ChemBioChem 2008, 9, 2633–2642. [Google Scholar] [CrossRef] [PubMed]
- Biasutto, L.; Sassi, N.; Mattarei, A.; Marotta, E.; Cattelan, P.; Toninello, A.; Garbisa, S.; Zoratti, M.; Paradisi, C. Impact of mitochondriotropic quercetin derivatives on mitochondria. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1797, 189–196. [Google Scholar] [CrossRef]
- Mattarei, A.; Sassi, N.; Durante, C.; Biasutto, L.; Sandonà, G.; Marotta, E.; Garbisa, S.; Gennaro, A.; Paradisi, C.; Zoratti, M. Redox Properties and Cytotoxicity of Synthetic Isomeric Mitochondriotropic Derivatives of the Natural Polyphenol Quercetin. Eur. J. Org. Chem. 2011, 2011, 5577–5586. [Google Scholar] [CrossRef]
- Sassi, N.; Biasutto, L.; Mattarei, A.; Carraro, M.; Giorgio, V.; Citta, A.; Bernardi, P.; Garbisa, S.; Szabò, I.; Paradisi, C.; et al. Cytotoxicity of a mitochondriotropic quercetin derivative: Mechanisms. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1817, 1095–1106. [Google Scholar] [CrossRef]
- León-González, A.J.; Auger, C.; Schini-Kerth, V.B. Pro-oxidant activity of polyphenols and its implication on cancer chemoprevention and chemotherapy. Biochem. Pharmacol. 2015, 98, 371–380. [Google Scholar] [CrossRef]
- Khajah, M.A.; Khushaish, S.; Luqmani, Y.A. Glucose deprivation reduces proliferation and motility, and enhances the anti-proliferative effects of paclitaxel and doxorubicin in breast cell lines in vitro. PLoS ONE 2022, 17, e0272449. [Google Scholar] [CrossRef] [PubMed]
- Biasutto, L.; Mattarei, A.; Paradisi, C. Synthesis and Testing of Novel Isomeric Mitochondriotropic Derivatives of Resveratrol and Quercetin. In Mitochondrial Medicine; Weissig, V., Edeas, M., Eds.; Springer: New York, NY, USA, 2021; pp. 141–160. [Google Scholar] [CrossRef]
- Grebowski, J.; Konopko, A.; Krokosz, A.; DiLabio, G.A.; Litwinienko, G. Antioxidant activity of highly hydroxylated fullerene C60 and its interactions with the analogue of α-tocopherol. Free Radic. Biol. Med. 2020, 160, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Jodko-Piórecka, K.; Sikora, B.; Kluzek, M.; Przybylski, P.; Litwinienko, G. Antiradical Activity of Dopamine, L-DOPA, Adrenaline, and Noradrenaline in Water/Methanol and in Liposomal Systems. J. Org. Chem. 2022, 87, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Konopko, A.; Litwinienko, G. Unexpected Role of pH and Microenvironment on the Antioxidant and Synergistic Activity of Resveratrol in Model Micellar and Liposomal Systems. J. Org. Chem. 2022, 87, 1698–1709. [Google Scholar] [CrossRef] [PubMed]
- Jodko-Piorecka, K.; Litwinienko, G. First Experimental Evidence of Dopamine Interactions with Negatively Charged Model Biomembranes. ACS Chem. Neurosci. 2013, 4, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Lewinska, A.; Adamczyk-Grochala, J.; Kwasniewicz, E.; Deregowska, A.; Semik, E.; Zabek, T.; Wnuk, M. Reduced levels of methyltransferase DNMT2 sensitize human fibroblasts to oxidative stress and DNA damage that is accompanied by changes in proliferation-related miRNA expression. Redox Biol. 2018, 14, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Lewińska, A.; Przybylski, P.; Adamczyk-Grochala, J.; Błoniarz, D.; Litwinienko, G.; Wnuk, M. Senolysis-Based Elimination of Chemotherapy-Induced Senescent Breast Cancer Cells by Quercetin Derivative with Blocked Hydroxy Groups. Cancers 2022, 14, 605. [Google Scholar] [CrossRef]
- Antoniak, M.A.; Pązik, R.; Bazylińska, U.; Wiwatowski, K.; Tomaszewska, A.; Kulpa-Greszta, M.; Adamczyk-Grochala, J.; Wnuk, M.; Maćkowski, S.; Lewińska, A.; et al. Multimodal polymer encapsulated CdSe/Fe3O4 nanoplatform with improved biocompatibility for two-photon and temperature stimulated bioapplications. Mater. Sci. Eng. C 2021, 127, 112224. [Google Scholar] [CrossRef]
- Bloniarz, D.; Adamczyk-Grochala, J.; Lewinska, A.; Wnuk, M. The lack of functional DNMT2/TRDMT1 gene modulates cancer cell responses during drug-induced senescence. Aging 2021, 13, 15833–15874. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Sturn, A.; Quackenbush, J.; Trajanoski, Z. Genesis: Cluster analysis of microarray data. Bioinformatics 2002, 18, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Deus, C.M.; Pereira, S.P.; Cunha-Oliveira, T.; Teixeira, J.; Simões, R.F.; Cagide, F.; Benfeito, S.; Borges, F.; Raimundo, N.; Oliveira, P.J. A mitochondria-targeted caffeic acid derivative reverts cellular and mitochondrial defects in human skin fibroblasts from male sporadic Parkinson’s disease patients. Redox Biol. 2021, 45, 102037. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.; Deus, C.M.; Borges, F.; Oliveira, P.J. Mitochondria: Targeting mitochondrial reactive oxygen species with mitochondriotropic polyphenolic-based antioxidants. Int. J. Biochem. Cell Biol. 2018, 97, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.J. (Ed.) Mitochondrial Biology and Experimental Therapeutics; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Hoye, A.T.; Davoren, J.E.; Wipf, P.; Fink, M.P.; Kagan, V.E. Targeting Mitochondria. Acc. Chem. Res. 2008, 41, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, N.; Victor, V.M. Molecular Strategies for Targeting Antioxidants to Mitochondria: Therapeutic Implications. Antioxid. Redox Signal. 2015, 22, 686–729. [Google Scholar] [CrossRef]
- Visioli, F.; Ingram, A.; Beckman, J.S.; Magnusson, K.R.; Hagen, T.M. Strategies to protect against age-related mitochondrial decay: Do natural products and their derivatives help? Free Radic. Biol. Med. 2022, 178, 330–346. [Google Scholar] [CrossRef]
- Jovanovic, S.V.; Steenken, S.; Hara, Y.; Simic, M.G. Reduction potentials of flavonoid and model phenoxyl radicals. Which ring in flavonoids is responsible for antioxidant activity? J. Chem. Soc. Perkin Trans. 2 1996, 2497. [Google Scholar] [CrossRef]
- Pietta, P.-G. Flavonoids as Antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, J.A.; Day, A.J.; Morgan, M.R.A. Experimental Determination of Octanol−Water Partition Coefficients of Quercetin and Related Flavonoids. J. Agric. Food Chem. 2005, 53, 4355–4360. [Google Scholar] [CrossRef] [PubMed]
- Ong, H.C.; Coimbra, J.T.S.; Kwek, G.; Ramos, M.J.; Xing, B.; Fernandes, P.A.; García, F. Alkyl vs. aryl modifications: A comparative study on modular modifications of triphenylphosphonium mitochondrial vectors. RSC Chem. Biol. 2021, 2, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Rokitskaya, T.I.; Aleksandrova, E.V.; Korshunova, G.A.; Khailova, L.S.; Tashlitsky, V.N.; Luzhkov, V.B.; Antonenko, Y.N. Membrane Permeability of Modified Butyltriphenylphosphonium Cations. J. Phys. Chem. B 2022, 126, 412–422. [Google Scholar] [CrossRef]
- Montecino-Garrido, H.; Sepúlveda, M.; Méndez, D.; Monroy-Cárdenas, M.; Alfaro, S.; González-Avendaño, M.; Caballero, J.; Urra, F.A.; Araya-Maturana, R.; Fuentes, E. Assessing mitochondria-targeted acyl hydroquinones on the mitochondrial platelet function and cytotoxic activity: Role of the linker length. Free Radic. Biol. Med. 2023, 208, 26–36. [Google Scholar] [CrossRef]
- Heimburg, T.; Jackson, A.D. The Thermodynamics of General Anesthesia. Biophys. J. 2007, 92, 3159–3165. [Google Scholar] [CrossRef]
- Storz, P. Forkhead Homeobox Type O Transcription Factors in the Responses to Oxidative Stress. Antioxid. Redox Signal. 2011, 14, 593–605. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS signaling under metabolic stress: Cross-talk between AMPK and AKT pathway. Mol. Cancer 2017, 16, 79. [Google Scholar] [CrossRef]
- Faubert, B.; Boily, G.; Izreig, S.; Griss, T.; Samborska, B.; Dong, Z.; Dupuy, F.; Chambers, C.; Fuerth, B.J.; Viollet, B.; et al. AMPK Is a Negative Regulator of the Warburg Effect and Suppresses Tumor Growth In vivo. Cell Metab. 2013, 17, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Kazyken, D.; Lentz, S.I.; Fingar, D.C. Alkaline intracellular pH (pHi) activates AMPK–mTORC2 signaling to promote cell survival during growth factor limitation. J. Biol. Chem. 2021, 297, 101100. [Google Scholar] [CrossRef] [PubMed]
- Gledhill, J.R.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Mechanism of inhibition of bovine F 1 -ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. USA 2007, 104, 13632–13637. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Lee, H.; Kim, S.; Park, J.; Ha, T. The anti-obesity effect of quercetin is mediated by the AMPK and MAPK signaling pathways. Biochem. Biophys. Res. Commun. 2008, 373, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Lee, J.O.; Kim, J.H.; Lee, S.K.; You, G.Y.; Park, S.H.; Park, J.M.; Kim, E.-K.; Suh, P.-G.; An, J.K.; et al. Quercetin suppresses HeLa cell viability via AMPK-induced HSP70 and EGFR down-regulation. J. Cell. Physiol. 2010, 223, 408–414. [Google Scholar] [CrossRef]
- Chellamuthu, A.; Gray, S.G. The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer. Cells 2020, 9, 1758. [Google Scholar] [CrossRef]
- Mei, L.; Shen, C.; Miao, R.; Wang, J.-Z.; Cao, M.-D.; Zhang, Y.-S.; Shi, L.-H.; Zhao, G.-H.; Wang, M.-H.; Wu, L.-S.; et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57Kip2 by an m5C-dependent manner. Cell Death Dis. 2020, 11, 270. [Google Scholar] [CrossRef]
- Yang, R.; Liang, X.; Wang, H.; Guo, M.; Shen, H.; Shi, Y.; Liu, Q.; Sun, Y.; Yang, L.; Zhan, M. The RNA methyltransferase NSUN6 suppresses pancreatic cancer development by regulating cell proliferation. eBioMedicine 2021, 63, 103195. [Google Scholar] [CrossRef]
- Bohnsack, K.; Höbartner, C.; Bohnsack, M. Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 2019, 10, 102. [Google Scholar] [CrossRef]
- Metodiev, M.D.; Spåhr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.-G.; Ruzzenente, B. NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly. PLoS Genet. 2014, 10, e1004110. [Google Scholar] [CrossRef] [PubMed]
- Childs, B.G.; Gluscevic, M.; Baker, D.J.; Laberge, R.-M.; Marquess, D.; Dananberg, J.; van Deursen, J.M. Senescent cells: An emerging target for diseases of ageing. Nat. Rev. Drug Discov. 2017, 16, 718–735. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kohli, J.; Demaria, M. Senescent Cells in Cancer Therapy: Friends or Foes? Trends Cancer 2020, 6, 838–857. [Google Scholar] [CrossRef]
- Wang, L.; Lankhorst, L.; Bernards, R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer 2022, 22, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tchkonia, T.; Pirtskhalava, T.; Gower, A.C.; Ding, H.; Giorgadze, N.; Palmer, A.K.; Ikeno, Y.; Hubbard, G.B.; Lenburg, M.; et al. The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell 2015, 14, 644–658. [Google Scholar] [CrossRef]
- Ly, J.D.; Grubb, D.R.; Lawen, A. The mitochondrial membrane potential (Δψm) in apoptosis; an update. Apoptosis 2003, 8, 115–128. [Google Scholar] [CrossRef]
- Webster, K.A. Mitochondrial membrane permeabilization and cell death during myocardial infarction: Roles of calcium and reactive oxygen species. Future Cardiol. 2012, 8, 863–884. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Przybylski, P.; Lewińska, A.; Rzeszutek, I.; Błoniarz, D.; Moskal, A.; Betlej, G.; Deręgowska, A.; Cybularczyk-Cecotka, M.; Szmatoła, T.; Litwinienko, G.; et al. Mutation Status and Glucose Availability Affect the Response to Mitochondria-Targeted Quercetin Derivative in Breast Cancer Cells. Cancers 2023, 15, 5614. https://doi.org/10.3390/cancers15235614
Przybylski P, Lewińska A, Rzeszutek I, Błoniarz D, Moskal A, Betlej G, Deręgowska A, Cybularczyk-Cecotka M, Szmatoła T, Litwinienko G, et al. Mutation Status and Glucose Availability Affect the Response to Mitochondria-Targeted Quercetin Derivative in Breast Cancer Cells. Cancers. 2023; 15(23):5614. https://doi.org/10.3390/cancers15235614
Chicago/Turabian StylePrzybylski, Paweł, Anna Lewińska, Iwona Rzeszutek, Dominika Błoniarz, Aleksandra Moskal, Gabriela Betlej, Anna Deręgowska, Martyna Cybularczyk-Cecotka, Tomasz Szmatoła, Grzegorz Litwinienko, and et al. 2023. "Mutation Status and Glucose Availability Affect the Response to Mitochondria-Targeted Quercetin Derivative in Breast Cancer Cells" Cancers 15, no. 23: 5614. https://doi.org/10.3390/cancers15235614
APA StylePrzybylski, P., Lewińska, A., Rzeszutek, I., Błoniarz, D., Moskal, A., Betlej, G., Deręgowska, A., Cybularczyk-Cecotka, M., Szmatoła, T., Litwinienko, G., & Wnuk, M. (2023). Mutation Status and Glucose Availability Affect the Response to Mitochondria-Targeted Quercetin Derivative in Breast Cancer Cells. Cancers, 15(23), 5614. https://doi.org/10.3390/cancers15235614








