MiR-21 Is Induced by Hypoxia and Down-Regulates RHOB in Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfections
2.2. Colony Forming Assay
2.3. Migration Assay by Boyden Chamber
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Protein Analysis
2.6. In Vivo Experiments
2.7. Databases
2.8. Statistics
3. Results
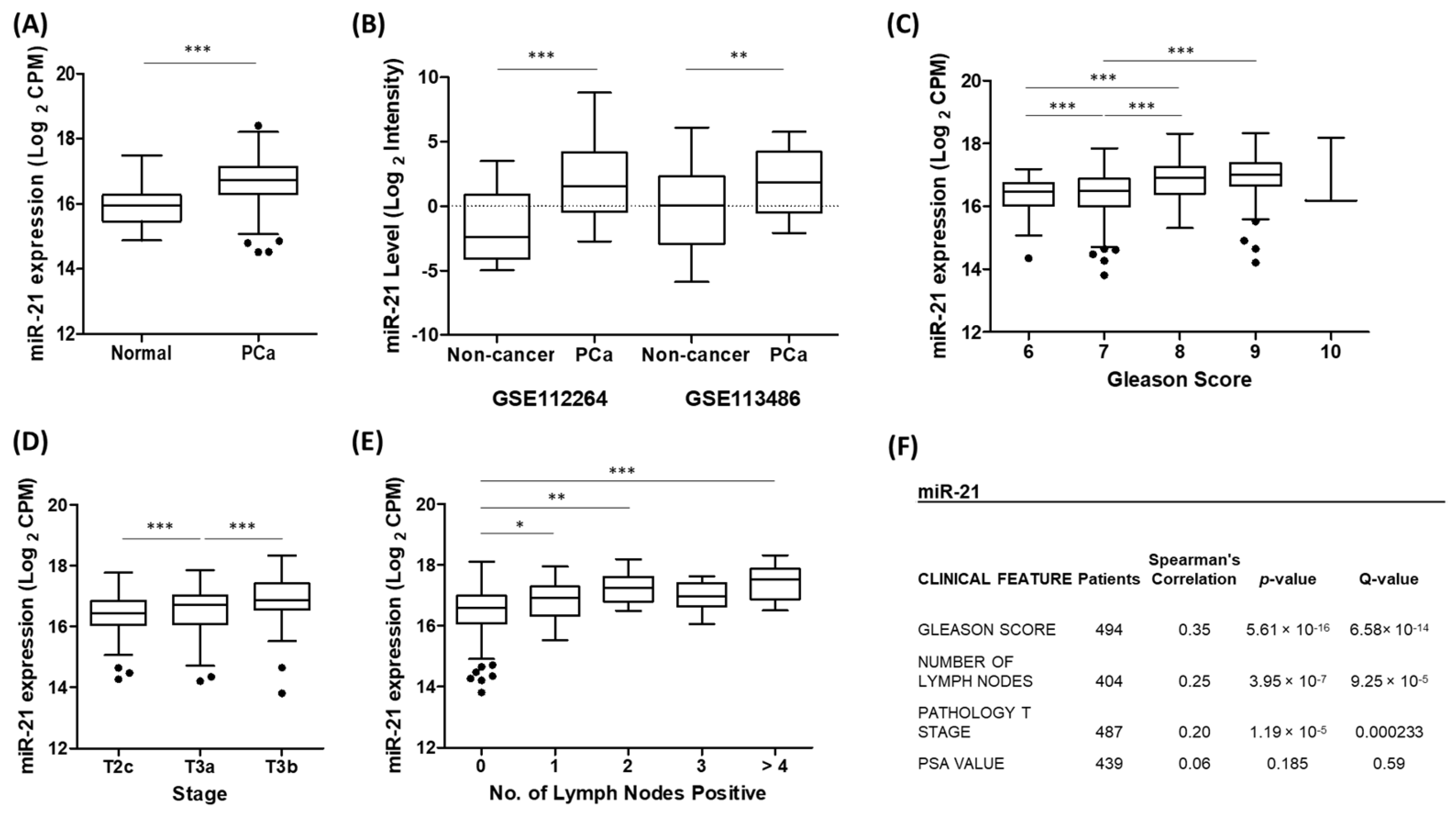
3.1. Up-Regulation of miR-21 Is Associated with Prostate Cancer
3.2. miR-21 Is Up-Regulated by Hypoxia in Prostate Cells
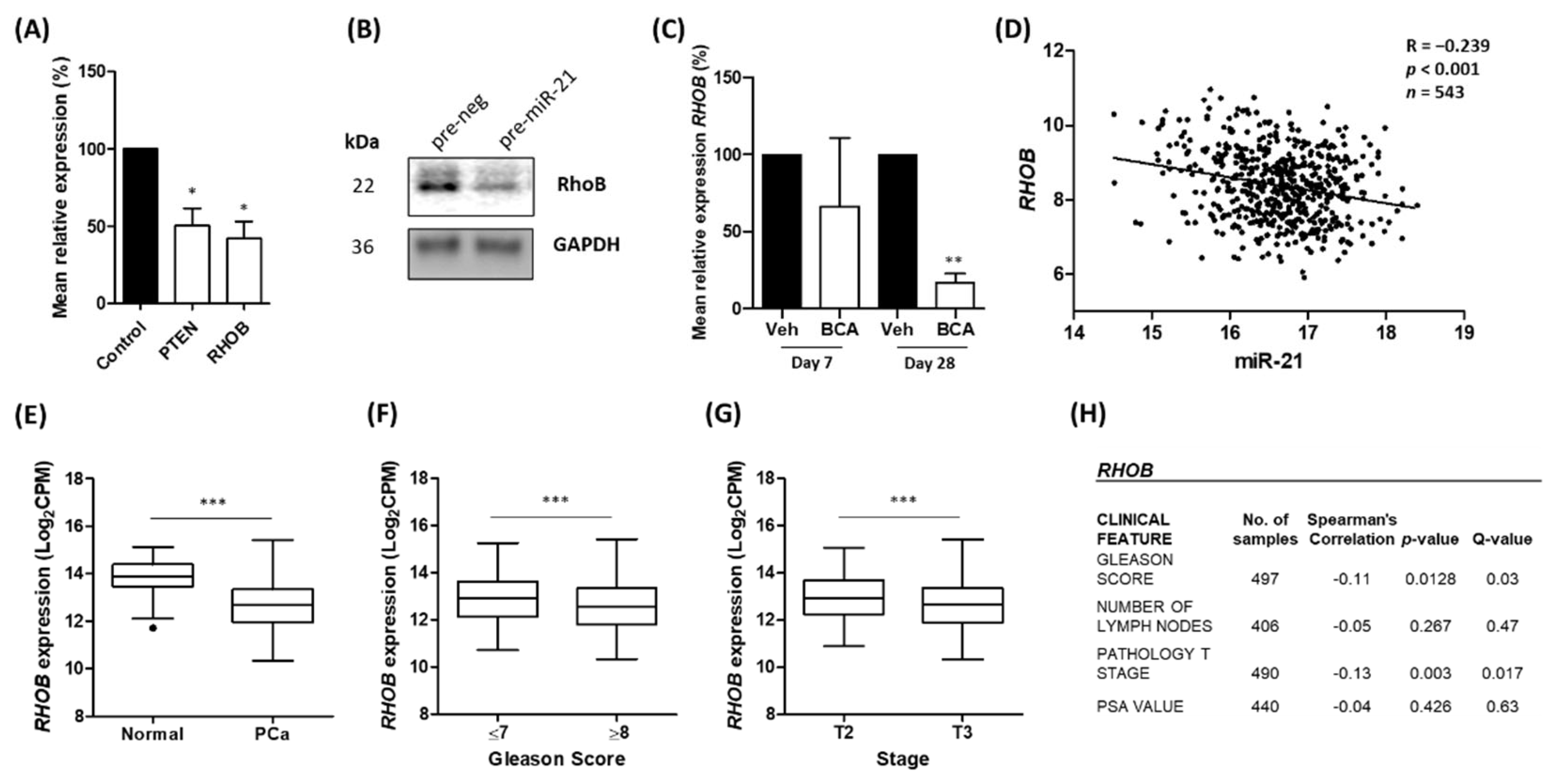
3.3. Ras Homolog Family Member B (RHOB) Is Down-Regulated by miR-21 in Prostate Cancer
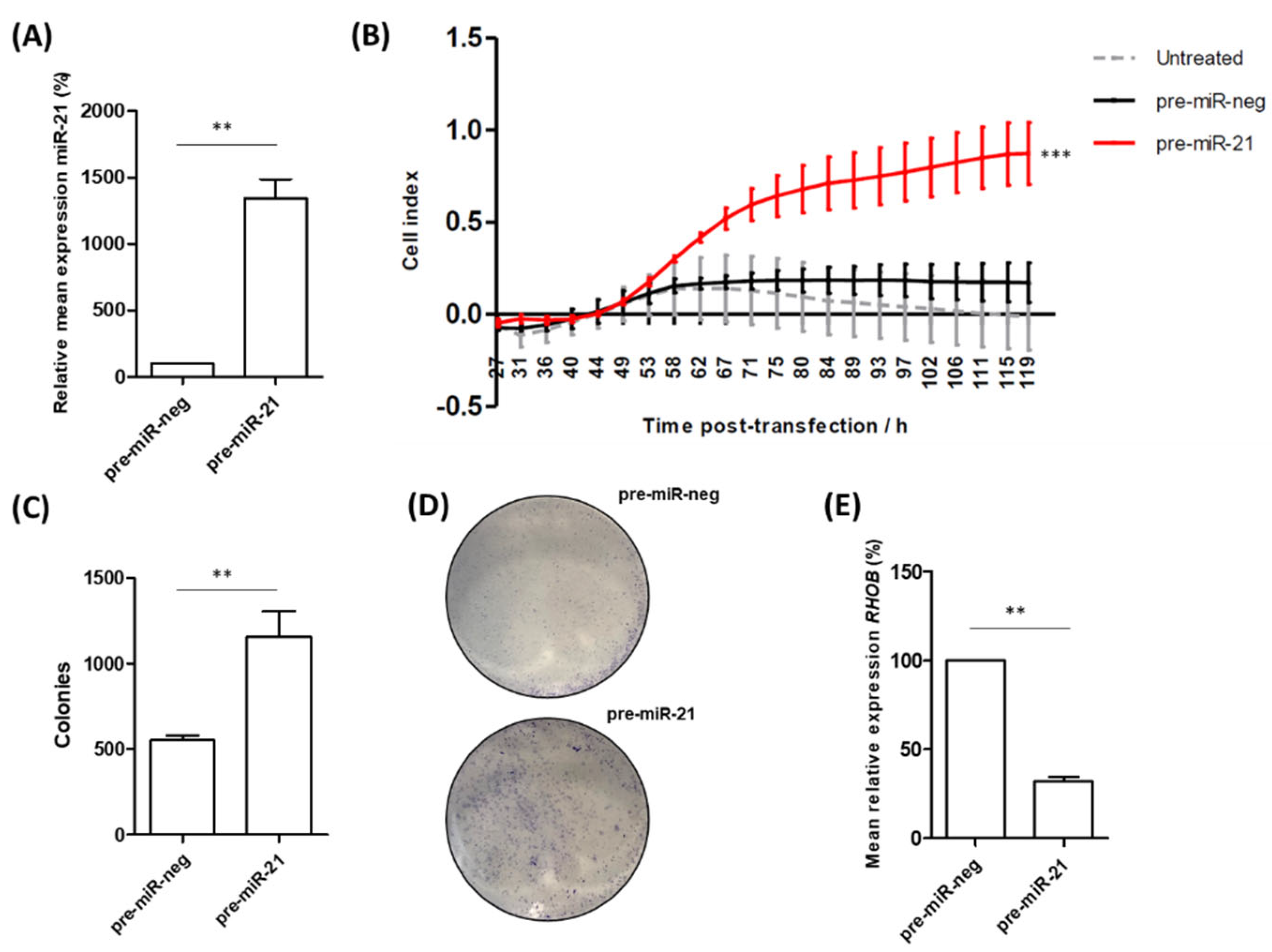
3.4. miR-21 Over-Expression Increases Migration and Colony-Forming Ability of RWPE-1 Cells
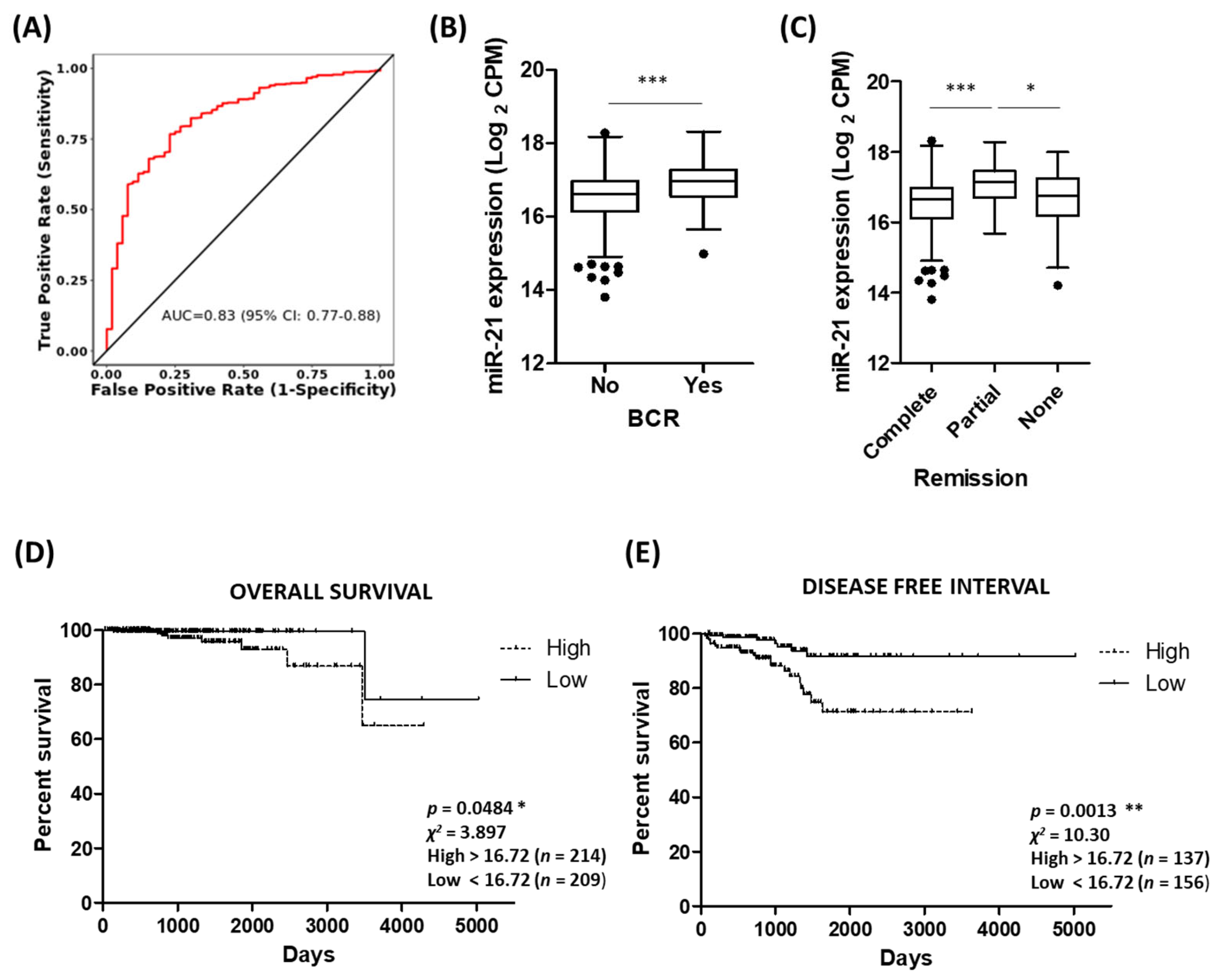
3.5. Potential of miR-21 as a Biomarker of Prostate Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours—Implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Araos, J.; Sleeman, J.P.; Garvalov, B.K. The role of hypoxic signalling in metastasis: Towards translating knowledge of basic biology into novel anti-tumour strategies. Clin. Exp. Metastasis 2018, 35, 563–599. [Google Scholar] [CrossRef] [PubMed]
- McKenna, D.J.; Errington, R.; Pors, K. Current challenges and opportunities in treating hypoxic prostate tumors. J. Cancer Metastasis Treat. 2018, 4, 11. [Google Scholar] [CrossRef]
- Macharia, L.W.; Wanjiru, C.M.; Mureithi, M.W.; Pereira, C.M.; Ferrer, V.P.; Moura-Neto, V. MicroRNAs, Hypoxia and the Stem-Like State as Contributors to Cancer Aggressiveness. Front. Genet. 2019, 10, 125. [Google Scholar] [CrossRef]
- Tapeh, B.E.; Alivand, M.R.; Solalii, S. Potential Interactions between miRNAs and Hypoxia: A New Layer in Cancer Hypoxia. Anti-Cancer Agents Med. Chem. 2021, 21, 2315–2326. [Google Scholar] [CrossRef]
- Sharma, N.; Baruah, M.M. The microRNA signatures: Aberrantly expressed miRNAs in prostate cancer. Clin. Transl. Oncol. 2019, 21, 126–144. [Google Scholar] [CrossRef]
- Kanwal, R.; Plaga, A.R.; Liu, X.; Shukla, G.C.; Gupta, S. MicroRNAs in prostate cancer: Functional role as biomarkers. Cancer Lett. 2017, 407, 9–20. [Google Scholar] [CrossRef]
- Kasomva, K.; Sen, A.; Paulraj, M.G.; Sailo, S.; Raphael, V.; Puro, K.-U.; Assumi, S.R.; Ignacimuthu, S. Roles of microRNA in prostate cancer cell metabolism. Int. J. Biochem. Cell Biol. 2018, 102, 109–116. [Google Scholar] [CrossRef]
- Bavelloni, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A Current Overview. Anticancer. Res. 2017, 37, 6511–6521. [Google Scholar] [CrossRef]
- Angel, C.Z.; Lynch, S.M.; Nesbitt, H.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. miR-210 is induced by hypoxia and regulates neural cell adhesion molecule in prostate cells. J. Cell. Physiol. 2020, 235, 6194–6203. [Google Scholar] [CrossRef]
- Bhandari, V.; Hoey, C.; Liu, L.Y.; Lalonde, E.; Ray, J.; Livingstone, J.; Lesurf, R.; Shiah, Y.-J.; Vujcic, T.; Huang, X.; et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 2019, 51, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, M.; Guan, Y.; Wu, Q. Hypoxia-Responsive Mir-301a and Mir-301b Promote Radioresistance of Prostate Cancer Cells via Downregulating NDRG2. Experiment 2016, 22, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liang, F.; Yue, J.; Liu, P.; Wang, J.; Wang, Z.; Li, H.; Cheng, D.; Du, J.; Zhang, K.; et al. MicroRNA-137 regulates hypoxia-mediated migration and epithelial-mesenchymal transition in prostate cancer by targeting LGR4 via the EGFR/ERK signaling pathway. Int. J. Oncol. 2020, 57, 540–549. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, D.; Wang, X.; Yao, X.; Ye, C.; Zhang, S.; Wang, H.; Chang, C.; Xia, H.; Wang, Y.-C.; et al. Hypoxia-inducible miR-182 enhances HIF1α signaling via targeting PHD2 and FIH1 in prostate cancer. Sci. Rep. 2015, 5, 12495. [Google Scholar] [CrossRef]
- Kumoğlu, G.; Döşkaya, M.; Iz, S.G. The biomarker features of miR-145-3p determined via meta-analysis validated by qRT-PCR in metastatic cancer cell lines. Gene 2019, 710, 341–353. [Google Scholar] [CrossRef]
- Singh, A.; Singh, A.K.; Giri, R.; Kumar, D.; Sharma, R.; Valis, M.; Kuca, K.; Garg, N. The role of microRNA-21 in the onset and progression of cancer. Future Med. Chem. 2021, 13, 1885–1906. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life Sci. 2018, 75, 3539–3551. [Google Scholar] [CrossRef]
- Bahreyni, A.; Rezaei, M.; Bahrami, A.; Khazaei, M.; Fiuji, H.; Ryzhikov, M.; Ferns, G.A.; Avan, A.; Hassanian, S.M. Diagnostic, prognostic, and therapeutic potency of microRNA 21 in the pathogenesis of colon cancer, current status and prospective. J. Cell. Physiol. 2019, 234, 8075–8081. [Google Scholar] [CrossRef]
- Anwar, S.L.; Sari, D.N.I.; Kartika, A.I.; Fitria, M.S.; Tanjung, D.S.; Rakhmina, D.; Wardana, T.; Astuti, I.; Haryana, S.M.; Aryandono, T. Upregulation of Circulating MiR-21 Expression as a Potential Biomarker for Therapeutic Monitoring and Clinical Outcome in Breast Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, S.; Fan, K.; Jiang, C. MicroRNA-21 and its impact on signaling pathways in cervical cancer (Review). Oncol. Lett. 2019, 17, 3066–3070. [Google Scholar] [CrossRef] [PubMed]
- Vila-Navarro, E.; Duran-Sanchon, S.; Vila-Casadesús, M.; Moreira, L.; Ginès, À.; Cuatrecasas, M.; Lozano, J.J.; Bujanda, L.; Castells, A.; Gironella, M. Novel Circulating miRNA Signatures for Early Detection of Pancreatic Neoplasia. Clin. Transl. Gastroenterol. 2019, 10, e00029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, D.; Zhang, R.; Gao, P.; Peng, R.; Li, J. The miR-21 potential of serving as a biomarker for liver diseases in clinical practice. Biochem. Soc. Trans. 2020, 48, 2295–2305. [Google Scholar] [CrossRef]
- Dioguardi, M.; Caloro, G.A.; Laino, L.; Alovisi, M.; Sovereto, D.; Crincoli, V.; Aiuto, R.; Coccia, E.; Troiano, G.; Muzio, L.L. Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis. Cancers 2020, 12, 936. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef]
- Kulshreshtha, R.; Davuluri, R.V.; Calin, G.; E Ivan, M. A microRNA component of the hypoxic response. Cell Death Differ. 2008, 15, 667–671. [Google Scholar] [CrossRef]
- Dong, X.; Pi, Q.; Yuemaierabola, A.; Guo, W.; Tian, H. Silencing LINC00294 Restores Mitochondrial Function and Inhibits Apoptosis of Glioma Cells under Hypoxia via the miR-21-5p/CASKIN1/cAMP Axis. Oxidative Med. Cell. Longev. 2021, 2021, 8240015. [Google Scholar] [CrossRef]
- Nijhuis, A.; Thompson, H.; Adam, J.; Parker, A.; Gammon, L.; Lewis, A.; Bundy, J.G.; Soga, T.; Jalaly, A.; Propper, D.; et al. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum. Mol. Genet. 2017, 26, 1552–1564. [Google Scholar] [CrossRef]
- Mace, T.A.; Collins, A.L.; Wojcik, S.E.; Croce, C.M.; Lesinski, G.B.; Bloomston, M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J. Surg. Res. 2013, 184, 855–860. [Google Scholar] [CrossRef]
- Dong, C.; Liu, X.; Wang, H.; Li, J.; Dai, L.; Li, J.; Xu, Z. Hypoxic non-small-cell lung cancer cell-derived exosomal miR-21 promotes resistance of normoxic cell to cisplatin. OncoTargets Ther. 2019, 12, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res 2016, 76, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wu, X.; Dai, L.; Ge, Z.; Gao, C.; Zhang, H.; Wang, F.; Zhang, X.-P.; Chen, B. Gambogenic Acid Exerts Antitumor Activity in Hypoxic Multiple Myeloma Cells by Regulation of miR-21. J. Cancer 2017, 8, 3278–3286. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Wang, F.; Nie, X.; Du, H.; Zhao, Y.; Yin, Z.; Li, H.; Fan, J.; Wen, Z.; Wang, D.W.; et al. The Cell Type–Specific Functions of miR-21 in Cardiovascular Diseases. Front. Genet. 2020, 11, 563166. [Google Scholar] [CrossRef]
- Bienertova-Vasku, J.; Novak, J.; Vasku, A. MicroRNAs in pulmonary arterial hypertension: Pathogenesis, diagnosis and treatment. J. Am. Soc. Hypertens. 2015, 9, 221–234. [Google Scholar] [CrossRef]
- Xu, X.; Kriegel, A.J.; Jiao, X.; Liu, H.; Bai, X.; Olson, J.; Liang, M.; Ding, X. miR-21 in ischemia/reperfusion injury: A double-edged sword? Physiol. Genom. 2014, 46, 789–797. [Google Scholar] [CrossRef]
- Song, J.; Sundar, K.; Gangaraju, R.; Prchal, J.T. Regulation of erythropoiesis after normoxic return from chronic sustained and intermittent hypoxia. J. Appl. Physiol. 2017, 123, 1671–1675. [Google Scholar] [CrossRef]
- Liu, L.-Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.-F.; Lai, L.; Jiang, B.-H. MiR-21 Induced Angiogenesis through AKT and ERK Activation and HIF-1α Expression. PLoS ONE 2011, 6, e19139. [Google Scholar] [CrossRef]
- Bao, B.; Ahmad, A.; Kong, D.; Ali, S.; Azmi, A.S.; Li, Y.; Banerjee, S.; Padhye, S.; Sarkar, F.H. Hypoxia Induced Aggressiveness of Prostate Cancer Cells Is Linked with Deregulated Expression of VEGF, IL-6 and miRNAs That Are Attenuated by CDF. PLoS ONE 2012, 7, e43726. [Google Scholar] [CrossRef]
- Workman, P.; Balmain, A.; Hickman, J.A.; McNally, N.J.; Rohas, A.M.; Mitchison, N.A.; Pierrepoint, C.G.; Raymond, R.; Rowlatt, C.; Stephens, T.C. UKCCCR guidelines for the welfare of animals in experimental neoplasia. Lab. Anim. 1988, 22, 195–201. [Google Scholar] [CrossRef]
- Kilkenny, C.; Browne, W.; Cuthill, I.C.; Emerson, M.; Altman, D.G.; NC3Rs Reporting Guidelines Working Group. Animal research: Reporting in vivo experiments: The ARRIVE guidelines. Br. J. Pharmacol. 2010, 160, 1577–1579. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qu, H.; Wang, S.; Chater, J.M.; Wang, X.; Cui, Y.; Yu, L.; Zhou, R.; Jia, Q.; Traband, R.; et al. CancerMIRNome: An interactive analysis and visualization database for miRNome profiles of human cancer. Nucleic Acids Res. 2022, 50, D1139–D1146. [Google Scholar] [CrossRef]
- TCGA Research Network; Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Urabe, F.; Matsuzaki, J.; Yamamoto, Y.; Kimura, T.; Hara, T.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Sakamoto, H.; et al. Large-scale Circulating microRNA Profiling for the Liquid Biopsy of Prostate Cancer. Clin. Cancer Res. 2019, 25, 3016–3025. [Google Scholar] [CrossRef]
- Ohno, M.; Matsuzaki, J.; Kawauchi, J.; Aoki, Y.; Miura, J.; Takizawa, S.; Kato, K.; Sakamoto, H.; Matsushita, Y.; Takahashi, M.; et al. Assessment of the Diagnostic Utility of Serum MicroRNA Classification in Patients With Diffuse Glioma. JAMA Netw. Open 2019, 2, e1916953. [Google Scholar] [CrossRef]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Kato, K.; et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2018, 110, 408–419. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, Y.-X.; Dong, B.; Du, X.; Wang, J.; Wang, X.; Gao, W.-Q.; Xue, W. Discovery of extracellular vesicles derived miR-181a-5p in patient’s serum as an indicator for bone-metastatic prostate cancer. Theranostics 2021, 11, 878–892. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qu, H.; Wang, S.; Wei, J.; Le Zhang, L.; Ma, R.; Lu, J.; Zhu, J.; Zhong, W.-D.; Jia, Z. GDCRNATools: An R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018, 34, 2515–2517. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.-P.; Meese, E.; et al. miRTargetLink 2.0—Interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef]
- Nesbitt, H.; Byrne, N.M.; Williams, S.N.; Ming, L.; Worthington, J.; Errington, R.J.; Patterson, L.H.; Smith, P.J.; McKeown, S.R.; McKenna, D.J. Targeting Hypoxic Prostate Tumors Using the Novel Hypoxia-Activated Prodrug OCT1002 Inhibits Expression of Genes Associated with Malignant Progression. Clin. Cancer Res. 2017, 23, 1797–1808. [Google Scholar] [CrossRef]
- Byrne, N.M.; Nesbitt, H.; Ming, L.; McKeown, S.R.; Worthington, J.; McKenna, D.J. Androgen deprivation in LNCaP prostate tumour xenografts induces vascular changes and hypoxic stress, resulting in promotion of epithelial-to-mesenchymal transition. Br. J. Cancer 2016, 114, 659–668. [Google Scholar] [CrossRef]
- Vega, F.M.; Ridley, A.J. The RhoB small GTPase in physiology and disease. Small GTPases 2018, 9, 384–393. [Google Scholar] [CrossRef]
- Ju, J.A.; Godet, I.; DiGiacomo, J.W.; Gilkes, D.M. RhoB is regulated by hypoxia and modulates metastasis in breast cancer. Cancer Rep. 2020, 3, e1164. [Google Scholar] [CrossRef]
- Huang, G.; Su, J.; Zhang, M.; Jin, Y.; Wang, Y.; Zhou, P.; Lu, J. RhoB regulates the function of macrophages in the hypoxia-induced inflammatory response. Cell. Mol. Immunol. 2017, 14, 265–275. [Google Scholar] [CrossRef]
- Connolly, E.C.; Van Doorslaer, K.; Rogler, L.E.; Rogler, C.E. Overexpression of miR-21 Promotes an In vitro Metastatic Phenotype by Targeting the Tumor Suppressor RHOB. Mol. Cancer Res. 2010, 8, 691–700. [Google Scholar] [CrossRef]
- Liu, M.; Tang, Q.; Qiu, M.; Lang, N.; Li, M.; Zheng, Y.; Bi, F. miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett. 2011, 585, 2998–3005. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Li, X.; Wu, S. Up-regulation of long non-coding RNA LOXL1-AS1 functions as an oncogene in cervical squamous cell carcinoma by sponging miR-21. Arch. Physiol. Biochem. 2020, 129, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhan, Y.; Chi, J.; Guo, S.; Zhong, X.; He, A.; Zheng, J.; Gong, Y.; Li, X.; Zhou, L. Using microRNAs as Novel Predictors of Urologic Cancer Survival: An Integrated Analysis. Ebiomedicine 2018, 34, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Rosenberg, A.Z.; Choi, S.M.; Fox-Talbot, K.; De Marzo, A.M.; Nonn, L.; Brennen, W.N.; Marchionni, L.; Halushka, M.K.; Lupold, S.E. Cell-type specific expression of oncogenic and tumor suppressive microRNAs in the human prostate and prostate cancer. Sci. Rep. 2018, 8, 7189. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Ding, S.; Li, X.; Wang, H.; Liu, S.; Wu, H.; Bi, D.; Ding, K.; Lu, J. Elevated expression of HIF-lα in actively growing prostate tissues is associated with clinical features of benign prostatic hyperplasia. Oncotarget 2016, 7, 12053–12062. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.N.T.; Kim, M.G.; Koo, B.; Liu, H.; Jang, Y.O.; Lee, H.J.; Kim, Y.; Park, Y.; Kim, H.S.; Kim, C.; et al. Chimeric nanocomposites for the rapid and simple isolation of urinary extracellular vesicles. J. Extracell. Vesicles 2022, 11, e12195. [Google Scholar] [CrossRef]
- Zavadil, J.; Juráček, J.; Čechová, B.; Andrašina, T.; Slabý, O.; Goldberg, N. Dynamic Changes in Circulating MicroRNA Levels in Liver Cancer Patients Undergoing Thermal Ablation and Transarterial Chemoembolization. Klin. Onkol. 2019, 32 (Suppl. S1), 164–166. [Google Scholar]
- Andrasina, T.; Juracek, J.; Zavadil, J.; Cechova, B.; Rohan, T.; Vesela, P.; Paldor, M.; Slaby, O.; Goldberg, S.N. Thermal Ablation and Transarterial Chemoembolization are Characterized by Changing Dynamics of Circulating MicroRNAs. J. Vasc. Interv. Radiol. 2021, 32, 403–411. [Google Scholar] [CrossRef]
- Siegal, T.; Charbit, H.; Paldor, I.; Zelikovitch, B.; Canello, T.; Benis, A.; Wong, M.L.; Morokoff, A.P.; Kaye, A.H.; Lavon, I. Dynamics of circulating hypoxia-mediated miRNAs and tumor response in patients with high-grade glioma treated with bevacizumab. J. Neurosurg. 2016, 125, 1008–1015. [Google Scholar] [CrossRef]
- Al-Rawaf, H.A.; Gabr, S.A.; Alghadir, A.H. Circulating Hypoxia Responsive microRNAs (HRMs) and Wound Healing Potentials of Green Tea in Diabetic and Nondiabetic Rat Models. Evid.-Based Complement. Altern. Med. 2019, 2019, 9019253. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-T.; Hsu, C.-H.; Huang, T.-L.; Tsai, Y.-C.; Chiang, C.-Y.; Chen, Z.-C.; Shih, J.-Y. MicroRNA-21 is Associated with the Severity of Right Ventricular Dysfunction in Patients with Hypoxia-Induced Pulmonary Hypertension. Acta Cardiol. Sin. 2018, 34, 511–517. [Google Scholar] [CrossRef]
- Xie, X.; Qu, P.; Wu, H.; Liu, P.; Luo, J.; Chi, J.; Liu, X.; Chen, X.; Xu, C. Circulating exosomal miR-21 mediates HUVEC proliferation and migration through PTEN/PI3K/AKT in Crohn’s disease. Ann. Transl. Med. 2022, 10, 258. [Google Scholar] [CrossRef]
- Whitehead, C.L.; Teh, W.T.; Walker, S.P.; Leung, C.; Larmour, L.; Tong, S. Circulating MicroRNAs in Maternal Blood as Potential Biomarkers for Fetal Hypoxia In-Utero. PLoS ONE 2013, 8, e78487. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 2021, 35, e21916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, J.; Ishihara, M.; Sharrow, A.C.; Flora, K.; He, Y.; Wu, L. The miRNA-21-5p Payload in Exosomes from M2 Macrophages Drives Tumor Cell Aggression via PTEN/Akt Signaling in Renal Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3005. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhang, Y.; Mu, J.; Yang, D.; Gu, X.; Zhang, J. Exosomal miR-21-5p contributes to ovarian cancer progression by regulating CDK6. Hum. Cell 2021, 34, 1185–1196. [Google Scholar] [CrossRef]
- Chang, J.; Li, H.; Zhu, Z.; Mei, P.; Hu, W.; Xiong, X.; Tao, J. microRNA-21-5p from M2 macrophage-derived extracellular vesicles promotes the differentiation and activity of pancreatic cancer stem cells by mediating KLF3. Cell Biol. Toxicol. 2022, 38, 577–590. [Google Scholar] [CrossRef]
- Tian, X.-P.; Wang, C.-Y.; Jin, X.-H.; Li, M.; Wang, F.-W.; Huang, W.-J.; Yun, J.-P.; Xu, R.-H.; Cai, Q.-Q.; Xie, D. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics 2019, 9, 1965–1979. [Google Scholar] [CrossRef]
- Wojciak-Stothard, B.; Zhao, L.; Oliver, E.; Dubois, O.; Wu, Y.; Kardassis, D.; Vasilaki, E.; Huang, M.; Mitchell, J.A.; Harrington, L.S.; et al. Role of RhoB in the Regulation of Pulmonary Endothelial and Smooth Muscle Cell Responses to Hypoxia. Circ. Res. 2012, 110, 1423–1434. [Google Scholar] [CrossRef]
- Vega, F.M.; Thomas, M.; Reymond, N.; Ridley, A.J. The Rho GTPase RhoB regulates cadherin expression and epithelial cell-cell interaction. Cell Commun. Signal. 2015, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, N.; Charalampopoulos, I.; Alevizopoulos, K.; Gravanis, A.; Stournaras, C. Rho/ROCK/actin signaling regulates membrane androgen receptor induced apoptosis in prostate cancer cells. Exp. Cell Res. 2008, 314, 3162–3174. [Google Scholar] [CrossRef] [PubMed]
- Vega, F.M.; Colomba, A.; Reymond, N.; Thomas, M.; Ridley, A.J. RhoB regulates cell migration through altered focal adhesion dynamics. Open Biol. 2012, 2, 120076. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Qiu, W.; Liu, Q.; Qian, M.; Wang, S.; Zhang, Z.; Gao, X.; Chen, Z.; Xue, H.; Li, G. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene 2018, 37, 4239–4259. [Google Scholar] [CrossRef]
- Jin, J.; Yu, G. Hypoxic lung cancer cell-derived exosomal miR-21 mediates macrophage M2 polarization and promotes cancer cell proliferation through targeting IRF1. World J. Surg. Oncol. 2022, 20, 241. [Google Scholar] [CrossRef]
- Guraya, S. Prognostic significance of circulating microRNA-21 expression in esophageal, pancreatic and colorectal cancers; a systematic review and meta-analysis. Int. J. Surg. 2018, 60, 41–47. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.; Zhu, W.; Gao, C.; Jiang, R.; Li, W.; Hu, Q.; Zhang, B. MicroRNA-21 and the clinical outcomes of various carcinomas: A systematic review and meta-analysis. BMC Cancer 2014, 14, 819. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Wei, F.; Zhang, X.; Yu, J.; Zhao, H.; Sun, Q.; Yan, F.; Yan, C.; Li, H.; et al. Diagnostic and prognostic value of circulating miR-21 for cancer: A systematic review and meta-analysis. Gene 2014, 533, 389–397. [Google Scholar] [CrossRef]
- Fu, X.; Han, Y.; Wu, Y.; Zhu, X.; Lu, X.; Mao, F.; Wang, X.; He, X.; Zhao, Y.; Zhao, Y. Prognostic role of microRNA-21 in various carcinomas: A systematic review and meta-analysis. Eur. J. Clin. Investig. 2011, 41, 1245–1253. [Google Scholar] [CrossRef]
- Stafford, M.C.; Willoughby, C.E.; Walsh, C.P.; McKenna, D.J. Prognostic value of miR-21 for prostate cancer: A systematic review and meta-analysis. Biosci. Rep. 2022, 42, BSR20211972. [Google Scholar] [CrossRef]
- Aghdam, A.M.; Amiri, A.; Salarinia, R.; Masoudifar, A.; Ghasemi, F.; Mirzaei, H. MicroRNAs as Diagnostic, Prognostic, and Therapeutic Biomarkers in Prostate Cancer. Crit. Rev. Eukaryot. Gene Expr. 2019, 29, 127–139. [Google Scholar] [CrossRef]
- Cozar, J.; Robles-Fernandez, I.; Rodriguez-Martinez, A.; Puche-Sanz, I.; Vazquez-Alonso, F.; Lorente, J.; Martinez-Gonzalez, L.; Alvarez-Cubero, M. The role of miRNAs as biomarkers in prostate cancer. Mutat. Res. Mol. Mech. Mutagen. 2019, 781, 165–174. [Google Scholar] [CrossRef]
- Fabris, L.; Ceder, Y.; Chinnaiyan, A.M.; Jenster, G.W.; Sorensen, K.D.; Tomlins, S.; Visakorpi, T.; Calin, G.A. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur. Urol. 2016, 70, 312–322. [Google Scholar] [CrossRef]
- Wang, J.; Ni, J.; Beretov, J.; Thompson, J.; Graham, P.; Li, Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol. 2020, 145, 102860. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Bryzgunova, O.E.; Laktionov, P.P. miRNAs and androgen deprivation therapy for prostate cancer. Biochim. et Biophys. Acta (BBA) Rev. Cancer 2021, 1876, 188625. [Google Scholar] [CrossRef]
- Salberg, U.B.; Skingen, V.E.; Fjeldbo, C.S.; Hompland, T.; Ragnum, H.B.; Vlatkovic, L.; Hole, K.H.; Seierstad, T.; Lyng, H. A prognostic hypoxia gene signature with low heterogeneity within the dominant tumour lesion in prostate cancer patients. Br. J. Cancer 2022, 127, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.; Li, Y.; Yan, Z.; He, Q.; Cheng, L.; Zhang, P.; Liu, B.; Liu, C.; Song, Y.; Xing, Y. Identification of ISG15 and ZFP36 as novel hypoxia- and immune-related gene signatures contributing to a new perspective for the treatment of prostate cancer by bioinformatics and experimental verification. J. Transl. Med. 2022, 20, 202. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Wang, J.; Guo, X.; Lv, Z.; Liu, J.; Yan, Q.; Liu, M.; Wang, J. Identification of a Hypoxia-Related Gene Signature for Predicting Systemic Metastasis in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 696364. [Google Scholar] [CrossRef]
- Yang, L.; Roberts, D.; Takhar, M.; Erho, N.; Bibby, B.A.; Thiruthaneeswaran, N.; Bhandari, V.; Cheng, W.-C.; Haider, S.; McCorry, A.M.; et al. Development and Validation of a 28-gene Hypoxia-related Prognostic Signature for Localized Prostate Cancer. Ebiomedicine 2018, 31, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Zedan, A.H.; Blavnsfeldt, S.G.; Hansen, T.F.; Nielsen, B.S.; Marcussen, N.; Pleckaitis, M.; Osther, P.J.S.; Sørensen, F.B. Heterogeneity of miRNA expression in localized prostate cancer with clinicopathological correlations. PLoS ONE 2017, 12, e0179113. [Google Scholar] [CrossRef]
- Haffner, M.C.; Zwart, W.; Roudier, M.P.; True, L.D.; Nelson, W.G.; Epstein, J.I.; De Marzo, A.M.; Nelson, P.S.; Yegnasubramanian, S. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2020, 18, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Flores-Téllez, T.d.N.; Baena, E. Experimental challenges to modeling prostate cancer heterogeneity. Cancer Lett. 2022, 524, 194–205. [Google Scholar] [CrossRef] [PubMed]
- McNally, C.J.; Ruddock, M.W.; Moore, T.; McKenna, D.J. Biomarkers That Differentiate Benign Prostatic Hyperplasia from Prostate Cancer: A Literature Review. Cancer Manag. Res. 2020, 12, 5225–5241. [Google Scholar] [CrossRef] [PubMed]
- McNally, C.J.; Watt, J.; Kurth, M.J.; Lamont, J.V.; Moore, T.; Fitzgerald, P.; Pandha, H.; McKenna, D.J.; Ruddock, M.W. A Novel Combination of Serum Markers in a Multivariate Model to Help Triage Patients Into “Low-” and “High-Risk” Categories for Prostate Cancer. Front. Oncol. 2022, 12, 837127. [Google Scholar] [CrossRef]
- Eklund, M.; Nordström, T.; Aly, M.; Adolfsson, J.; Wiklund, P.; Brandberg, Y.; Thompson, J.; Wiklund, F.; Lindberg, J.; Presti, J.C.; et al. The Stockholm-3 (STHLM3) Model can Improve Prostate Cancer Diagnostics in Men Aged 50–69 yr Compared with Current Prostate Cancer Testing. Eur. Urol. Focus 2018, 4, 707–710. [Google Scholar] [CrossRef]
- Punnen, S.; Pavan, N.; Parekh, D.J. Finding the Wolf in Sheep’s Clothing: The 4Kscore Is a Novel Blood Test That Can Accurately Identify the Risk of Aggressive Prostate Cancer. Rev. Urol. 2015, 17, 3–13. [Google Scholar]
- Schröder, F.H.; Hugosson, J.; Roobol-Bouts, M.J.; Tammela, T.L.J.; Ciatto, S.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Lilja, H.; Zappa, M.; et al. Screening and Prostate-Cancer Mortality in a Randomized European Study. N. Engl. J. Med. 2009, 360, 1320–1328. [Google Scholar] [CrossRef]

| Gene Set | Gene Set ID | Description | Count/Total | Adjusted p-Value 1 | Gene Symbol |
|---|---|---|---|---|---|
| KEGG | hsa05206 | MicroRNAs in cancer | 35/612 | 1.32 × 10−7 | CDC25A; BCL2; SPRY2; TIMP3; RECK; E2F2; PTEN; E2F1; MARCKS; TPM1; CDK6; PDCD4; SERPINB5; BMPR2; MYC; ERBB2; HNRNPK; TP63; EGFR; NFKB1; VEGFA; MDM4; TGFB2; PIK3R1; MMP9; BRCA1; PRKCE; APC; CCNG1; STAT3; DICER1; E2F3; ABCB1; BMI1; SOCS1 |
| hsa05215 | Prostate cancer | 16/612 | 8.95 × 10−6 | BCL2; E2F2; PTEN; E2F1; ERBB2; EGFR; PLAT; NFKB1; RB1; PDGFD; PIK3R1; MMP9; AKT2; IGF1R; E2F3; FOXO1 | |
| Disease Ontology | DOID:10283 | prostate cancer | 41/612 | 2.03 × 10−5 | BCL2; SPRY2; PTEN; HIPK3; FAS; BMPR2; MYC; ERBB2; TOPORS; MSH2; EGFR; ICAM1; SP1; SMARCA4; NFKB1; SOD3; SMAD7; MMP2; VEGFA; TGFB1; RB1; PDGFD; MUC1; PIK3R1; RPS6KA3; MMP9; BRCA1; PTK2; SKP2; PBX1; WNT5A; MAP3K1; PURA; HIF1A; CXCL10; IGF1R; SET; KLK2; CEBPB; BMI1; CASP8 |
| DOID:10286 | prostate carcinoma | 13/612 | 9.53 × 10−3 | BCL2; PTEN; MYC; ERBB2; MSH2; EGFR; NFKB1; MMP2; VEGFA; PDGFD; PTK2; IGF1R; CASP8 | |
| DisGeNET | umls:C0936223 | Metastatic Prostate Carcinoma | 24/612 | 5.11 × 10−6 | JAG1; PTEN; MYC; ERBB2; EGFR; IL1B; NFKB1; NTF3; DTX3L; MMP2; VEGFA; PARP1; TGFB1; ACAT1; SUZ12; MUC1; MMP9; PARP9; WNK1; TNFRSF11B; SATB1; WWP1; HIF1A; CLU |
| umls:C0007112 | Adenocarcinoma of prostate | 16/612 | 4.22 × 10−4 | BCL2; PTEN; ERBB2; EGFR; PPARA; PLPP1; VEGFA; TGFB1; RB1; MMP9; PRKCE; MIB1; TLR4; OLR1; STAT3; HIF1A | |
| umls:C1328504 | Hormone refractory prostate cancer | 11/612 | 4.44 × 10−4 | BCL2; PTEN; ERBB2; EGFR; PARP1; TGFB1; APC; STAT3; CLU; HMGB1; CASP8 | |
| umls:C1654637 | androgen independent prostate cancer | 15/612 | 9.38 × 10−4 | BCL2; PTEN; ERBB2; MEF2C; EGFR; RASGRP3; MMP9; PBX1; AGO2; AKT2; FOXO3; HIF1A; CLU; COX2; ABCB1 |
| Gene Set | Gene Set ID | Description | Count/ Total | Adjusted p-Value 1 | Gene Symbol |
|---|---|---|---|---|---|
| KEGG | hsa04066 | HIF-1 signaling pathway | 15/612 | 8.10 × 10−5 | BCL2; PDHA2; ERBB2; EGFR; NFKB1; VEGFA; MKNK2; PIK3R1; TLR4; AKT2; STAT3; VHL; HIF1A; IGF1R; EGLN1 |
| Gene Ontology- Biological Process | GO:0001666 | response to hypoxia | 29/612 | 2.31 × 10−4 | BCL2; REST; TGFBR2; PTEN; E2F1; APAF1; MYC; TGFBR3; ICAM1; PLAT; PPARA; SOD3; MMP2; VEGFA; TGFB1; MDM4; DDAH1; TGFB2; APOLD1; PRKCE; IRAK1; VHL; FOXO3; HIF1A; SIRT2; DNM1L; STUB1; EGLN1; PSMD9 |
| GO:0071456 | cellular response to hypoxia | 17/612 | 4.70 × 10−3 | BCL2; PTEN; E2F1; MYC; ICAM1; VEGFA; MDM4; DDAH1; PRKCE; IRAK1; VHL; FOXO3; HIF1A; SIRT2; STUB1; EGLN1; PSMD9 | |
| GO:0070482 | response to oxygen levels | 32/612 | 9.66 × 10−5 | BCL2; REST; TGFBR2; PTEN; E2F1; APAF1; FAS; MYC; TGFBR3; ICAM1; PLAT; PPARA; SOD3; MMP2; VEGFA; TGFB1; MDM4; DDAH1; TGFB2; APOLD1; PRKCE; IRAK1; VHL; FOXO3; HIF1A; SIRT2; DNM1L; STUB1; EGLN1; PSMD9; OXTR; FOXO1 | |
| GO:0036293 | response to decreased oxygen levels | 30/612 | 1.66 × 10−4 | BCL2; REST; TGFBR2; PTEN; E2F1; APAF1; MYC; TGFBR3; ICAM1; PLAT; PPARA; SOD3; MMP2; VEGFA; TGFB1; MDM4; DDAH1; TGFB2; APOLD1; PRKCE; IRAK1; VHL; FOXO3; HIF1A; SIRT2; DNM1L; STUB1; EGLN1; PSMD9; OXTR | |
| GO:0071453 | cellular response to oxygen levels | 20/612 | 1.37 × 10−3 | BCL2; PTEN; E2F1; FAS; MYC; ICAM1; VEGFA; MDM4; DDAH1; PRKCE; IRAK1; VHL; FOXO3; HIF1A; SIRT2; DNM1L; STUB1; EGLN1; PSMD9; FOXO1 | |
| GO:0036294 | cellular response to decreased oxygen levels | 18/612 | 3.27 × 10−3 | BCL2; PTEN; E2F1; MYC; ICAM1; VEGFA; MDM4; DDAH1; PRKCE; IRAK1; VHL; FOXO3; HIF1A; SIRT2; DNM1L; STUB1; EGLN1; PSMD9 | |
| GO:0034599 | cellular response to oxidative stress | 21/612 | 7.61 × 10−3 | BCL2; REST; TPM1; RHOB; EIF2S1; PPIF; EGFR; TNFAIP3; SOD3; MMP2; PARP1; PDGFD; PKD2; PLEKHA1; MMP9; TLR4; FOXO3; HIF1A; SIRT2; PCGF2; FOXO1 | |
| GO:0006979 | response to oxidative stress | 26/612 | 1.90 × 10−2 | BCL2; REST; TPM1; SESN1; RHOB; EIF2S1; PPIF; EGFR; TNFAIP3; SP1; SOD3; MMP2; PARP1; PDGFD; MYEF2; PKD2; PLEKHA1; MMP9; TLR4; FOXO3; HIF1A; SIRT2; CCR7; EGLN1; PCGF2; FOXO1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angel, C.Z.; Stafford, M.Y.C.; McNally, C.J.; Nesbitt, H.; McKenna, D.J. MiR-21 Is Induced by Hypoxia and Down-Regulates RHOB in Prostate Cancer. Cancers 2023, 15, 1291. https://doi.org/10.3390/cancers15041291
Angel CZ, Stafford MYC, McNally CJ, Nesbitt H, McKenna DJ. MiR-21 Is Induced by Hypoxia and Down-Regulates RHOB in Prostate Cancer. Cancers. 2023; 15(4):1291. https://doi.org/10.3390/cancers15041291
Chicago/Turabian StyleAngel, Charlotte Zoe, Mei Yu Cynthia Stafford, Christopher J. McNally, Heather Nesbitt, and Declan J. McKenna. 2023. "MiR-21 Is Induced by Hypoxia and Down-Regulates RHOB in Prostate Cancer" Cancers 15, no. 4: 1291. https://doi.org/10.3390/cancers15041291
APA StyleAngel, C. Z., Stafford, M. Y. C., McNally, C. J., Nesbitt, H., & McKenna, D. J. (2023). MiR-21 Is Induced by Hypoxia and Down-Regulates RHOB in Prostate Cancer. Cancers, 15(4), 1291. https://doi.org/10.3390/cancers15041291






